Abstract
The intra-S-phase checkpoint in yeast responds to stalled replication forks by activating the ATM-like kinase Mec1 and the CHK2-related kinase Rad53, which in turn inhibit spindle elongation and late origin firing and lead to a stabilization of DNA polymerases at arrested forks. A mutation that destabilizes the second subunit of the Origin Recognition Complex, orc2-1, reduces the number of functional replication forks by 30% and severely compromises the activation of Rad53 by replication stress or DNA damage in S phase. We show that the restoration of the checkpoint response correlates in a dose-dependent manner with the restoration of pre-replication complex formation in G1. Other forms of DNA damage can compensate for the reduced level of fork-dependent signal in the orc2-1 mutant, yet even in wild-type cells, the amount of damage required for Rad53 activation is higher in S phase than in G2. Our data suggest the existence of an S-phase-specific threshold that may be necessary to allow cells to tolerate damage-like DNA structures present at normal replication forks.
Keywords: Replication checkpoint; Rad53; ORC; yeast, DNA damage; S phase
As cells proliferate, the genome must be replicated and segregated with high fidelity. Surveillance mechanisms, or checkpoints, detect damaged DNA or on-going replication and block cell cycle progression in order to allow adequate time to repair the damaged DNA or to complete replication (Weinert 1998; Zhou and Elledge 2000). Failure to delay the cell cycle can convert an easily reparable DNA lesion into one more deleterious, resulting in chromosomal rearrangements or loss. The importance of this mechanism is underscored by a large number of heritable human diseases that arise from defects in checkpoint or DNA-damage repair functions (for review, see Kastan 1997).
Genetic analysis of the DNA damage checkpoint pathway in yeast has allowed classification of its components into sensors, which detect DNA damage, adaptors, which integrate and transmit the signal, and effector kinases, which promote downstream functions including the induction of repair genes, suppression of cell cycle progression, the arrest of replication polymerases, down-regulation of late origins and of sister chromatid segregation (for review, see Zhou and Elledge 2000; Melo and Toczyski 2002). In G1 and G2 phases of the budding yeast cell cycle, a DNA-bound complex of Rad17/Mec3/Ddc1 and Rad24 proteins, as well as a complex containing the ATM homolog Mec1p and its cofactor Ddc2p, play crucial roles as sensors. These communicate through Rad9p to activate the major effector kinases, Rad53p (CHK2 in other species) and Chk1p. These components and their roles are conserved from yeast to man (Melo and Toczyski 2002).
The response of the cell to DNA damage or fork arrest during S phase is distinct from the G1/S or G2/M checkpoints, both at the level of sensors and with respect to the cellular response to Rad53p activation. It is useful to consider this intra-S-phase response with respect to two operationally distinct lesions. The first arises from replication forks stalled by high concentrations of hydroxyurea (HU), a drug that reduces dNTP pools through inhibition of ribonucleotide reductase. In this case, Rad53p activation is at least partially dependent on replication enzymes at the fork, such as DNA polymerase ɛ, its cofactor Dpb11p, Replication factor C, and the RecQ-family helicase, Sgs1p, which appear to signal that forks are stalled (for review, see Foiani et al. 2000). In the S-phase response to DNA alkylation by methylmethane sulfonate (MMS), and the resulting single or double-strand breaks, Rad53p activation is achieved primarily through Rad24p and the Rad17/Mec3/Ddc1 complex (Lydall and Weinert 1997; Paulovich et al. 1997; Frei and Gasser 2000; Myung and Kolodner 2002). The two pathways not only act in parallel, but cross-talk; DNA alkylation can stall replication forks (mimicking the effects of dNTP depletion), and HU-arrested forks are prone to strand breakage. Mrc1p is a highly charged, cell cycle-regulated protein that may serve as the primary adaptor for checkpoint kinase activation in response to HU (Alcasabas et al. 2001; Tanaka and Russell 2001), whereas Rad9p has a secondary role, helping modulate Rad53p activation in S phase in response to MMS.
