Abstract
Two homeodomain proteins, Yox1 and Yhp1, act as repressors at early cell cycle boxes (ECBs) to restrict their activity to the M/G1 phase of the cell cycle in budding yeast. These proteins bind to Mcm1 and to a typical homeodomain binding site. The expression of Yox1 is periodic and directly correlated with its binding to, and repression of, ECB activity. The absence of Yox1 and Yhp1 or the constitutive expression of Yox1 leads to the loss of cell-cycle regulation of ECB activity. Therefore, the cell-cycle-regulated expression of these repressors defines the interval of ECB-dependent transcription. Twenty-eight genes, including MCM2-7, CDC6, SWI4, CLN3, and a number of genes required during late M phase have been identified that are coordinately regulated by this pathway.
Keywords: ECB, cell cycle, SWI4, CLN3, MCM2-7, transcription
Cyclin-dependent kinases (Cdks) drive the cell cycle in all eukaryotic cells. In budding yeast, Cdk1 (Cdc28) expression is constant, but cyclin transcription, stability, and activity are regulated across the cell cycle (Miller and Cross 2001). These multiple levels of regulation result in the ordered appearance of different G1 (Cln)- and B-type (Clb) cyclins, which direct the phase-specific localization and/or substrate specificity of the kinase. There is a critical distinction between G1 phase and the rest of the cell cycle, in that G1 is expandable in response to the environment (Rupe 2002). The length of G1 is influenced by age, growth conditions, and the size of the cell (Hartwell and Unger 1977; Johnston et al. 1979). In contrast, once the cells exit G1, the length of the rest of the cycle is fairly constant (Jagadish and Carter 1977), even after severe nutrient limitation (Johnston et al. 1977). Accumulation of G1 cyclins (Clns) is rate-limiting for the G1 to S transition, and Clns are regulated at virtually every level (Wittenberg et al. 1990; Gallego et al. 1997; Polymenis and Schmidt 1997; MacKay et al. 2001; Newcomb et al. 2002). However, one of the great remaining mysteries is what triggers the rapid accumulation of Clns and causes the irreversible transition into S phase in the normal mitotic cycle.
Entry into G1 requires that Clb kinase activity be eliminated (Zachariae and Nasmyth 1999). Clb kinase activity decays due to cessation of CLB transcription, targeted proteolysis of the Clbs by the anaphase-promoting complex (APC), and the M/G1-specific expression of an inhibitory subunit, Sic1, which inactivates Clb/Cdk complexes. Low Clb kinase activity allows the nuclear localization and assembly of Cdc6 and Mcm2-7 onto origin DNA to form the prereplication complexes (PRCs; Tye 1999). These PRC components are transcribed coordinately at the M/G1 boundary, and the assembly of this highly conserved complex sets the stage for DNA replication. Once the PRCs are formed, Clb kinases are required to initiate replication. This is brought about by the accumulation of Cln/Cdk complexes, which phosphorylate and promote the degradation of Sic1 (Schneider et al. 1996; Tyers 1996; Nash et al. 2001) and restore Clb kinase activity.
Accumulation of the G1 cyclins requires the activation of Cln3/Cdk. This kinase is uniquely capable of activating two late G1-specific transcription complexes (SBF and MBF; Dirick et al. 1995; Stuart and Wittenberg 1996). Once activated, SBF and MBF cause a burst of transcription of the late G1 cyclins CLN1 and CLN2, and many other genes required for S phase. The burst of CLN1 and CLN2 transcription is delayed under conditions that prolong G1 (Sillje et al. 1997). This indicates that Cln3/Cdk and/or the transcription factors (SBF and MBF) are the likely targets of G1 regulation.
Swi4, which is the DNA-binding component of SBF, and Cln3 are both rate-limiting for the transition to S phase (Cross 1988; Nash et al. 1988; McInerny et al. 1997). Heterozygotes at one or both of these loci delay S phase, and overproduction of either Cln3 or Swi4 speeds the transition to S phase. The fact that Cln3 and Swi4 are gene dose-dependent activators of G1 progression suggests that their levels are limiting and potentially regulated during G1. The first evidence of regulation is at the level of transcription. CLN3 and SWI4 mRNA levels peak at the M/G1 boundary in mixed populations of mothers and daughters (McInerny et al. 1997), and in mid-G1 in elutriated daughters (MacKay et al. 2001). Early cell cycle box (ECB) elements, which confer M/G1-specific transcription, are necessary for the normal expression of both CLN3 and SWI4. Moreover, the transition to S phase is delayed and misregulated in cells in which ECB elements have been deleted from the CLN3 and SWI4 promoters (MacKay et al. 2001).
The ECB includes a binding site for Mcm1, which is required for activity (McInerny et al. 1997) and is bound constitutively by Mcm1 (Mai et al. 2002). Mcm1 belongs to the MADS family of transcription factors. These proteins contain a conserved DNA binding and dimerization domain named the MADS box after the four founding members of the family: Mcm1, Agamous, Deficiens, and serum response factor (SRF; Treisman and Ammerer 1992). In Saccharomyces cerevisiae, Mcm1 is required for the expression of many genes, including genes involved in arginine metabolism, mating-type specification, and cell-cycle regulation (Johnson 1995). In every known instance, Mcm1 partners with other transcription factors to achieve regulatory specificity. In higher eukaryotes, MADS box proteins are known to specifically interact with members of the paired class of homeodomain proteins (Grueneberg et al. 1992). In these instances, the MADS protein provides DNA binding specificity and the homeodomain proteins confer regulatory properties (Bondos and Tan 2001). Here we show conservation of this interaction in budding yeast.
This paper reports the identification of two homeodomain proteins: Yox1 and Yhp1, which bind to Mcm1 and to a sequence adjacent to the Mcm1-binding site in ECB elements. Yox1 and Yhp1 are repressors that restrict ECB-mediated transcription to the M/G1 interval of the cell cycle. Yox1 expression is also cell cycle-regulated, and its expression is directly correlated with its binding to ECB elements and determines the timing of ECB activity. Twenty-eight genes repressed by Yox1 and/or Yhp1 have been identified. These genes are involved in late mitotic events, formation of the PRC, and initiating the transcriptional cascade that triggers S phase.
Results
Yox1 and Yhp1 bind to Mcm1
In an attempt to identify the proteins that act in concert with Mcm1 to confer M/G1-specificity to the ECB elements, we followed up an observation made a decade ago that the human homeodomain protein Phox1 interacts with the conserved MADS domain of Mcm1 and SRF (Grueneberg et al. 1992). The interaction between Phox1 and the MADS box requires only the homeodomain of Phox1, so we sought yeast proteins with high homology to the Phox1 homeodomain sequence as potential regulators of Mcm1 activity.
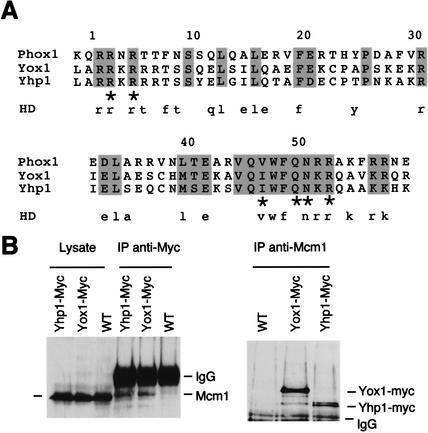
The closest matches to the Phox1 homeodomain sequence were found in two yeast proteins: Yox1 and Yhp1, whose sequences are 38% identical overall and 75% identical within the homeodomain (Fig. 1A). To determine whether Yox1 and Yhp1 bind Mcm1, we immunoprecipitated cells carrying either Yox1 or Yhp1 tagged with myc epitopes with anti-myc or anti-Mcm1 antibodies. The precipitates were then immunoblotted with antibodies to Mcm1 and myc, respectively. Figure 1B shows that Yox1–myc and Yhp1–myc coimmunoprecipitate with Mcm1 by both strategies. These tagged proteins are active (see Materials and Methods), and they are expressed from their native promoters. Thus, we conclude that Mcm1 interacts with both of these homeodomain proteins in vivo.
