Abstract
Many nuclear proteins are inactivated during mitotic entry, presumably as a prerequisite to chromatin condensation and cell division. C2H2 zinc fingers define the largest transcription factor family in the human proteome. The linker separating finger motifs is highly conserved and resembles TGEKP in more than 5000 occurrences. However, the reason for this conservation is not fully understood. We demonstrate that all three linkers in the DNA-binding domain of Ikaros are phosphorylated during mitosis. Phosphomimetic substitutions abolished DNA-binding and pericentromeric localization. A linker within Sp1 was also phosphorylated, suggesting that linker phosphorylation provides a global mechanism for inactivation of the C2H2 family.
Keywords: Zinc finger, mitosis, phosphorylation, Ikaros, cell cycle
It has long been known that entry into mitosis is accompanied by the cessation of transcription (Prescott and Bender 1962). Transcription by RNA polymerase II is inhibited by phosphorylation of TFIIH, TFIID, and RNA polymerase II itself (Segil et al. 1996; Bellier et al. 1997; Akoulitchev and Reinberg 1998; Long et al. 1998). Phosphorylation also inhibits the activities of other protein complexes that are generally important for transcription, including the SWI/SNF nucleosome remodeling complex (Sif et al. 1998).
Although inactivation of the general machinery should be sufficient for transcriptional shutdown, several gene-specific transcription factors are also known to be inactivated during the G2/M transition. For some factors, reduced DNA-binding activities have been observed in extracts from mitotic cells (Segil et al. 1991; Caelles et al. 1995; Martínez-Balbás et al. 1995; Gottesfeld and Forbes 1997). For others, normal DNA-binding activities were observed, but the factors were displaced from chromatin during mitosis via unknown mechanisms (Martínez-Balbás et al. 1995; Gottesfeld and Forbes 1997). The reason for the mitotic inactivation of gene-specific transcription factors is not known. However, one can speculate that inactivation is necessary for chromatin condensation, cell division, and/or the re-establishment of gene expression patterns as cells exit mitosis.
The C2H2 zinc finger is the most prevalent protein motif in mammalian cells and defines the largest family of sequence-specific DNA-binding proteins (Lander et al. 2001; Tupler et al. 2001). The C2H2 motif is characterized by conserved cysteines, histidines, and hydrophobic residues, which stabilize the three-dimensional structure consisting of a two-stranded antiparallel β-sheet and α-helix surrounding a central zinc ion (Wolfe et al. 2000). Although the three hydrophobic and four zinc-coordinating residues are the only highly conserved residues in an individual C2H2 motif, an additional region is highly conserved in C2H2 DNA-binding domains, which always contain more than one finger: the 5-amino acid linker separating the individual finger motifs (Wolfe et al. 2000). The linker sequence matches or resembles TGEKP in the vast majority of zinc finger linkers encoded by the human genome (Wolfe et al. 2000; Lander et al. 2001).
The existence of a highly conserved linker has led to considerable interest in its significance. Structural studies revealed that the linker is flexible when the protein is free in solution, but becomes rigid and well-ordered upon DNA binding (Clemens et al. 1994; Wuttke et al. 1997; Bowers et al. 1999; Laity et al. 2000). These studies also revealed that the conserved threonine plays a critical role in stablizing the α-helix within the preceding finger motif. This role is supported by measurements of the DNA-binding affinities of mutant proteins (Thukral et al. 1991; Wilson et al. 1992; Choo and Klug 1993). The mutant studies revealed that linker residues other than the threonine can make contributions to binding affinity. However, as emphasized by Choo and Klug (1993), the contributions to binding affinity are often minimal and do not fully explain the remarkable conservation of the linker.
The Ikaros protein contains four C2H2 zinc finger motifs near its N terminus that contribute to sequence-specific DNA binding (Hahm et al. 1994; Molnár and Georgopoulos 1994). Ikaros is expressed in most hematopoietic cells and plays essential roles in the development of the immune system and in an immune response (Cortes et al. 1999). Several lines of evidence suggest that Ikaros contributes to the heritable silencing of developmentally regulated genes (Georgopoulos 2002; Smale and Fisher 2002). Consistent with this hypothesis, Ikaros is predominantly targeted to foci of pericentromeric heterochromatin in interphase nuclei through direct binding to satellite repeat sequences (Brown et al. 1997; Cobb et al. 2000). Ikaros has also been implicated in the regulation of cell cycle progression (Cortes et al. 1999) and its subnuclear localization varies at different cell cycle stages (Brown et al. 1997, 1999; Kim et al. 1999). In particular, Ikaros was found to dissociate from chromatin during early stages of mitosis (Brown et al. 1997).