When DNA replication is blocked by HU or by MMS-induced damage, Rad53p activation blocks initiation at unfired origins, stabilizes stalled forks, and prevents progression through S phase, as monitored by spindle elongation and DNA content (Paulovich and Hartwell 1995; Marini et al. 1997; Santocanale and Diffley 1998; Shirahige et al. 1998; Lopes et al. 2001; Tercero and Diffley 2001). The conserved effector kinase Chk1p is required to prevent entry into mitosis in response to DNA damage or incomplete DNA replication (the G2/M or S/M checkpoint), yet it is not necessary for the intra-S-phase response described above (Sanchez et al. 1999). Similar pathways have been shown to lead to intra-S-phase checkpoint activation in mammalian cells (Falck et al. 2002), although the pathways may differ in early embryonic systems that do not impose temporal control on origin firing. Xenopus embryonic extracts, for example, respond to replicative stress by activating an S/M checkpoint through Claspin (an Mrc1p homolog) and Chk1 kinase (Kumagai and Dunphy 2000).
Surprisingly, in yeast, the induction of a double-strand break (DSB), or the DNA damage induced by low levels of bleomycin is not sufficient to provoke checkpoint activation in S phase, whereas in G2-phase cells, similar lesions are sufficient to activate Rad53p and retard mitotic progression (Sandell and Zakian 1993; Pellicioli et al. 2001; see below). This suggests that the processing, detection, or response to DNA-strand breaks differ between S and G2 phases. Similarly, in mammalian cells, it appears that the p53 response to DNA damage is functionally impaired in S phase (Gottifredi et al. 2001).
Among the yeast mutants that alter the DNA-damage response in S phase are mutations in the Origin Recognition Complex (ORC, Shirahige et al. 1998; Weinberger et al. 1999). Although no mechanism was proposed, it was noted recently that the orc2-1 mutation increases cell death in the presence of the DNA-damaging drug adozelesin (Weinberger et al. 1999), and impairs full suppression of late-firing origins on MMS (Shirahige et al. 1998). These studies failed to clarify whether orc2-1 acted upstream or downstream of checkpoint kinase activation. In the absence of damage, the primary function of the six-subunit ORC is to nucleate an initiation competent pre-Replication Complex at origins in early G1 phase (preRC; for review, see Bell 2002), by recruiting Cdc6p and Cdt1p, which in turn load the hexameric Mini Chromosome Maintenance (MCM) complex. The weak helicase activity associated with this latter complex is essential for both the initiation of DNA replication and fork elongation (Labib et al. 2000).
We have explored the mechanism that renders orc2-1 cells hypersensitive to DNA-damaging agents in S phase. We find that orc2-1 cells have reduced levels of Orc2p and are impaired for Rad53p activation in response to HU, MMS, or bleomycin treatment at permissive temperature. We show that ORC acts upstream of Rad53p, in a dose-dependent manner, and that its function in intra-S-phase checkpoint activation reflects its inefficiency at establishing replication competent origins in G1 phase. However, unlike the loss of MCM function (Labib et al. 2000, 2001), depleting Orc2p after preRC formation does not impair the initiation nor the elongation steps of DNA replication. Similarly, once sufficient replication origins have fired, Orc2p appears to have no further role in either the induction or the maintenance of Rad53p activation. Finally, we find that DNA damage and stalled fork signals can be integrated to reach a threshold sufficient for checkpoint activation in both wild-type and orc2-1 cells. Our data suggest the existence of an S-phase-specific threshold for Rad53p kinase activation, which allows normal fork progression.