Figure 1.
(A) Alignment of homeodomains of human Phox1 and its two closest budding yeast relatives, Yox1 and Yhp1. Shaded residues are conserved in all three sequences. Asterisks indicate residues important for DNA binding (Mann 1995). Lowercase residues below represent those that are typically conserved across the domain in most homeodomain proteins (Galliot et al. 1999). (B) Yox1 and Yhp1 both coimmunoprecipitate with Mcm1. (Left panel) The left three lanes show crude lysates from cells carrying the wild-type (WT) or the myc-tagged allele of Yox1 or Yhp1 as indicated. The right three lanes show the same lysates after immunoprecipitation with polyclonal antibodies to the myc tag. Samples were resolved by SDS-PAGE and immunoblotted with antibodies to Mcm1. “IgG” indicates immunoglobulin (55kD). (Right panel) The same strains were immunoprecipitated with Mcm1 antibodies and immunoblotted with antibodies to the myc epitope.
Yox1 and Yhp1 influence cell-cycle kinetics
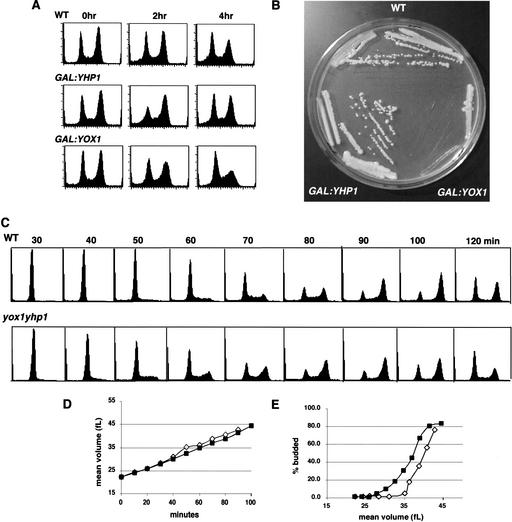
To further define the roles of Yox1 and Yhp1, we generated deletion mutants and overproduction constructs. In agreement with previous reports (Kaufman 1993; Kunoh et al. 2000), we found that cells carrying deletions of YOX1, YHP1, or both YOX1 and YHP1 were viable and grew at fairly comparable rates. However, overproduction of Yhp1 produces a notable but transient shift in the population from predominantly G1 cells to predominantly G2 (Fig. 2A). This shift did not significantly affect the overall transit time of the cell cycle, as indicated by colony size (Fig. 2B). Overproduction of Yox1 results in a dramatic slowing of growth. In liquid culture, cells overproducing Yox1 are large and heterogeneous in shape with a DNA content indicative of primarily G1- and S-phase cells (Fig. 2A). These cells are viable, but grow very slowly (Fig. 2B).
Figure 2.
Constitutive overproduction of Yox1 is highly deleterious to cells. (A) FACS profiles of wild-type cells, and cells after 0, 2, and 4 h of induction of Yhp1 or Yox1 overproduction from the GAL promoter. (B) Colony formation on a YEP galactose plate of wild-type cells compared to GAL:YOX1 and GAL:YHP1 cells. (C) Wild-type and yox1yhp1 G1 daughter cells were collected by elutriation, and their DNA content was followed by FACS analysis. (D) Mean cell volume (in femtoliters) was measured with a Coulter Z2 size analyzer and plotted as a function of time. (E) The kinetics of budding were followed and plotted as a function of mean cell volume. Wild-type (open symbols), yox1yhp1 (closed symbols).
We then followed the cell-cycle kinetics of wild type and the yox1yhp1 double mutant, starting with elutriated G1 cells, and compared their FACS profiles as they progressed from G1 to G2 DNA content. Figure 2C shows that loss of these two homeodomain proteins speeds the G1 to S transition in G1 daughter cells. DNA replication begins at least 10 min earlier in the double mutant than in the wild-type cells. This is not due to differences in the starting cell size or the rate of growth of these cells (Fig. 2D). Rather, it indicates a speeding of the G1 to S transition. Budding also occurs at a smaller cell size in the yox1yhp1 double mutant (Fig. 2E).
Yox1 and/or Yhp1 are repressors of M/G1-specific genes
Because Yox1 and Yhp1 are likely to be transcription factors, we surveyed their genome-wide effects on transcription. Using microarray analysis, we observed a reproducible twofold or greater repression of 184 transcripts when Yox1 was overproduced (data not shown). Using an algorithm that specifically identifies transcripts which oscillate during the cell cycle, 1106 genes have been identified which show significant periodicity (Zhao et al. 2001) in at least one of the three data sets that follow genome-wide transcript levels through the cell cycle (Cho et al. 1998; Spellman et al. 1998). Using this criterion, we found that 112 of the 184 repressed transcripts (61%) were potentially cell-cycle-regulated, and most of these transcripts peak in late M and early G1. Moreover, all of the known ECB-regulated genes and the MCM family were included in this group.
The cell-cycle-regulated genes repressed by Yox1 overproduction could be indirectly affected by the stalling of the cell cycle in G1 or S phase (Fig. 2A). Another possibility is that the excess Yox1 sequesters and inactivates Mcm1. Many M- and M/G1-specific genes require Mcm1 for their transcription (Lydall et al. 1991; Maher et al. 1995; McInerny et al. 1997). However, there are other Mcm1-regulated genes, for example, those involved in arginine metabolism (ARG3, ARG5, CAR1, and CAR2), that are unaffected by Yox1 overproduction (data not shown), so some Mcm1 activity persists in these cells. A third possibility is that Yox1 is a transcriptional repressor and some of these genes are its direct targets.
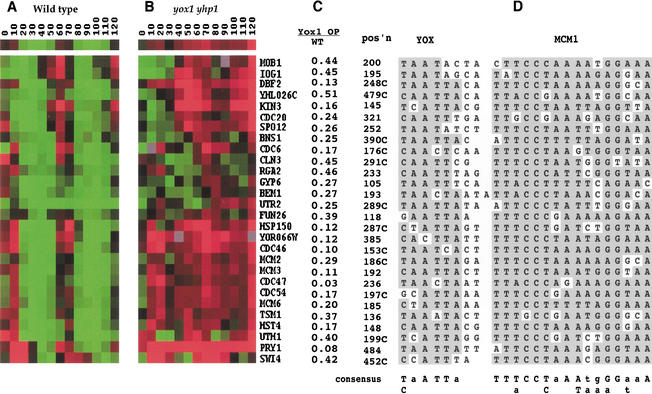
In vivo targets of Yox1-mediated repression should be derepressed in the absence of Yox1. To identify such genes, and to avoid the potential complication of redundancy between Yox1 and Yhp1, we compared transcript profiles of the wild-type and the yox1yhp1 double mutant. These two strains were synchronized with α-factor and followed through two cell cycles. Figure 3 shows the transcript profiles for 28 genes that are cell-cycle-regulated in wild-type and derepressed in the yox1yhp1 cells. Interestingly, all of these periodically transcribed genes peak at about the same time in the wild-type cell cycle. They differ in their capacity to be activated during the first cycle after release from the α-factor arrest, but all 28 transcripts peak 60–70 min after release. This represents the late M, early G1 phase of the cell cycle. In the yox1yhp1 mutant, the transcript levels also vary initially, but all 28 transcripts show significant derepression through the cell cycle. In addition, all 28 genes were identified as being repressed by Yox1 overproduction (Fig. 3C).
Figure 3.
Twenty-eight genes negatively regulated by Yox1 and/or Yhp1 share a common promoter element. Genome-wide transcript analysis of wild-type (A) and yox1yhp1 (B) cells through the cell cycle. Cells were synchronized with α-factor and monitored at 10-min intervals through two cell cycles. The compiled data are represented in rows for each gene and columns for each timepoint. Red indicates increased abundance and green indicates decreased abundance of the time course cDNA with respect to the invariant control cDNA. Black indicates the ratio of time course cDNA to control of 1. The first row represents the value for each timepoint averaged over all 28 genes. (C) Transcripts elevated in yox1yhp1 cells are repressed two- to thirtyfold by overproduction of Yox1. Transcript levels of these 28 genes after 4 h of overproduction of Yox1 were compared by microarray analysis to those of wild-type cells grown under the same conditions. These values, for each gene listed, are expressed as a ratio of the level in the Yox1 overproducer over the wild type. (D) The position (in bases upstream from the translational start site) and the sequences of the YOX- and Mcm1-binding sites in each putative M/G1-specific ECB element are shown, with shading to indicate bases that conform to the consensus sequence below. “C” in the position column indicates reverse orientation to that listed.