To explore the mechanisms underlying the dynamic changes in Ikaros localization, we studied its dissociation from pericentromeric heterochromatin during the G2/M transition. The initial results suggested that a G2/M-specific phosphorylation event that inhibits DNA binding is responsible for its release from heterochromatin. Surprisingly, phosphopeptide mapping experiments revealed that the G2/M-specific phosphoacceptors are within the three linkers separating the four zinc finger motifs. G2/M-specific phosphorylation of a C2H2 linker in Sp1 was also observed, suggesting that linker phosphorylation is a common mechanism for mitotic inactivation of C2H2 zinc finger proteins. Thus, the results suggest that the conserved linker serves dual functions in stabilizing DNA binding and in providing a common recognition sequence for a kinase that is responsible for mitotic inactivation.
Results and Discussion
Inhibition of DNA binding and pericentromeric targeting in G2/M-arrested cells
To explore the mechanisms underlying the dynamic changes in Ikaros localization, its release from pericentromeric foci in mitotic cells was monitored in the murine thymocyte line, VL3-3M2 (Groves et al. 1995). In asynchronous cells or in cells treated for 12 h with the DNA replication inhibitor, mimosine, the majority of cells were at the G1 or S stages of the cell cycle, as determined by flow cytometric analysis of DNA content (Fig. 1A, G1). Confocal immunofluorescence revealed that Ikaros was localized to distinct foci in these cells (Fig. 1B), consistent with the pericentromeric localization documented previously in both G1 and G2 (Brown et al. 1997). Following incubation with the drug vinblastine (Wendell et al. 1993), which blocks the G2/M transition by disrupting the mitotic spindle apparatus, the DNA content of the vast majority of cells was consistent with G2/M arrest (Fig. 1A, G2/M). In these cells, Ikaros localized diffusely and appeared to be excluded from the propidium iodide-stained DNA (Fig. 1B). These results are consistent with previous observations in mitotic cells isolated by elutriation (Brown et al. 1997).
Figure 1.
Mitotic inactivation of Ikaros. (A) DNA content of exponentially growing VL3-3M2 cells (left) and mimosine- and vinblastine-treated cells (middle and right) was determined by flow cytometry. (B) The subnuclear localization of Ikaros was analyzed in asynchronous (AS) and vinblastine-arrested (G2/M) VL3-3M2 cells by confocal microscopy. DNA was visualized using propidium iodide. (C) Ikaros concentrations in asynchronous and vinblastine-arrested samples were compared by Western blot (lanes 1,2). DNA-binding activities were compared by gel shift in the absence (lanes 3,5) and presence (lanes 4,6) of calf-intestine alkaline phosphatase (20 U). (D) VL3-3M2 cells were grown in the presence of 32P-labeled orthophosphate. Asynchronous and vinblastine-arrested samples were analyzed by immunoprecipitation using Ikaros antibodies. (E) Phosphopeptide maps were generated for Ikaros from asynchronous and vinblastine-arrested VL3-3M2 cells. The five phosphopeptides that were never detected in asynchronous cells are numbered on the G2/M map.
Western blot analysis revealed that similar concentrations of Ikaros isoforms V and VI (Hahm et al. 1994) were present in asynchronous and G2/M-arrested cells (Fig. 1C, lanes 1,2). In contrast, gel-shift analyses revealed that the DNA-binding activity of Ikaros was greatly reduced in the extracts from G2/M cells (Fig. 1C, lanes 3,5). Because the direct binding of Ikaros to satellite repeats is essential for targeting to pericentromeric foci (Cobb et al. 2000), the loss of DNA binding is probably responsible for altering subnuclear localization. Phosphatase treatment of nuclear extracts from G2/M-arrested cells resulted in a dramatic increase in DNA-binding activity (Fig. 1C, lanes 5,6), suggesting that mitotic inactivation of DNA binding may be due to direct phosphorylation.
G2/M-specific phosphorylation of Ikaros
To determine whether Ikaros is specifically phosphorylated in G2/M cells, asynchronous and G2/M-arrested VL3-3M2 cells were incubated with 32P-labeled orthophosphate to label endogenous, phosphorylated proteins. Immunoprecipitation of Ikaros from cell lysates, followed by SDS-PAGE and exposure to film, revealed that the Ikaros isoforms were phosphorylated in both samples (Fig. 1D). Two-dimensional phosphopeptide mapping of endogenous Ikaros isoform VI revealed several radiolabeled tryptic peptides (Fig. 1E). Some phosphopeptides were detectable in both the asynchronous and G2/M-arrested samples. Some of these were equally abundant in the two samples, whereas others were more abundant in one of the samples (Fig. 1E). In contrast, only five phosphopeptides detected in G2/M cells were never detected in asynchronous cells in six independent experiments. Three of these spots (1–3) were consistently intense, whereas the other two (4 and 5) were much weaker, suggesting less efficient phosphorylation.