Results
Mutation of ORC2 compromises the S-phase checkpoint response
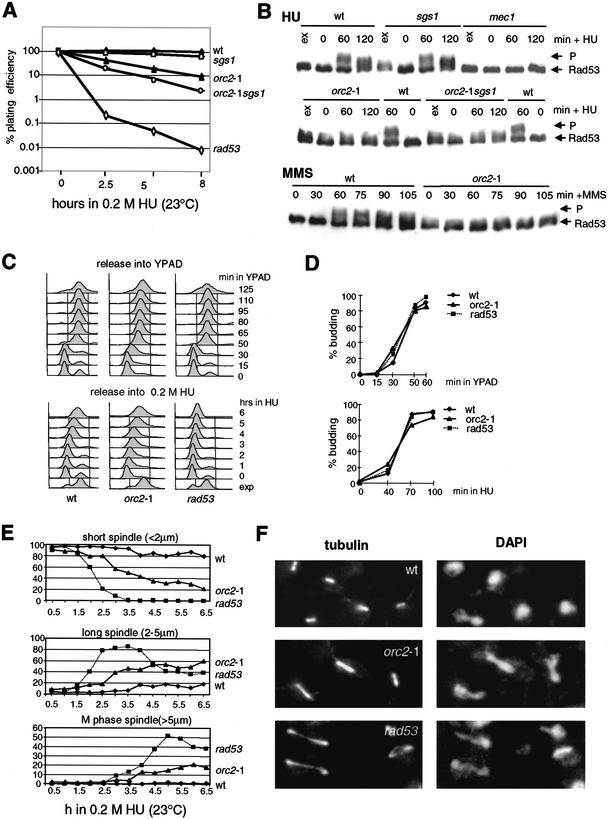
It was reported recently that yeast strains bearing a temperature-sensitive mutation in ORC2, which encodes the second largest subunit of the ORC complex, fail to slow the progression of DNA synthesis when exposed to MMS at semipermissive temperature (Shirahige et al. 1998). The defect was less pronounced than in rad53-deficient cells and it was not clear whether Orc2p was required for Rad53p activation or whether origins were simply de-regulated such that they no longer respond to Rad53p activation (Shirahige et al. 1998). To examine this further, we tested whether the orc2-1 mutation compromises the cellular response to HU. We made use of the published orc2-1 allele (Foss et al. 1993), after repeated backcrossing of the published strain to its wild-type parental background to remove potential secondary mutations (see Materials and Methods). The viability of the orc2-1 mutant (GA-1254) and a strain lacking Sgs1p, a DNA helicase also implicated in the S-phase checkpoint response (hereafter sgs1Δ, Frei and Gasser 2000) was compared with an isogenic wild-type strain after exposure to HU (Fig. 1A,D).Whereas sgs1Δ strains are slightly more sensitive to HU-induced fork arrest, the viability of the orc2-1 mutant is significantly compromised, even at permissive temperature (2.5 h, 23°C). The effect of combining the orc2-1 and sgs1Δ mutations is additive, although the double mutant is less sensitive to HU than the rad53 strain, which fails to elicit the checkpoint response altogether (rad53 = mec2-1; Fig. 1A).
Figure 1.
orc2-1 cells are defective in S-phase checkpoint. (A) orc2-1 cells are hypersensitive to HU. Isogenic strains carrying the orc2-1 mutation, a deletion of sgs1, both mutations, or rad53-11 (mec2-1) were synchronized by α-factor, then released into 0.2 M HU for the indicated times at 23°C. Washed cells were plated in triplicate on YPAD, and plating efficiency was determined after 3 d. Values for each strain are normalized to survival at time 0. ♦, wild-type (GA-180); ▴, orc2-1 (GA-1254); □, sgs1∷TRP1 (GA-734); ○, orc2-1 sgs1∷TRP1 (GA-1387); ⋄, rad53-11 (mec2-1; GA-1230). (B) Exponentially growing cultures (ex) were blocked in G1 with α-factor and released in 0.2 M HU (+ HU) or 0.015% MMS (+ MMS) for the indicated times at 23°C. Western blots of whole-cell extracts monitor phosphorylation of Rad53-Myc with the 9E10 Mab. P indicates the fully phosphorylated form of Rad53p. Isogenic strains were as follows: wild-type (wt, GA-1829, A364a background); sgs1∷LEU2 (sgs1, GA-1830); mec1-1 sml1 (mec1, GA-1048); orc2-1 (GA-1831); orc2-1 sgs1∷LEU2 (orc2-1 sgs1, GA-1832). (C) Wild-type, orc2-1 and rad53-11 strains as in A were cultured and blocked with α-factor at 23°C. Cells were released in YPAD with or without 0.2 M HU at 23°C, and samples analyzed by FACS at the indicated times. (D) Bud emergence was scored microscopically on 100 cells of cultures examined in C. (E) Isogenic strains carrying an integrated GFP–TUB1 fusion (tubulin) were blocked with α-factor and released into 0.2 M HU. At the indicated times, spindle length was monitored by live fluorescence microscopy. For each genotype and each time point, 100 cells were measured. Strains used are as follows: wild-type (♦, GA-1535), orc2-1 (▴, GA-1533), and rad53 (mec2-1; ■, GA-1499). (F) The strains used in A were synchronized by α-factor, then released into 0.2 M HU for 5 h at 23°C. Tubulin is visualized with anti-TAT1 antibody and DNA with DAPI.