Identification of a shared sequence in Yox1 and Yhp1-regulated genes
The computer program Consensus (Hertz and Stormo 1999) was used to look for common motifs within 500 base pairs (bp) of the translational start sites (ATG) of the 15 genes most affected by changes in Yox1 and/or Yhp1 levels. This search readily identified the 16-bp palindrome to which Mcm1 is known to bind in all 15 promoters. We then used Co-Bind (GuhaThakurta and Stormo 2001) to look for other common motifs within 40 bp of the Mcm1-binding sites. This search identified the sequence (T/CaATTa) which resides within 3 bp of the Mcm1-binding site. We will refer to this site as a YOX site. Both Yox1 and Yhp1 were previously identified for their ability to bind DNA, and a Yhp1-binding site was identified which is in agreement with this YOX consensus site (Kaufman 1993; Kunoh et al. 2000). We then used Patser (G. Stormo and G. Hertz, http://ural.wustl.edu/∼jhc1/consensus) to find other adjacent YOX- and Mcm1-binding sites occurring within 500 bp of the ATG of all yeast genes. Figure 3D shows the alignment of these sites for all 28 genes that are repressed by Yox1 overproduction and derepressed in yox1yhp1 cells.
Different patterns of response to combined loss of Yox1 and Yhp1 activity
Close inspection of the cell-cycle transcription profiles of Yox1-repressed genes shows that there are differing degrees of deregulation in the absence of Yox1 and Yhp1. Figure 4A and B shows representative transcript profiles for the most affected class, which are transcribed at a high constitutive level in yox1yhp1 cells. This class includes all six MCM genes. Clearly, Yox1 and/or Yhp1 provide most of the cell-cycle regulation to these promoters.
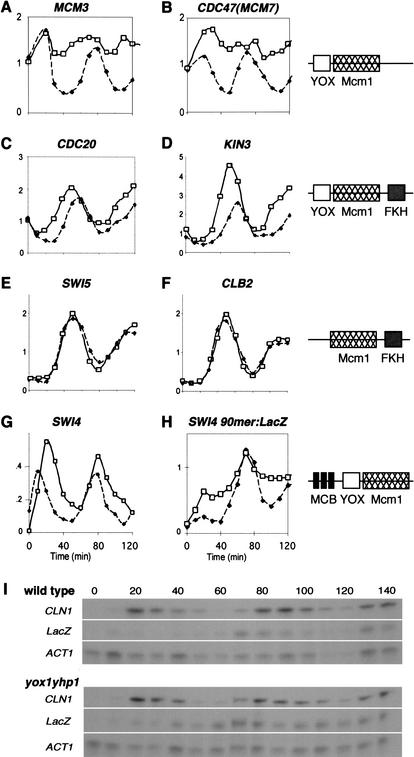
Figure 4.
Transcription is derepressed during the cell cycle in yox1yhp1 cells. The profiles of seven cell-cycle-regulated transcripts taken from the microarray experiment (A–F) or as measured by S1 protection (G,H) are graphed, as indicated, from wild-type (solid symbols) and yox1yhp1 (open symbols) cells. (I) The primary data for panel H, which monitors SWI4 90mer: lacZ transcription through the cell cycle. CLN1 mRNA is monitored as a control for cell-cycle synchrony. SWI4 and lacZ levels normalized to ACT1, an invariant transcript, are graphed in G and H.
The CDC20, SPO12, IQG1, and KIN3 transcripts remain highly periodic but are expressed for a broader interval of time in yox1yhp1 cells (Fig. 4C,D). These genes contain hybrid promoter elements in that there is a YOX site on one side of the Mcm1-binding site, and a forkhead (FKH)-binding site on the other side (Zhu and Davis 1998). Mcm1- and adjacent FKH-binding sites confer M-specific transcription to a family of genes (Koranda et al. 2000; Kumar et al. 2000; Zhu et al. 2000). The YOX/MCM1/FKH promoter elements confer a distinct pattern of transcription in that they are activated after the M-specific genes, SWI5 and CLB2 (Fig. 4E,F) and before the M/G1-specific genes (Fig. 4A,B) in wild-type cells. Transcription of the M-specific genes is unaffected in yox1yhp1 cells, indicating that Yox1 and/or Yhp1 do not affect Mcm1 activity at these sites. In contrast, the YOX/MCM1/FKH promoters are activated prematurely in cells lacking Yox1 and Yhp1. Thus, Yox1 and/or Yhp1 serve to delay transcription of this subset of genes until late M phase, after the bulk of M-specific transcripts have been made.
SWI4 shows a different pattern of deregulation, in which the mRNA levels continue to oscillate, but peak transcription persists about 10 min longer in the absence of Yox1 and Yhp1 compared to wild-type cells. This pattern may be explained by the fact that the SWI4 promoter also contains three MCB elements (Foster et al. 1993), which are known to confer late G1-specific transcription. When the SWI4 ECB was analyzed in isolation (Fig. 4H,I), we found that its transcription is highly deregulated throughout the cycle in yox1yhp1 cells. This suggests that the ECB elements in SWI4 and MCM2-7 are similarly derepressed in the absence of Yox1 and Yhp1, but this deregulation can be obscured or compensated for by other cell-cycle regulatory elements.
Yox1 and Yhp1 are required for cell-cycle-regulated transcription driven by ECB elements
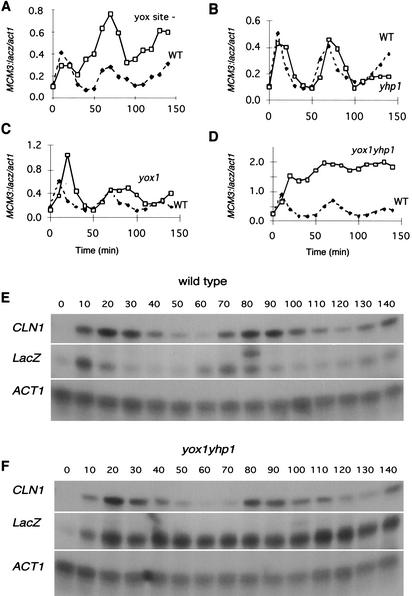
To discern the relative contribution of Yox1, Yhp1, and the YOX site to ECB regulation, we cloned a fragment of the MCM3 promoter, including the YOX- and Mcm1-binding sites and 10 bp of flanking sequence, into a lacZ reporter construct. Figure 5A and E shows that this minimal ECB activates M/G1-specific transcription of lacZ. From previous studies, we know that mutation of the Mcm1-binding site eliminates ECB transcriptional activity (McInerny et al. 1997). To determine the role of the YOX site, we made two substitutions in the YOX site of MCM3 ECB:lacZ and followed its transcriptional activity across the cell cycle. Figure 5A shows that loss of the YOX site leads to a dramatic increase in lacZ transcript after the first peak of expression. In subsequent cycles, peak expression still occurs at the M/G1 boundary, but the transcript level always exceeds the peak level of the transcript driven by the wild-type element. This confirms the repressing function of the YOX site in the context of an ECB element. However, it is also clear that loss of the YOX site does not eliminate M/G1-specific activation of this ECB or its repression during α-factor arrest.
Figure 5.
Yox1 or Yhp1 is required to restrict transcription of ECB elements to the M/G1 interval of the cell cycle. (A) Transcription driven by the wild-type ECB element of MCM3 through the cell cycle (solid symbols), or the equivalent reporter (open symbols) lacking the YOX site. (B–D) Parallel analyses of the MCM3:lacZ transcription in yhp1, yox1, or yox1yhp1 cells (open symbols) compared to wild type (solid symbols). The primary S1 protection data for the MCM3:lacZ reporter analyzed in wild-type and yox1yhp1 cells are provided in E and F, respectively.