The G2/M-specific phosphorylation sites correspond to the conserved linkers
To identify the residues that are specifically phosphorylated at G2/M, Ikaros isoform VI was expressed ectopically in HEK 293T cells. Phosphopeptide maps were generated following vinblastine treatment, revealing five phosphopeptides resembling those observed in VL3-3M2 cells (Fig. 2, WT). These phosphopeptides, which were not observed in asynchronous cells, comigrated with the VL3-3M2 peptides when the 293T and VL3-3M2 samples were loaded together (data not shown).
Figure 2.
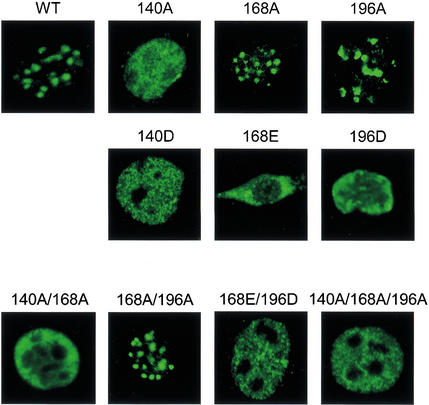
G2/M-specific phosphorylation of the three C2H2 linkers. (Top) Amino acid sequences of the N-terminal zinc fingers of murine Ikaros are shown, along with the linker consensus. (Bottom) Phosphopeptide maps generated with wild-type and mutant Ikaros proteins expressed in HEK 293T cells. Phosphopeptides that are absent with each mutant protein are indicated by a dashed circle. Simultaneous loading of 140A and 168A, or 168A and 196A, restores all phosphopeptides.
An analysis of deletion mutants spanning the entire Ikaros protein (Cobb et al. 2000) revealed that the five G2/M-specific phosphopeptides were in the vicinity of the N-terminal zinc finger DNA-binding domain (data not shown). An examination of potential phosphoacceptors within this region led to the hypothesis that the serines and threonines within the three linkers separating the four zinc finger motifs might be phosphorylated (Fig. 2, top). To test this hypothesis, the potential phosphoacceptor within each linker was changed to an alanine. Phosphopeptide mapping revealed that mutation of threonine 140 (linker 1) abolished phosphopeptide 2 (Fig. 2, 140A), whereas mutation of serine 168 (linker 2) abolished phosphopeptides 1 and 3 (Fig. 2, 168A). (The presence of two tryptic peptides containing serine 168 was presumably due to inefficient cleavage at Lys 171.) Simultaneous loading of the 140A and 168A samples restored all of the phosphopeptides observed with the wild-type protein (140A + 168A). Phosphopeptides 4 and 5 were lost when serine 196 (linker 3) was mutated (196A). When this sample and a 168A sample (which is different from the sample analyzed with 140A) were loaded simultaneously, the wild-type map was again restored (Fig. 2, 168A + 196A). These results demonstrate that the five G2/M-specific phosphopeptides result from phosphorylation of the three conserved linkers. It is interesting to note that the least abundant phosphopeptides (4 and 5) were derived from linker 3, which diverges from the canonical TGEKP linker sequence to the greatest extent (Fig. 2, top). Thus, the efficiency of phosphorylation appears to increase with increasing similarity to the canonical sequence.
Analysis of phosphomimetic mutations
To determine whether linker phosphorylation can account for the mitotic inactivation of Ikaros, gel-shift experiments were performed with nuclear extracts from 293T cells containing overexpressed wild-type or mutant Ikaros proteins. Two radiolabeled DNA probes were tested, one containing a consensus Ikaros-binding site (IK bs4; Molnár and Georgopoulos 1994) and the other containing a high-affinity binding site from the γ satellite repeat sequence, which appears to be responsible for Ikaros targeting to pericentromeric foci (Cobb et al. 2000). Alanine substitutions at positions 168 and 196 had no effect on protein–DNA complex abundance (relative to the wild-type protein) at either of two extract concentrations (Fig. 3, lanes 1,5,7,9,12,14). In contrast, the protein–DNA complex was abolished by an alanine substitution at position 140 (Fig. 3, lanes 3,10). Although binding affinities were not quantified, these results suggest that binding affinity is enhanced by threonine 140 of the wild-type protein, but not by serines 168 and 196. These results are consistent with expectations, as a threonine is required for the α-helix capping function that has been attributed to the conserved linker (Laity et al. 2000; Wolfe et al. 2000). Most importantly, phosphomimetic substitutions (to aspartate or glutamate) at either of the three positions significantly reduced binding affinity (Fig. 3, lanes 4,6,8,11,13,15). With the phosphomimetic substitutions at positions 168 and 196, the protein–DNA complex was undetectable in the presence of low extract concentrations (Fig. 3, lanes 13,15) and was reduced in the presence of higher concentrations (Fig. 3, lanes 6,8). When the phosphomimetic substitutions were introduced simultaneously at two or at all three positions, the protein–DNA complex was abolished in the presence of low or high-extract concentrations (Fig. 3, lanes 16–26).