To monitor the intra-S checkpoint response of the orc2-1 cells, we scored the length of intranuclear microtubules and the phosphorylation status of Rad53p on HU (Sanchez et al. 1996; Sun et al. 1996). In wild-type cells synchronized in G1 and then released into S phase in the presence of HU, Rad53p becomes maximally phosphorylated by 60 min after release, in a Mec1p-dependent manner (Fig. 1B). This level of modification is never reached in orc2-1-deficient cells, even after 120 min at 23°C (Fig. 1B). The defect in Rad53p activation is dominant over the sgs1Δ mutation, yet in the absence of HU, we detect a low level of Rad53p phosphorylation in the orc2-1 strain at permissive temperature. This is Rad24p dependent (data not shown), and confirms that orc2-1 cells sustain a low-level DNA damage response. However, they are unable to hyperphosphorylate Rad53p, which is necessary for activation of the intra-S-phase checkpoint (Fig. 1B).
Sgs1p acts on the same pathway as DNA pol ɛ, but in parallel to Rad24p, to activate Rad53p in response to HU or MMS (Frei and Gasser 2000). In contrast to sgs1Δ cells, in which a drop in Rad53p activation can only be detected in the absence of Rad24p, orc2-1 cells fail to activate Rad53p even with Rad24p intact (Fig. 1B). This is also true for damage induced by MMS (Fig. 1B), showing that the orc2-1 deficiency is upstream of both the stalled fork and the DNA-damage activation pathways. The defect is not due to a delayed entry into S phase or of bud emergence (Fig. 1C,D). Intriguingly, orc2-1 cells do have a delayed transit through mitosis even in the absence of MMS or HU, which correlates with a partial activation of Rad53p very late in G2 (data not shown).
Spindle-length analysis shows that instead of the short intranuclear spindles that characterize an S-phase arrest in wild-type cells, ∼80% of the orc2-1 cells have either moderately or fully elongated spindles on HU (Fig. 1E,F). Consistent with a slight delay in the metaphase-to-anaphase transition, we see that most orc2-1 cells that have microtubules of ∼3 μm instead of >5 μm, as monitored in the rad53 strain (Fig. 1E,F). Nonetheless, the orc2-1 cells have clearly lost the intra-S-phase response that arrests spindle elongation.
A mutation in the E1 ubiquitin activating enzyme suppresses orc2-1 lethality at 30°C
In current models for the intra-S-phase checkpoint, proteins that are positioned at the replication fork such as DNA pol ɛ, Dpb11p, and RFC, are thought to detect aberrant DNA structures that form when forks stall, and to signal Rad53p activation (for review, see Foiani et al. 2000). However, it is more difficult to imagine such a role for ORC, which being origin bound, is ill placed to contact stalled forks in mid-S phase. To understand better the orc2-1 deficiency, we sequenced the mutant allele. Two point mutations were detected as follows: a silent A-to-G mutation in amino acids 1047, and a C-to-T transition that substitutes leucine for a conserved proline at amino acids 1808, 18 residues from the termination codon.