We also monitored MCM3 ECB:lacZ transcription across the cell cycle in yhp1, yox1, and yox1yhp1 cells (Fig. 5B–D). Loss of Yhp1 activity has little or no effect upon the level or the periodicity of the ECB-driven transcript. In the absence of Yox1 activity, the transcript continues to oscillate but the period of transcriptional activity is prolonged by at least 20 min. This indicates that Yox1 activity is required to repress ECB activity during late G1 and perhaps early S phase. However, at later timepoints, repression is established in the yox1 cells and the transcript level drops to the trough level of the wild-type strain. This late repression in yox1 cells requires Yhp1, because when both Yox1 and Yhp1 activities are eliminated, ECB-mediated transcription is high across the cell cycle with no evidence of periodicity. Thus, it appears that Yox1 and Yhp1 share the capacity to repress ECB activity late in the cell cycle, but Yox1 alone represses ECB function in late G1. The finding that loss of Yox1 and Yhp1 activities leads to high-level constitutive activation of the ECB reporter indicates that Yox1 and Yhp1 are required for, and may be the sole source of, cell-cycle regulation conferred upon the MCM3 ECB.
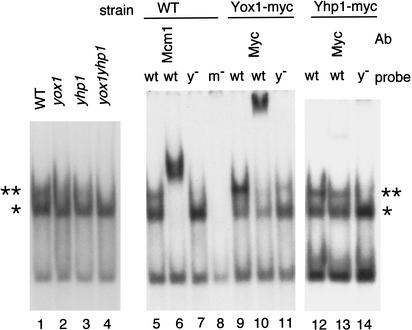
To confirm that Yox1 and/or Yhp1 bind to the YOX site in the MCM3 ECB, gel retardation assays were performed (Fig. 6). With the wild-type ECB, we observe two prominent ECB-specific complexes, which are marked in the figure with asterisks. The upper complex (**) is diminished in yox1 and yhp1 extracts and eliminated in the double mutant, suggesting that either of these two homeodomain proteins may bind to the ECB and give rise to this low-mobility complex. The lower specific complex (*) is not affected by the yox1 or yhp1 mutations, indicating that neither protein is required for this complex. However, both specific complexes contain Mcm1, because Mcm1-specific antibodies retard their mobilities. The YOX site is important for formation of the upper complex, as this complex is much less abundant when the YOX site is mutated. Mutation of the Mcm1 site precludes formation of either complex. A strain carrying a myc-epitope-tagged Yox1 forms a more prominent upper complex with slightly reduced mobility, consistent with the increased size of the tagged protein. Most of this upper complex is shifted to a higher molecular weight by the addition of myc antibodies, indicating that Yox1–myc is present in most of the upper complexes formed in vitro. Again, this upper complex is diminished but not eliminated by mutation of the YOX site. Extracts made from a strain carrying Yhp1–myc also show a prominent upper complex, but only a fraction of these complexes are supershifted by myc antibodies. This could reflect reduced expression of Yhp1–myc or difficulty in detecting the tagged protein. However, the finding that nearly all of the upper complexes formed in Yox1–myc-containing cells can be supershifted by myc antibodies suggests that Yox1 is the predominant binding partner under the conditions of this assay.
Figure 6.
Mcm1, Yox1, and Yhp1 bind to ECB elements. (Lanes 1–4) Gel retardation assays were carried out using the MCM3 ECB element and cell extracts from wild type (WT), yox1, yhp1, or yox1yhp1, as indicated. (Lanes 5–14) Gel retardation assays with the wild-type MCM3 ECB or the same sequence carrying multiple mutations in either the YOX site (y−) or the Mcm1 site (m−) as a probe. Extracts were used from wild-type cells or cells carrying myc-tagged Yox1 or Yhp1 as indicated above. Antibodies to Mcm1 (lane 6) or to the myc tag (lanes 10,13) were also added. ** indicates the upper complex, containing Mcm1, Yox1, and/or Yhp1. * marks the complex of Mcm1 with DNA.
Association of Yox1 with ECB elements directly correlates with repression
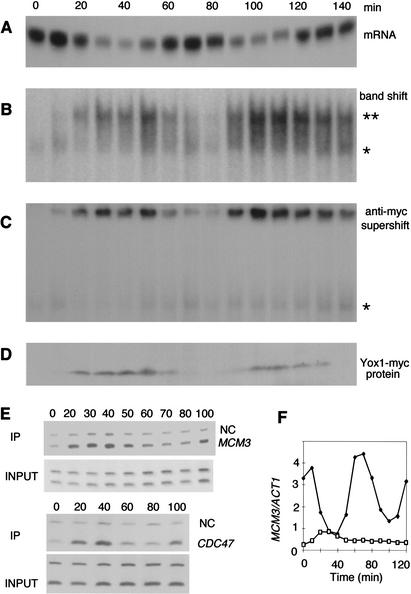
The Yox1-containing complex appears to be the predominant complex in wild-type cells, and it was of interest to determine whether this complex varies during the cell cycle and whether Yox1 binding correlates with ECB repression. Figure 7A shows the transcript profile for the MCM3 ECB:lacZ construct across two cell cycles. Figure 7B shows the results of the gel retardation assay with samples taken from Yox1–myc-containing cells across the same time course. From this analysis it is clear that the largest complexes vary in intensity across the cell cycle and peak from 20 to 60 min after release from the arrest. The upper complex (Fig. 7B,**) is almost undetectable during the first 10 min and then again at 70–80 min. These upper complexes contain Yox1, as indicated by the further retardation of its mobility when anti-myc antibodies are added to the reaction (Fig. 7C). Comparison of Yox1 binding in vitro to the pattern of ECB-driven transcription shows that the association of Yox1 with ECB complexes correlates with repression of ECB-driven transcription.
Figure 7.
Cell-cycle regulation of Yox1 expression results in the cell-cycle regulation of ECB activity. (A) α-factor-synchronized cells were analyzed at 10-min intervals for lacZ transcript driven by the MCM3 ECB element. (B) DNA binding to the MCM3 ECB probe. Asterisks denote ECB-specific complexes as in Figure 6. (C) Gel retardation assay as in B to which antibodies to the myc tag were added. (D) Immunoblot of the cell extracts used in B and C detecting Yox1–myc with anti-myc antibodies. (E) CHIP assays performed with anti-myc polyclonal antibodies on the MCM3 and CDC47 promoters with extracts from synchronized Yox1–myc cells. “NC” indicates the negative control. PCR was performed on chromatin fragments before (INPUT) and after IP. (F) Genomic MCM3 mRNA was monitored from α-factor synchronized wild-type cells (solid symbols) and compared to the same transcript from yox1yhp1 cells expressing Yox1 constitutively from the GALs:YOX1 construct (open symbols).
To verify that the same pattern of Yox1 binding occurs in vivo, we carried out a series of chromatin immunoprecipitations (CHIPs). Using anti-myc antibodies, we verified that Yox1–myc forms complexes on the genomic CDC47 and MCM3 promoters that are cell cycle-specific (Fig. 7E). Binding is not apparent in early G1 when ECB transcription is high. Maximum Yox1–myc-binding occurs from 20 to 40 min and then again at 100 min, mirroring the pattern observed with the gel retardation assay. These data suggest that Yox1 exerts its repressive function by binding to ECB complexes during the interval from late G1 to M phase.