Figure 3.
Phosphomimetic substitutions reduce DNA binding. Nuclear extracts were prepared from 293T cells expressing wild-type (WT) and mutant Ikaros proteins. Gel shifts were performed with two extract concentrations (3×, 6 μg per reaction, lanes 1–8,16–21; 1×, 2 μg per reaction, lanes 9–15,22–26). Two radiolabeled probes were examined, one containing a consensus recognition sequence (IK bs4, bottom), and the second containing a variant derived from the murine γ-satellite repeat (γ sat A, top).
To determine whether the in vitro effects on DNA binding are sufficient to alter subnuclear localization, the wild-type and mutant Ikaros proteins were expressed in NIH 3T3 cells by retroviral transduction. Confocal immunofluorescence was performed using Ikaros antibodies. Confocal images of individual, representative cells are shown in Figure 4. Wild-type Ikaros localized to foci that were shown previously to correspond to pericentromeric heterochromatin (Cobb et al. 2000). Consistent with the in vitro gel-shift results, pericentromeric targeting was retained with mutants 168A and 196A, but was lost with mutant 140A. Pericentromeric targeting was also lost with the three phosphomimetic mutants (140D, 168E, and 196D) and with all proteins containing two or three substitutions, with the exception of mutant 168A/196A. Taken together, these results strongly suggest that phosphorylation of the conserved linkers within the N-terminal zinc finger domain of Ikaros is responsible for the loss of pericentromeric localization during the G2/M transition.
Figure 4.
Phosphomimetic substitutions abolish pericentromeric targeting. Confocal immunofluorescence was used to examine the subnuclear localization of wild-type (WT) and mutant Ikaros, following retroviral transduction into NIH 3T3 cells. Single-cell images are shown.
Linker phosphorylation of Sp1
A central question raised by the above results is whether linker phosphorylation is restricted to Ikaros or to proteins positioned at pericentromeric heterochromatin. To determine whether pericentromeric localization is required for phosphorylation, an Ikaros mutant was examined that alters a zinc finger amino acid required for base recognition (Cobb et al. 2000). This mutation, 159A, disrupts DNA binding and pericentromeric targeting, but should have no significant effect on the structure of the zinc finger domain. Phosphopeptide mapping experiments with this mutant revealed that the three linkers were phosphorylated as efficiently as in the wild-type protein (data not shown), demonstrating that pericentromeric localization is not required.
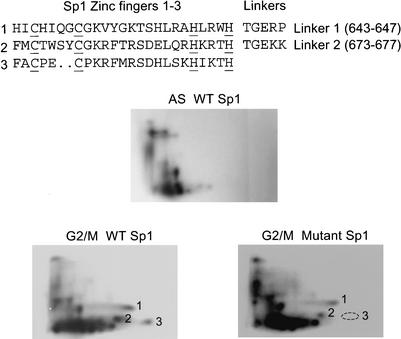
To determine whether linker phosphorylation can be observed with other C2H2 zinc finger proteins, the ubiquitous transcription factor Sp1 (Kadonaga et al. 1987) was examined. Previous studies showed that the DNA-binding activity of Sp1 is greatly reduced in extracts from mitotic cells (Martínez-Balbás et al. 1995). Phosphopeptide mapping of flag-tagged Sp1 expressed in 293T cells revealed multiple phosphopeptides that were restricted to G2/M-arrested cells (Fig. 5, G2/M WT). Importantly, one of three phosphopeptides in the lower right quadrant was absent in four independent experiments when the threonines within the two linkers were replaced by alanine (Fig. 5, G2/M Mutant, phosphopeptide 3). This result suggests that at least one of the Sp1 linkers can be phosphorylated.
Figure 5.
Linker phosphorylation of Sp1. (Top) Sequence of the DNA-binding domain of human Sp1 is shown. Wild-type and mutant Sp1 proteins were expressed in 293T cells. The mutant protein contained alanine substitutions at the phosphoacceptors in both linkers. Phosphopeptide maps were generated for the wild-type protein in asynchronous and vinblastine-arrested cells and, for the mutant protein, in vinblastine-arrested cells. Phosphopeptide 3 was absent in four independent experiments.
Conclusions
One reason for studying the dynamic changes in subnuclear localization of Ikaros was to further explore its potential contributions to the heritable silencing of developmentally regulated genes. The finding that DNA binding is disrupted during mitosis strongly suggests that Ikaros does not remain associated with silent genes through mitosis and, therefore, is unlikely to be an epigenetic mark that propagates the silent state. A hypothesis that is more consistent with these and previous results is that Ikaros initiates silencing and perhaps the pericentromeric recruitment of inactive genes (Smale and Fisher 2002). Ikaros may also help maintain the silent state during interphase (Brown et al. 1999; Smale and Fisher 2002). The mitotic inactivation of proteins implicated in heritable silencing is not without precedent, as polycomb group proteins are often displaced from chromatin during mitosis (Yamamoto et al. 1997; Buchenau et al. 1998; Dietzel et al. 1999; Voncken et al. 1999). In contrast, HP-1α and the histone methyltransferase SUV39H1 remain associated with centromeric foci through mitosis (Minc et al. 1999; Aagaard et al. 2000).
The precise function of the linker sequence that separates C2H2 zinc fingers has been of considerable interest because of its remarkable conservation. Although structural studies suggest that each residue within a canonical linker can contribute to the stability of the protein–DNA complex, mutagenesis studies revealed that some residues made little contribution to binding affinity, especially when alanine substitutions were tested rather than radical substitutions (Thurkal et al. 1991; Wilson et al. 1992; Choo and Klug 1993; Clemens et al. 1994). The minimal effects led to speculation that another selective pressure might contribute to the strong conservation of the linker sequence (Choo and Klug 1993). Several potential contributions were considered, all of which would serve to enhance the DNA-binding or transcriptional activation properties of zinc finger proteins (Choo and Klug 1993).
The results presented here strongly support the hypothesis that a second selective pressure is responsible for the conservation of the linker. However, the second function does not lead to enhanced binding or transactivation, but rather to the disruption of DNA binding during mitosis. It is interesting to note that the most significant contribution toward DNA-binding affinity is provided by a threonine in the first position of the linker (Wolfe et al. 2000). This critical structural role explains why a threonine phosphoacceptor is far more common than a serine. Other conserved linker residues are likely to be required for kinase recognition. The glycine, glutamate, and lysine are the most attractive candidates because they made the weakest contributions to binding affinity in an alanine-scanning analysis (Thurkal et al. 1991). The identity of the kinase responsible for linker phosphorylation remains unknown, as the conserved linker does not match the recognition sequence for cdc2 or any other well-characterized kinase. Furthermore, direct tests of several potential candidates failed to identify the relevant kinase (data not shown). Identification of the kinase may therefore require biochemical purification.
Although several sequence-specific DNA-binding proteins are known to be phosphorylated and/or inactivated during mitosis (Gottesfeld and Forbes 1997), the phosphorylated amino acids have been identified in only two other endogenous, sequence-specific DNA-binding proteins, Oct-1 and GHF-1 (Segil et al. 1991; Caelles et al. 1995). Interestingly, both of these proteins are members of the POU domain family and are phosphorylated on the same conserved amino acid within the POU motif. These results, along with the current findings, suggest that each family of DNA-binding proteins may have evolved a common mechanism for mitotic inactivation. The unusual size of the C2H2 zinc finger family and the prevalence of the canonical linker within the family suggests that mitotic inactivation may be extremely common among sequence-specific DNA-binding proteins.
Finally, it is worth noting a likely benefit during eukaryotic evolution of this mitotic inactivation strategy. A comparison of the genome sequences from several eukaryotic organisms revealed that the C2H2 zinc finger family has expanded at an unusually rapid rate, presumably through gene duplication (Lander et al. 2001). If newly evolved proteins lacking mitotic inactivation mechanisms are inherently detrimental to the cell, the benefit of an intrinsic inactivation target that coincides with an important structural determinant can easily be envisioned.
Materials and methods
Cell culture and flow cytometry
To arrest the murine VL3-3M2 thymocyte line (Groves et al. 1995) at G1 or G2/M, cells were incubated for 12 h at 37°C in growth medium supplemented with 0.2 mM mimosine or 0.2 μM vinblastine (Wendell et al. 1993), respectively. HEK 293T cells were incubated for 24 h in 0.3 mM mimosine (G1) or 0.2 μM vinblastine (G2/M). DNA content was determined using propidium iodide (PI). Stained cells were analyzed on a FACSCalibur flow cytometer (Becton Dickinson).