Yox1 expression is periodic, and this determines the interval of ECB repression
Because Yox1 is a periodically expressed gene, it was of interest to see whether Yox1 protein levels were correlated with Yox1 binding to ECB elements. Figure 7D shows that the pattern of Yox1 expression is directly correlated with ECB binding and repression. The simplest interpretation of this finding is that Yox1 is regulated at the level of transcription, and when it is expressed, it binds and represses its target genes. If this is the case, constitutive production of Yox1 should result in constitutive repression of target genes. That would explain the initial finding that GAL-induced overexpression of Yox1 is highly deleterious to cell growth, since many of the Yox1 target genes are essential. To determine whether YOX1, transcribed constitutively at a moderate level, was sufficient to repress its target genes throughout the cell cycle, we constructed a GALs:YOX1 strain wherein GAL promoter activity was attenuated to about one-tenth of the normal GAL-induced level (Mumberg et al. 1994). This strain grows well in galactose. Figure 7F shows the pattern of MCM3 transcription from a wild-type cell compared to that of the yox1yhp1 strain carrying GALs:YOX1. Wild-type cells show a tenfold oscillation in MCM3 transcript levels. In contrast, the constitutive expression of Yox1 represses MCM3 down to trough levels and maintains it at that low level throughout the cell cycle. This is not due to loss of synchrony, as judged by the synchronous budding profile of these cells (data not shown). Rather, the transcription of Yox1 throughout the cell cycle specifically eliminates the cell-cycle regulation of MCM3.
Discussion
Homeodomain proteins play prominent roles in development. Their expression is tissue-specific and often defines cell identity by directing the expression of genes that drive specific developmental decisions. As a result, ectopic expression of homeodomain proteins can also lead to dramatic homeotic transformations of one cell type to another (Bondos and Tan 2001). In the unicellular eukaryote, S. cerevisiae, we observe a variation on this theme, in that the transcription of two homeodomain proteins, Yox1 and Yhp1, is temporally restricted to specific intervals of the cell cycle, serving to limit the activity of a constitutively expressed transcription factor and induce phase-specific transcription of a battery of genes.
Yox1 and Yhp1 bind to Mcm1, and to the same DNA sequence as Phox1, their closest relative. The binding site is a typical homeodomain binding site (T/CaATTa), and as expected, Yox1 and Yhp1 share residues with Phox1 that are critical for DNA binding (Mann 1995). They also share a number of other residues within the homeodomain which are likely to be important for binding MADS boxes. However, there is no obvious similarity between Phox1 and the two yeast proteins outside of the homeodomain, and thus they are unlikely to have other common properties beyond those conferred via the homeodomain. Yox1 and Yhp1 share a core of homology, but they diverge over about half of their sequence. This leaves open the possibility that they may interact with other DNA-binding proteins and regulate a more diverse set of genes (Horak et al. 2002).
The role of Yox1 and Yhp1 as transcriptional repressors was suggested by the identification of a group of genes that are repressed by overexpression of Yox1 and derepressed throughout the cell cycle in cells lacking Yox1 and Yhp1. The genes are transcribed specifically during the late M/early G1 interval of the cell cycle, and each contains at least one close match to an ECB element in its promoter, to which Yox1 and Yhp1 were shown to bind. ECB elements were previously found in the SWI4, CLN3, CDC47, and CDC6 promoters and were shown to confer M/G1-specific transcription (McInerny et al. 1997). It now appears that at least two features are important for the function of an ECB element. The first is the 16-bp palindrome to which Mcm1 is known to bind. The second is the ability to bind either Yox1 or Yhp1. A consensus binding site for Yox1 and Yhp1 was identified adjacent to the Mcm1-binding site. However, we have not explored the importance of the spacing between these sites, nor can we conclude that these are the only two sequences that are important for ECB function. For example, we know that a sequence flanking the Mcm1-binding site but on the opposite side to the YOX site influences the level and the timing of transcription of the SWI4 ECB (Mai et al. 2002). Additional isolated YOX- and/or Mcm1-binding sites can also be found within the promoters of Yox1-regulated genes. Indeed, we searched yeast intergenic DNA, using a weight matrix derived from the YOX sites shown in Figure 3, and we found that YOX sites occur about every 50 bp. It seems unlikely that this very common sequence serves as a regulatory site on its own.
Alternatively, it is possible that the presence of a YOX site is less important than the ability of Yox1 to gain access to Mcm1 in any given promoter context. There is considerable evidence that Yox1 and/or Yhp1 can repress Mcm1 activity in the absence of an adjacent YOX site. In vitro, binding of Yox1 or Yhp1 to Mcm1 is detectable on DNA lacking the YOX site. In addition, Yox1 overproduction represses transcription from an ECB reporter about tenfold, compared to fivefold repression of the same ECB with the YOX site mutated (S. Miles and L. Breeden, unpubl.). These data indicate that the interaction between these homeodomain proteins and Mcm1 is strong enough to tether these repressors to the ECB complex even in the absence of their DNA-binding site. Consistent with this, the transcription of the ECB reporter construct lacking the YOX site is derepressed across the cell cycle, but it still peaks at the M/G1 boundary. This residual regulation is likely to be mediated by Yox1 and/or Yhp1, because the ECB reporter shows no M/G1-specific activation in the yox1yhp1 mutant. The simplest explanation for these data is that Yox1 and/or Yhp1 can bind to Mcm1 and repress transcription in the absence of their DNA-binding site. The ability to confer transcriptional regulation in the absence of DNA binding is a property that has been documented for a number of homeodomain proteins, including Phox1 (Grueneberg et al. 1992; Catron et al. 1995; Copeland et al. 1996). If this is also true for Yox1 and Yhp1, their ability to restrict Mcm1 activity to the M/G1 boundary may only require access to Mcm1. Interaction with Mcm1 may be strengthened by the presence of an adjacent YOX site, and it may be prevented by the presence of binding sites for other Mcm1 partners.
The hybrid promoter elements we identified have Mcm1-binding sites flanked on one side by a YOX site and on the other by a forkhead (FKH)-binding site. Yox1 and/or Yhp1 clearly restrict the transcriptional activity of these promoters, but they do not delay their activation to coincide with other YOX-regulated genes. Rather, these hybrid promoters are activated for a unique interval that we refer to as late M. Mcm1 and Fkh proteins are bound to the early M-specific promoters throughout the cell cycle, and a third protein, Ndd1, is recruited to activate transcription (Koranda et al. 2000). The late M-specific transcription conferred by the hybrid promoter elements may be due to competition between Yox1 and Ndd1.
Yox1 and Yhp1 are redundant in that either can repress ECB function in the absence of the other. However, Yox1 has the unique ability to repress transcription of about 200 genes and retard the growth rate of cells when it is constitutively overproduced from the GAL promoter. One possible explanation is that Yox1 is expressed at a much higher level than Yhp1 under these conditions. Yhp1–myc is difficult to detect in immunoblots (data not shown), so it may be specifically targeted for proteolysis. It is also possible that Yox1 has a stronger interaction with Mcm1 or that it is better able to recruit corepressors. The other well studied case in which Mcm1 and another homeodomain protein, alpha2, repress a-specific gene expression involves the recruitment of Ssn6 and Tup1 (Wahi and Johnson 1995). Possible corepressors of Yox1-mediated repression are being investigated.
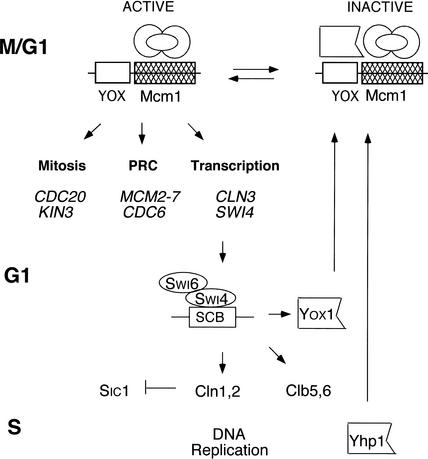
Mcm1 is bound to ECB elements throughout the cell cycle, but Yox1 binds transiently from late G1 to M phase when ECB activity is repressed. Moreover, the absence of Yox1 and Yhp1 or the constitutive expression of Yox1 results in loss of cell-cycle regulation of ECB activity. These data support the view that the M/G1-specificity of ECB elements is conferred solely by the regulated expression of the Yox1 and Yhp1 repressors. Figure 8 highlights the transcriptional circuitry that controls G1 progression and the contributions made by ECB-regulated genes. ECBs are activated by Mcm1 and possibly other unknown proteins, and their activity is sustained until Yox1 or Yhp1 is expressed. YOX1 is transcribed in late G1. Chromatin immunoprecipitations (IPs; Iyer et al. 2001; Simon et al. 2001) and microarrays performed on cells overproducing Swi4 (J. Sidorova and L. Breeden, unpubl.) suggest that Swi4/Swi6 complexes are responsible for the late G1-specific transcription of YOX1. Interestingly, SWI4 is an ECB-regulated and Yox1-repressed gene, so this sets up a classic negative feedback loop, where Yox1 halts its own synthesis by repressing synthesis of its activator. This negative feedback has the effect of sustaining ECB-activated transcription until Yox1 reaches the threshold required to effect repression. The coordination of YOX1 transcription with other Swi4-regulated genes, for example, CLN1 and CLN2, enables these cyclins to accumulate to the threshold required to drive cells into S phase, and then be turned off. Swi4 also binds YHP1 (Horak et al. 2002), but the significance of this is unknown. YHP1 is transcribed in late S phase and shares the capacity to repress ECB activity late in the cell cycle. Elimination of both Yox1 and Yhp1 is required to reactivate ECBs for the next G1.