Transfection, retroviral transduction, and confocal microscopy
The pcDNA3-based expression plasmids (InVitrogen) and pMSCV pac-based retroviral expression vectors (Hawley et al. 1994) for HA epitope-tagged Ikaros isoform VI and the isoform VI deletion mutants were described previously (Cobb et al. 2000). Isoform VI substitution mutants were generated by a standard two-step PCR sewing protocol. A human Sp1 cDNA containing 696 C-terminal amino acids (Kadonaga et al. 1987) was cloned into the pcDNA3 vector along with sequences encoding an N-terminal FLAG epitope. HEK 293T cells were transfected with pcDNA3-based plasmids and NIH 3T3 cells were transduced with retroviruses as described (Cobb et al. 2000). Confocal microscopy was performed as described using Ikaros antibodies (Cobb et al. 2000; Trinh et al. 2001).
Biochemical procedures
Nuclear extracts were prepared as described (Cobb et al. 2000; Trinh et al. 2001). Western blots and gel-shift experiments were performed as described (Cobb et al. 2000). VL3-3M2 and 293T cells arrested with mimosine were washed twice with phosphate-free medium and then incubated for 3 (VL3-3M2) or 4 (293T) h with 0.8 mCi/mL 32P-labeled orthophosphate (NEN) in phosphate-free medium in the presence of mimosine. During vinblastine arrest, 1 mCi/mL 32P-labeled orthophosphate was added 4 h before the cells were harvested. Nuclear extracts were prepared and incubated with Ikaros CTS antibodies (Cobb et al. 2000) for 1 h at 4°C. The resulting complexes were bound to protein A-Sepharose (Pharmacia), extensively washed, and subjected to SDS-PAGE, followed by autoradiography. For immunoprecipitation of flag-tagged Sp1, nuclear extracts were incubated with anti-FLAG M2-agarose (Sigma). Bands corresponding to Ikaros or Sp1 were excised, digested with trypsin and chymotrypsin, and analyzed on two-dimensional thin layer cellulose plates.
Acknowledgments
We thank Joel Gottesfeld for helpful advice. This work was supported by NIH grants DK43726 (S.T.S.), HD07512 (S.D.), and CA82430 (S.D.), and by NIH Training Grant CA009120 (R.F.). S.D. was also supported by the Variety Club-D. Barry Reardon Endowment, a Laura and Greg Norman Research Fellowship, a Stop Cancer New Generation Seed Grant Award, a Pennington Scholar Award, and a Gwynne Hazen Cherry Memorial Foundation Award. S.T.S. is an Investigator of the Howard Hughes Medical Institute.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL steves@hhmi.ucla.edu; FAX (310) 206-8623.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1040502.
References
- Aagaard L, Schmid M, Warburton P, Jenuwein T. Mitotic phosphorylation of SUV39H1, a novel component of active centromeres, coincides with transient accumulation at mammalian centromeres. J Cell Sci. 2000;113:817–829. doi: 10.1242/jcs.113.5.817. [DOI] [PubMed] [Google Scholar]
- Akoulitchev S, Reinberg D. The molecular mechanism of mitotic inhibition of TFIIH is mediated by phosphorylation of CDK7. Genes & Dev. 1998;12:3541–3550. doi: 10.1101/gad.12.22.3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellier S, Dubois MF, Nishida E, Almouzni G, Bensaude O. Phosphorylation of the RNA polymerase II largest subunit during Xenopus laevis oocyte maturation. Mol Cell Biol. 1997;17:1434–1440. doi: 10.1128/mcb.17.3.1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers PM, Schaufler LE, Klevit RE. A folding transition and novel zinc finger accessory domain in the transcription factor ADR1. Nat Struct Biol. 1999;6:478–485. doi: 10.1038/8283. [DOI] [PubMed] [Google Scholar]
- Brown KE, Guest SS, Smale ST, Hahm K, Merkenschlager M, Fisher AG. Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell. 1997;91:845–854. doi: 10.1016/s0092-8674(00)80472-9. [DOI] [PubMed] [Google Scholar]
- Brown KE, Baxter J, Graf D, Merkenschlager M, Fisher AG. Dynamic repositioning of genes in the nucleus of lymphocytes preparing for cell division. Mol Cell. 1999;3:207–217. doi: 10.1016/s1097-2765(00)80311-1. [DOI] [PubMed] [Google Scholar]