Figure 8.
Regulation of ECB-driven transcription and its role in G1 progression. Activation of ECB elements occurs in M/G1 phase of the cell cycle, involving constitutively bound Mcm1 and possibly another activator. Examples of the three prominent classes of ECB-regulated genes are listed. Yox1, transcribed in late G1 by Swi4 and Swi6, rises to the level required to repress ECB function, thereby inhibiting its own synthesis. This negative feedback sustains ECB-activated transcription until Yox1 reaches the threshold required to affect repression. YOX1, CLN1, CLN2, CLB5, and CLB6 are coordinately transcribed. This coordination enables these cyclins to accumulate to the threshold required to drive cells into S phase, and then be turned off rapidly. Yhp1 is made in late S phase. Either Yox1 or Yhp1 can bind to ECBs late in the cycle and repress their activity. Elimination of Yox1 and Yhp1 is required to reactivate the ECB elements late in M phase.
One outstanding question is why two different repressors, with different kinetics of expression, have evolved to regulate M/G1-specific genes. It may be that Yhp1 serves solely as a back-up for Yox1, or that Yhp1 is the primary regulator of a subset of the genes identified in the study of yox1yhp1 cells. A third, more interesting possibility is that these two waves of repressor activity evolved to allow differential regulation of ECB-dependent genes under different conditions. Loss of both Yox1 and Yhp1 speeds the G1 to S transition. Differential effects on the early or late wave of repressor activity could influence the cell cycle by altering the timing of the establishment or maintenance of the repressed state.
Materials and methods
Strains, plasmids, and growth conditions
Cells were grown at 30°C in yeast peptone media (YEP) supplemented with 2% galactose or glucose as indicated. W303 (MATa ade2-1 trp1-1 can1-100 leu2-3,115 his3-11 ura3 ho ssd1-d) and its derivatives were used in all of the experiments. YOX1 and YHP1 were deleted using pFA6a-HIS3MX6 and pFA6a-TRP1, respectively, as described (Longtine et al. 1998). GALs:YOX1 plasmid was constructed by inserting the YOX1 coding sequence into p416 GALS (Mumberg et al. 1994).
Strains with GAL:YOX1 or GAL:YHP1 integrated at the YOX1 and YHP1 loci were constructed using pFA6a-HIS3MX6-pGAL1 as described (Longtine et al. 1998). The C-terminal Myc tagging of Yox1 and Yhp1 was done with the same method, using pFA6a-13Myc-HIS3MX6 and pFA6a-13Myc-TRP1, respectively, to insert the myc tag into the native loci. Yox1–myc and Yhp1–myc were tested for function in a yhp1 and a yox1 background, respectively. Defects in the tagged proteins would lead to constitutive transcription of MCM3 through the cell cycle (see Fig. 5). Yox1–myc was proven to be fully functional; however, the tagged Yhp1 had only partial activity.
The MCM3 ECB:lacZ reporter and mutant derivatives were generated with oligonucleotides with 5′ XhoI and 3′ NotI ends cloned into pSH144, a LacZ reporter vector and integrated at URA3. The wild-type sequence (GGTAGAAGAAACAATTA CTTTTCCTAAATGGGTAAAAACTCGTG), or the equivalent sequence with YOX site or the Mcm1-binding site mutated at positions indicated in bold serve as the only upstream activation sequence to the lacZ gene in this vector.
Cells were synchronized with either 5 μg/mL α-factor (United Biochemical Research; Breeden 1997) or by elutriation (Johnston and Johnson 1997) into fresh medium. Synchrony was followed by counting buds and by flow cytometry on a Becton-Dickinson FACScan 2 with Sytox (Foss 2001).
Immunoprecipitations
Cells were lysed by vortexing with glass beads for 4 × 30 sec, level 4.5, on a Fast Prep FP120 (Savant BIO/CAN Scientific) in lysis buffer (100 mM NaCl, 20 mM Tris, at pH 7.5, 5% glycerol, 1 mM EDTA, 1 mM MgCl2, 0.1% NP-40). Next, 1.5 mg of whole cell protein was immunoprecipitated with anti-c-myc (Rabbit polyclonal IgG, Santa Cruz Biotechnology) at 4°C for 2 h. Protein G coupled to Dynabeads (Dynal) was used to pull down the complex.
Transcript measurements
For microarray analysis through the cell cycle, the yox1yhp1 cells were synchronized with α-factor in YEP-containing 2% glucose. RNA was extracted from cell samples taken out every 10 min after arrest release. Thirty micrograms of total RNA was used for cDNA synthesis. The cDNA was coupled to Cy5 using an amino-allyl dye-coupling procedure (http://cmgm.stanford.edu/pbrown/protocols/aadUTPCouplingProcedure.htm). RNA from an asynchronous population of W303a cells was used as a control, and its cDNA was labeled with Cy3 dye. Cy5-labeled cDNA from each timepoint was mixed with Cy3-labeled control cDNA, and hybridized to yeast cDNA microarrays as described (Fazzio et al. 2001). Arrays were analyzed with GenePix Pro software (Axon Instruments).
S1 protection assays were performed using oligonucleotide probes as described (Mai et al. 2002), except that probes were purified with a G-25 Sephadex column, phenol/chloroform extracted, and ethanol-precipitated.
DNA binding assays
Gel retardation assays were performed as described (Mai et al. 2002). Binding to the ECB element from the MCM3 promoter (from region −211 to −160) or to the same oligonucleotides containing either the YOX site or Mcm1 site mutated (as above) was assayed using 40 μg of crude cell protein. Complexes were allowed to form at room temperature for 30 min. Supershifts were performed with 0.6 μg of antibody (monoclonal anti c-myc; clone 9E10, Roche Diagnostics), or polyclonal anti-Mcm1 (Jarvis et al. 1989).
CHIPs were performed with modifications to those previously described (Strahl-Bolsinger et al. 1997; Dudley et al. 1999). Cells were cross-linked with 1% formaldehyde at room temperature for 15 min. The cross-linker was quenched by addition of glycine (125 mM) and incubated for an additional 5 min. Cells were washed 2× and then broken with glass beads with two 30-sec pulses in a Mini-BeadBeater 8 (Biospec Products) in a lysis buffer containing 50 mM Hepes-KOH, at pH 7.6, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, and 0.1% Na deoxycholate. The chromatin was washed 2× in the lysis buffer and sonicated 5 × 10 sec. Four hundred micrograms of sheared chromatin was used for the IP. The IPs were washed sequentially 3× each with lysis buffer, lysis buffer with 500 mM NaCl, and the CHIP wash buffer (10 mM Tris-HCl, at pH 8.0, 0.25 M LiCl, 0.5% NP-40, 0.5% Na deoxycholate, and 1 mM EDTA). The final wash was performed with TE (10 mM Tris-HCl, at pH 8.0, 1 mM EDTA), and the precipitate was eluted from the beads by incubating for 15 min at 65°C with 50 mM Tris-HCl, at pH 8.0, 10 mM EDTA, and 1% SDS. Cross-links were reversed by overnight incubation at 65°C. Proteins were digested with Proteinase K. DNA was phenol-extracted and ethanol-precipitated, then resuspended in 50 μL TE and RNase-digested. For input samples, 40 μg was made up to 250 μL with TE/1% SDS. Cross-links were reversed and the DNA purified as indicated above.