- Buchenau P, Hodgson J, Strutt H, Arndt-Jovin DJ. The distribution of polycomb-group proteins during cell division and development in Drosophila embryos: Impact on models for silencing. J Cell Biol. 1998;141:469–481. doi: 10.1083/jcb.141.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caelles C, Hennemann H, Karin M. M-phase-specific phosphorylation of the POU transcription factor GHF-1 by a cell cycle-regulated protein kinase inhibits DNA binding. Mol Cell Biol. 1995;15:6694–6701. doi: 10.1128/mcb.15.12.6694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo Y, Klug A. A role in DNA binding for the linker sequences of the first three fingers of TFIIIA. Nucleic Acids Res. 1993;21:3341–3346. doi: 10.1093/nar/21.15.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens KR, Zhang P, Liao X, McBryant SJ, Wright PE, Gottesfeld JM. Relative contributions of the zinc fingers of transcription factor IIIA to the energetics of DNA binding. J Mol Biol. 1994;244:23–35. doi: 10.1006/jmbi.1994.1701. [DOI] [PubMed] [Google Scholar]
- Cobb BS, Morales-Alcelay S, Kleiger G, Brown KE, Fisher AG, Smale ST. Targeting of Ikaros to pericentromeric heterochromatin by direct DNA binding. Genes & Dev. 2000;14:2146–2160. doi: 10.1101/gad.816400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes M, Wong E, Koipally J, Georgopoulos K. Control of lymphocyte development by the Ikaros gene family. Curr Opin Immunol. 1999;11:167–171. doi: 10.1016/s0952-7915(99)80028-4. [DOI] [PubMed] [Google Scholar]
- Dietzel S, Niemann H, Bruckner B, Maurange C, Paro R. The nuclear distribution of Polycomb during Drosophila melanogaster development shown with a GFP fusion protein. Chromosoma. 1999;108:83–94. doi: 10.1007/s004120050355. [DOI] [PubMed] [Google Scholar]
- Georgopoulos K. Haematopoietic cell-fate decisions, chromatin regulation and ikaros. Nat Rev Immunol. 2002;2:162–174. doi: 10.1038/nri747. [DOI] [PubMed] [Google Scholar]
- Gottesfeld JM, Forbes DJ. Mitotic repression of the transcriptional machinery. Trends Biochem Sci. 1997;22:197–202. doi: 10.1016/s0968-0004(97)01045-1. [DOI] [PubMed] [Google Scholar]
- Groves T, Katis P, Madden Z, Manickam K, Ramsden D, Wu G, Guidos CJ. In vitro maturation of clonal CD4+CD8+ cell lines in response to TCR engagement. J Immunol. 1995;154:5011–5022. [PubMed] [Google Scholar]
- Hahm K, Ernst P, Lo K, Kim GS, Turck C, Smale ST. The lymphoid transcription factor LyF-1 is encoded by specific, alternatively spliced mRNAs derived from the Ikaros gene. Mol Cell Biol. 1994;14:7111–7123. doi: 10.1128/mcb.14.11.7111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley RG, Lieu FH, Fong AZ, Hawley TS. Versatile retroviral vectors for potential use in gene therapy. Gene Therapy. 1994;1:136–138. [PubMed] [Google Scholar]
- Kadonaga JT, Carner KR, Masiarz FR, Tjian R. Isolation of cDNA encoding transcription factor Sp1 and functional analysis of the DNA binding domain. Cell. 1987;51:1079–1090. doi: 10.1016/0092-8674(87)90594-0. [DOI] [PubMed] [Google Scholar]
- Kim J, Sif S, Jones B, Jackson A, Koipally J, Heller E, Winandy S, Viel A, Sawyer A, Ikeda T, et al. Ikaros DNA-binding proteins direct formation of chromatin remodeling complexes in lymphocytes. Immunity. 1999;10:345–355. doi: 10.1016/s1074-7613(00)80034-5. [DOI] [PubMed] [Google Scholar]
- Laity JH, Dyson HJ, Wright PE. DNA-induced α-helix capping in conserved linker sequences is a determinant of binding affinity in Cys2-His2 zinc fingers. J Mol Biol. 2000;295:719–727. doi: 10.1006/jmbi.1999.3406. [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton M, Birren B, Nusbaum C, Zody MC, Baldwin J, Devan K, Dewar K, Doyle M, Pitzhugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Long JJ, Leresche RW, Kriwaski RW, Gottesfeld JM. Repression of TFIIH transcriptional activity and TFIIH-associated cdk7 kinase activity at mitosis. Mol Cell Biol. 1998;18:1467–1476. doi: 10.1128/mcb.18.3.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Balbás MA, Dey A, Rabindran SK, Ozato K, Wu C. Displacement of sequence-specific transcription factors from mitotic chromatin. Cell. 1995;83:29–38. doi: 10.1016/0092-8674(95)90231-7. [DOI] [PubMed] [Google Scholar]
- Minc E, Allory Y, Worman HJ, Courvalin JC, Buendia B. Localization and phosphorylation of HP1 proteins during the cell cycle in mammalian cells. Chromosoma. 1999;108:220–234. doi: 10.1007/s004120050372. [DOI] [PubMed] [Google Scholar]
- Molnár A, Georgopoulos K. The Ikaros gene encodes a family of functionally diverse zinc finger DNA-binding proteins. Mol Cell Biol. 1994;14:8292–8303. doi: 10.1128/mcb.14.12.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott DM, Bender MA. Synthesis of RNA and protein during mitosis in mammalian tissue culture cells. Exp Cell Res. 1962;26:260–268. doi: 10.1016/0014-4827(62)90176-3. [DOI] [PubMed] [Google Scholar]
- Segil N, Roberts SB, Heintz N. Mitotic phosphorylation of the Oct-1 homeodomain and regulation of Oct-1 DNA binding activity. Science. 1991;254:1814–1816. doi: 10.1126/science.1684878. [DOI] [PubMed] [Google Scholar]
- Segil N, Guermah M, Hoffmann A, Roeder RG, Heintz N. Mitotic regulation of TFIID inhibition of activator-dependent transcription and changes in subcellular localization. Genes & Dev. 1996;10:2389–2400. doi: 10.1101/gad.10.19.2389. [DOI] [PubMed] [Google Scholar]
- Sif S, Stukenberg PT, Kirschner MW, Kingston RE. Mitotic inactivation of a human SWI/SNF chromatin remodeling complex. Genes & Dev. 1998;12:2842–2851. doi: 10.1101/gad.12.18.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smale ST, Fisher AG. Chromatin structure and gene activation in the immune system. Annu Rev Immunol. 2002;20:427–462. doi: 10.1146/annurev.immunol.20.100301.064739. [DOI] [PubMed] [Google Scholar]
- Thukral SK, Morrison ML, Young ET. Alanine scanning site-directed mutagenesis of the zinc fingers of transcription factor ADR1: Residues that contact DNA and that transactivate. Proc Natl Acad Sci. 1991;88:9188–9192. doi: 10.1073/pnas.88.20.9188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinh LA, Ferrini R, Cobb BS, Weinmann AS, Hahm K, Ernst P, Garraway IP, Merkenschlager M, Smale ST. Down-regulation of TDT transcription in CD4+CD8+ thymocytes by Ikaros proteins in direct competition with an Ets activator. Genes & Dev. 2001;15:1817–1832. doi: 10.1101/gad.905601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tupler R, Perini G, Green MR. Expressing the human genome. Nature. 2001;409:832–833. doi: 10.1038/35057011. [DOI] [PubMed] [Google Scholar]
- Voncken JW, Schweizer D, Aagaard L, Sattler L, Jantsch MF, van Lohuizen M. Chromatin-association of the Polycomb group protein BMI1 is cell cycle-regulated and correlates with its phosphorylation status. J Cell Sci. 1999;112:4627–4639. doi: 10.1242/jcs.112.24.4627. [DOI] [PubMed] [Google Scholar]
- Wendell KL, Wilson L, Jordan MA. Mitotic block in HeLa cells by vinblastine: Ultrastructural changes in kinetochore-microtubule attachment and in centrosomes. J Cell Sci. 1993;104:261–274. doi: 10.1242/jcs.104.2.261. [DOI] [PubMed] [Google Scholar]
- Wilson TE, Day ML, Pexton T, Padgett KA, Johnston M, Milbrandt J. In vivo mutational analysis of the NGFI-A zinc fingers. J Biol Chem. 1992;267:3718–3724. [PubMed] [Google Scholar]
- Wolfe SA, Nekludova L, Pabo CO. DNA recognition by Cys2His2 zinc finger proteins. Annu Rev Biophys Biomol Struct. 2000;29:183–212. doi: 10.1146/annurev.biophys.29.1.183. [DOI] [PubMed] [Google Scholar]
- Wuttke DS, Foster MP, Case DA, Gottesfeld JM, Wright PE. Solution structure of the first three zinc fingers of TFIIIA bound to the cognate DNA sequence: Determinants of affinity and sequence specificity. J Mol Biol. 1997;273:183–206. doi: 10.1006/jmbi.1997.1291. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Girard F, Bello B, Affolter M, Gehring WJ. The cramped gene of Drosophila is a member of the Polycomb-group, and interacts with mus209, the gene encoding Proliferating Cell Nuclear Antigen. Development. 1997;124:3385–3394. doi: 10.1242/dev.124.17.3385. [DOI] [PubMed] [Google Scholar]