For PCR, 2 μL of IP or appropriately diluted input DNA was used. Two sets of primers were used in each reaction. Primer sets were designed to flank and amplify the ECB elements in MCM3 and CDC47 or part of the ACT1 coding sequence, which serves as a negative control. Primer sequences are available upon request. PCR involved an initial denaturation of 3 min at 94°C, then 26 cycles at: 94°C for 10 sec, 57°C for 5 sec, and 72°C for 10 sec, then a final extension at 72°C for 1 min. Control PCR reactions were carried out with dilutions of the input DNA to make sure that we were in the linear range of the assay.
Acknowledgments
We sincerely thank Gary Stormo for his valuable suggestions, and the use of programs Patser, Consensus, and Co-Bind. We also appreciate the many helpful discussions with members of the Breeden, Biggins, and Tsukiyama laboratories and the excellent technical support of J. Delrow and C. Neal in the FHCRC microarray facility. Thanks also to S. Biggins and R. Eisenman for helpful comments on the manuscript. This research was supported by grants from the NIH (GM41073 to L.B. and HG00249 to G.S.)
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL lbreeden@fhcrc.org; FAX (206) 667-6526.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1034302.
References
- Bondos SE, Tan XX. Combinatorial transcriptional regulation: The interaction of transcription factors and cell signaling molecules with homeodomain proteins in Drosophila development. Crit Rev Eukaryot Gene Expr. 2001;11:145–171. [PubMed] [Google Scholar]
- Breeden LL. Alpha factor synchronization of budding yeast. Methods Enzymol. 1997;283:332–341. doi: 10.1016/s0076-6879(97)83027-3. [DOI] [PubMed] [Google Scholar]
- Catron KM, Zhang H, Marshall SC, Inostroza JA, Wilson JM, Abate C. Transcriptional repression by Msx-1 does not require homeodomain DNA-binding sites. MolCell Biol. 1995;15:861–871. doi: 10.1128/mcb.15.2.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho RJ, Campbell MJ, Winzeler EA, Steinmetz L, Conway A, Wodicka L, Wolfsberg TG, Gabrielian AE, Landsman D, Lockhart DJ, et al. A genome-wide transcriptional analysis of the mitotic cell cycle. Mol Cell. 1998;2:65–73. doi: 10.1016/s1097-2765(00)80114-8. [DOI] [PubMed] [Google Scholar]
- Copeland JW, Nasiadka A, Dietrich BH, Krause HM. Patterning of the Drosophila embryo by a homeodomain-deleted Ftz polypeptide. Nature. 1996;379:162–165. doi: 10.1038/379162a0. [DOI] [PubMed] [Google Scholar]
- Cross FR. DAF1, a mutant gene affecting size control, pheromone arrest, and cell cycle kinetics of Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:4675–4684. doi: 10.1128/mcb.8.11.4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirick L, Bohm T, Nasmyth K. Roles and regulation of Cln-Cdc28 kinases at the start of the cell cycle of Saccharomyces cerevisiae. EMBO J. 1995;14:4803–4813. doi: 10.1002/j.1460-2075.1995.tb00162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley AM, Rougeulle C, Winston F. The Spt components of SAGA facilitate TBP binding to a promoter at a post-activator-binding step in vivo. Genes & Dev. 1999;13:2940–2945. doi: 10.1101/gad.13.22.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazzio TG, Kooperberg C, Goldmark JP, Neal C, Basom R, Delrow J, Tsukiyama T. Widespread collaboration of Isw2 and Sin3-Rpd3 chromatin remodeling complexes in transcriptional repression. Mol Cell Biol. 2001;21:6450–6460. doi: 10.1128/MCB.21.19.6450-6460.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foss EJ. Poflp regulates DNA damage responses during S phase in Saccharomyces cerevisiae. Genetics. 2001;157:567. doi: 10.1093/genetics/157.2.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster R, Mikesell GE, Breeden L. Multiple Swi6-dependent cis-acting elements control SWI4 transcription through the cell cycle. Mol Cell Biol. 1993;13:3792–3801. doi: 10.1128/mcb.13.6.3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego C, Gari E, Colomina N, Herrero E, Aldea M. The Cln3 cyclin is down-regulated by translational repression and degradation during the G1 arrest caused by nitrogen deprivation in budding yeast. EMBO J. 1997;16:7196–7206. doi: 10.1093/emboj/16.23.7196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galliot B, de Vargas C, Miller D. Evolution of homeobox genes: Q50 paired-like genes founded the paired class. Dev Genes Evol. 1999;209:186–197. doi: 10.1007/s004270050243. [DOI] [PubMed] [Google Scholar]
- Grueneberg DA, Natesan S, Alexandre C, Gilman MZ. Human and Drosophila homeodomain proteins that enhance the DNA-binding activity of serum response factor. Science. 1992;257:1089–1095. doi: 10.1126/science.257.5073.1089. [DOI] [PubMed] [Google Scholar]
- GuhaThakurta D, Stormo GD. Identifying target sites for cooperatively binding factors. Bioinformatics. 2001;17:608–621. doi: 10.1093/bioinformatics/17.7.608. [DOI] [PubMed] [Google Scholar]
- Hartwell LH, Unger MW. Unequal division in Saccharomyces cerevisiaeand its implications for the control of cell cycle. J Cell Biol. 1977;75:422–435. doi: 10.1083/jcb.75.2.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertz GZ, Stormo GD. Identifying DNA and protein patterns with statistically significant alignments of multiple sequences. Bioinformatics. 1999;15:563–577. doi: 10.1093/bioinformatics/15.7.563. [DOI] [PubMed] [Google Scholar]
- Horak, C.E., Luscombe, N.M., Qian, J., Bertone, P., Piccirrillo, S., Gerstein, M., and Snyder, M. 2002. Complex transcriptional circuitry at the G1/S transition in Saccharomyces cerevisiae. Genes & Dev. (this issue). [DOI] [PMC free article] [PubMed]
- Iyer VR, Horak CE, Scafe CS, Botstein D, Snyder M, Brown PO. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature. 2001;409:533–538. doi: 10.1038/35054095. [DOI] [PubMed] [Google Scholar]
- Jagadish MN, Carter BL. Genetic control of cell division in yeast cultured at different growth rates. Nature. 1977;269:145–147. doi: 10.1038/269145a0. [DOI] [PubMed] [Google Scholar]
- Jarvis EE, Clark KL, Sprague GF. The yeast transcription activator PRTF, a homolog of the mammalian serum response factor, is encoded by the MCM1 gene. Genes & Dev. 1989;3:936–945. doi: 10.1101/gad.3.7.936. [DOI] [PubMed] [Google Scholar]
- Johnson AD. Molecular mechanisms of cell-type determination in budding yeast. Curr Opin Genet Dev. 1995;5:552–558. doi: 10.1016/0959-437x(95)80022-0. [DOI] [PubMed] [Google Scholar]
- Johnston LH, Johnson AL. Elutriation of budding yeast. Methods Enzymol. 1997;283:342–350. doi: 10.1016/s0076-6879(97)83028-5. [DOI] [PubMed] [Google Scholar]
- Johnston GC, Pringle JR, Hartwell LH. Coordination of growth with cell division in the yeast Saccharomyces cerevisiae. Exp Cell Res. 1977;105:79–98. doi: 10.1016/0014-4827(77)90154-9. [DOI] [PubMed] [Google Scholar]
- Johnston GC, Ehrhardt CW, Lorincz A, Carter BL. Regulation of cell size in the yeast Saccharomyces cerevisiae. J Bacteriol. 1979;137:1–5. doi: 10.1128/jb.137.1.1-5.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman E. In vitro binding to the leucine tRNA gene identifies a novel yeast homeobox gene. Chromosoma. 1993;102:174–179. doi: 10.1007/BF00387732. [DOI] [PubMed] [Google Scholar]
- Koranda M, Schleiffer A, Endler L, Ammerer G. Forkhead-like transcription factors recruit Ndd1 to the chromatin of G2/M-specific promoters. Nature. 2000;406:94–97. doi: 10.1038/35017589. [DOI] [PubMed] [Google Scholar]
- Kumar R, Reynolds DM, Shevchenko A, Goldstone SD, Dalton S. Forkhead transcription factors, Fkh1p and Fkh2p, collaborate with Mcm1p to control transcription required for M-phase. Curr Biol. 2000;10:896–906. doi: 10.1016/s0960-9822(00)00618-7. [DOI] [PubMed] [Google Scholar]
- Kunoh T, Kaneko Y, Harashima S. YHP1 encodes a new homeoprotein that binds to the IME1 promoter in Saccharomyces cerevisiae. Yeast. 2000;16:439–449. doi: 10.1002/(SICI)1097-0061(20000330)16:5<439::AID-YEA536>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cervisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Lydall D, Ammerer G, Nasmyth K. A new role for MCM1 in yeast: Cell cycle regulation of SWI5 transcription. Genes & Dev. 1991;5:2405–2419. doi: 10.1101/gad.5.12b.2405. [DOI] [PubMed] [Google Scholar]
- MacKay V, Mai B, Waters L, Breeden L. Early cell cycle box-mediated transcription of CLN3 and SWI4 contributes to the proper timing of the G1-to-S transition in budding yeast. Mol Cell Biol. 2001;21:4140–4148. doi: 10.1128/MCB.21.13.4140-4148.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher M, Cong F, Kindelberger D, Nasmyth K, Dalton S. Cell cycle-regulated transcription of the CLB2 gene is dependent on Mcm1 and a ternary complex factor. Mol Cell Biol. 1995;15:3129–3137. doi: 10.1128/mcb.15.6.3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mai B, Miles S, Breeden LL. Characterization of the ECB binding complex responsible for the M/G1-specific transcription of CLN3 and SWI4. Mol Cell Biol. 2002;22:430–441. doi: 10.1128/MCB.22.2.430-441.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann RS. The specificity of homeotic gene function. Bioessays. 1995;17:855–863. doi: 10.1002/bies.950171007. [DOI] [PubMed] [Google Scholar]
- McInerny CJ, Partridge JF, Mikesell GE, Creemer DP, Breeden LL. A novel Mcm1-dependent promoter element in the SWI4, CLN3, CDC6 and CDC47 promoters activates M/G1-specific transcription. Genes & Dev. 1997;11:1277–1288. doi: 10.1101/gad.11.10.1277. [DOI] [PubMed] [Google Scholar]
- Miller ME, Cross FR. Cyclin specificity: How many wheels do you need on a unicycle? J Cell Sci. 2001;114:1811–1820. doi: 10.1242/jcs.114.10.1811. [DOI] [PubMed] [Google Scholar]
- Mumberg D, Muller R, Funk M. Regulatable promoters of Saccharomyces cerevisiae: Comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 1994;22:5767–5768. doi: 10.1093/nar/22.25.5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash R, Tokiwa G, Anand S, Erickson K, Futcher AB. The WHI1 gene of Saccharomyces cerevisiae tethers cell division to cell size and is a cyclin homolog. EMBO J. 1988;7:4335–4346. doi: 10.1002/j.1460-2075.1988.tb03332.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash P, Tang X, Orlicky S, Chen Q, Gertler FB, Mendenhall MD, Sicheri F, Pawson T, Tyers M. Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature. 2001;414:514–521. doi: 10.1038/35107009. [DOI] [PubMed] [Google Scholar]
- Newcomb LL, Hall DD, Heideman W. AZF1 is a glucose-dependent positive regulator of CLN3 transcription in Saccharomyces cerevisiae. Mol Cell Biol. 2002;22:1607–1614. doi: 10.1128/mcb.22.5.1607-1614.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polymenis M, Schmidt EV. Coupling of cell division to cell growth by translational control of the G1 cyclin CLN3 in yeast. Genes & Dev. 1997;11:2522–2531. doi: 10.1101/gad.11.19.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupe I. Checking cell size in yeast. Trends Genet. 2002;8:479–485. doi: 10.1016/s0168-9525(02)02745-2. [DOI] [PubMed] [Google Scholar]
- Schneider BL, Yang Q-H, Futcher AB. Linkage of replication to start by the Cdk inhibitor Sic1. Science. 1996;272:560–562. doi: 10.1126/science.272.5261.560. [DOI] [PubMed] [Google Scholar]
- Sillje HH, ter Schure EG, Rommens AJ, Huls PG, Woldringh CL, Verkleij AJ, Boonstra J, Verrips CT. Effects of different carbon fluxes on G1 phase duration, cyclin expression, and reserve carbohydrate metabolism in Saccharomyces cerevisiae. J Bacteriol. 1997;179:6560–6565. doi: 10.1128/jb.179.21.6560-6565.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon I, Barnett J, Hannett N, Harbison CT, Rinaldi NJ, Volkert TL, Wyrick JJ, Zeitlinger J, Gifford DK, Jaakkola TS, et al. Serial regulation of transcriptional regulators in the yeast cell cycle. Cell. 2001;106:697–708. doi: 10.1016/s0092-8674(01)00494-9. [DOI] [PubMed] [Google Scholar]
- Spellman PT, Sherlock G, Zhang MQ, Iyer VR, Anders K, Eisen MB, Brown PO, Botstein D, Futcher B. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol Biol Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl-Bolsinger S, Hecht A, Luo K, Grunstein M. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes & Dev. 1997;11:83–93. doi: 10.1101/gad.11.1.83. [DOI] [PubMed] [Google Scholar]
- Stuart D, Wittenberg C. CLN3, not positive feedback, determines the timing of CLN2 transcription in cycling cells. Genes & Dev. 1996;9:2780–2794. doi: 10.1101/gad.9.22.2780. [DOI] [PubMed] [Google Scholar]
- Treisman R, Ammerer G. The SRF and MCM1 transcription factors. Curr Opin Genet Dev. 1992;2:221–226. doi: 10.1016/s0959-437x(05)80277-1. [DOI] [PubMed] [Google Scholar]
- Tye BK. MCM Proteins in DNA Replication. Annu Rev Biochem. 1999;68:649–686. doi: 10.1146/annurev.biochem.68.1.649. [DOI] [PubMed] [Google Scholar]
- Tyers M. The cyclin-dependent kinase inhibitor p40SIC1 imposes the requirement for Cln G1 cyclin function at start. Proc Natl Acad Sci. 1996;93:7772–7776. doi: 10.1073/pnas.93.15.7772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahi M, Johnson AD. Identification of genes required for alpha2 repression in Saccharomyces cerevisiae. Genetics. 1995;140:79–90. doi: 10.1093/genetics/140.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittenberg C, Sugimoto K, Reed SI. G1-specific cyclins of S. cerevisiae: Cell cycle periodicity, regulation by mating pheromone, and association with the p34CDC28 protein kinase. Cell. 1990;62:225–237. doi: 10.1016/0092-8674(90)90361-h. [DOI] [PubMed] [Google Scholar]
- Zachariae W, Nasmyth K. Whose end is destruction: Cell division and the anaphase-promoting complex. Genes & Dev. 1999;13:2039–2058. doi: 10.1101/gad.13.16.2039. [DOI] [PubMed] [Google Scholar]
- Zhao LP, Prentice R, Breeden L. Statistical modeling of large microarray data sets to identify stimulus-response profiles. Proc Natl Acad Sci. 2001;98:5631–5636. doi: 10.1073/pnas.101013198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G, Davis TN. The fork head transcription factor Hcm1p participates in the regulation of SPC110, which encodes the calmodulin-binding protein in the yeast spindle pole body. Biochim Biophys Acta. 1998;1448:236–244. doi: 10.1016/s0167-4889(98)00135-9. [DOI] [PubMed] [Google Scholar]
- Zhu G, Spellman PT, Volpe T, Brown PO, Botstein D, Davis TN, Futcher B. Two yeast forkhead genes regulated the cell cycle and pseudohyphal growth. Nature. 2000;406:90–93. doi: 10.1038/35017581. [DOI] [PubMed] [Google Scholar]