The explanation of how the orc2-1 allele compromises the DNA-replication checkpoint arose from an analysis of a spontaneous mutation that suppresses the orc2-1-associated cell death at 30°C. In contrast to published data, we found that the orc2-1 strain YJL861 (also called JRY4125, gift of J. Li, University of California, San Francisco) was viable at 30°C, although the characteristic orc− lethality was manifest at 34°C and 37°C. By backcrossing the strain to the parental W303 background, we isolated an independently segregating mutation that was responsible for the suppression of lethality at 30°C. The orc2-1 segregates that lack the second mutation grew at 23°C, but died at ≥30°C, whereas spores carrying either the suppressor mutation alone or both mutations, grew readily at both temperatures (Supplementary Fig. 1). Although this second mutation suppresses the orc2-1 defect at 30°C, it is lethal on its own at 37°C, a phenotype used to clone the wild-type allele of the gene in question. Genetic and sequence analyses localized the suppressor to Chr XI and identified the complementing ORF as UBA1. UBA1 is an essential gene encoding the unique E1 ubiquitin-activating enzyme in yeast (McGrath et al. 1991).
A single copy vector expressing UBA1 restores lethality at 30°C in the orc2-1 uba1 double mutant, and the double mutant survives at all temperatures in the presence of both UBA1- and ORC2-bearing plasmids (see Supplementary Fig. 1B). Because UBA1 is essential for ubiquitin-mediated protein degradation, the suppressor mutation suggested that a labile factor was responsible for the orc2-1 phenotype. The combined mutations pre1-1 and pre2-2, which are each partially deficient in proteasome function (Hilt et al. 1993), also suppress orc2-1 lethality at 30°C (data not shown).
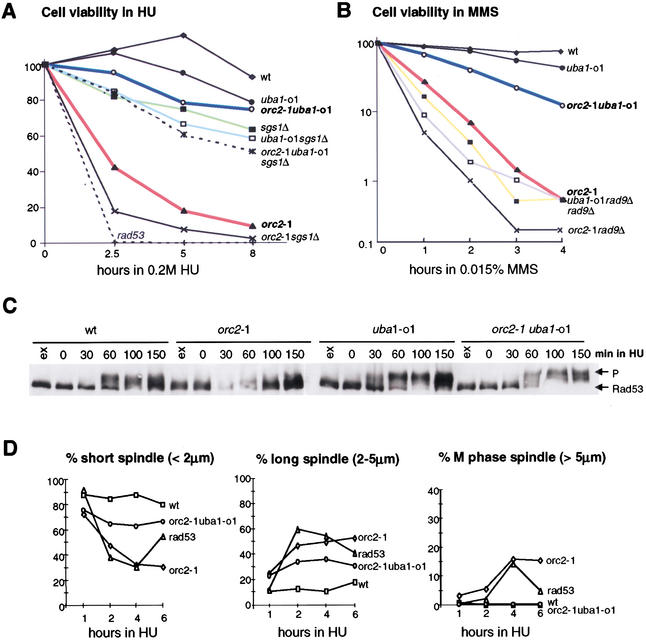
To see whether the uba1 allele (henceforth uba1-o1) also restores the S-phase-checkpoint response in the orc2-1 background, we examined the response to HU and MMS in the orc2-1 uba1-o1 double mutant. Strains carrying single and double mutations were synchronized in G1, released at 23°C for indicated times in medium containing either HU or MMS, and then plated in the absence of drug. Whereas the orc2-1 strain rapidly loses viability in the presence of HU, the orc2-1 uba1-o1 double mutant resists as well as uba1-o1 cells (>80% wild-type values; Fig. 2A). The suppression of the HU sensitivity by uba1-o1 is specific to orc2-1, having no effect when combined with sgs1Δ (Fig. 2A). Similarly, on MMS, the uba1-o1 mutation suppresses the sensitivity of an orc2-1, but not that of a rad9Δ strain (Fig. 2B).
Figure 2.
The S-phase checkpoint response is restored in an orc2-1 uba1-o1 double mutant. (A,B) The uba1-o1 mutation suppresses the hypersensitivity of orc2-1 cells to HU (A) and MMS (B). Viability assays were performed as in Figure 1A after indicated times in 0.2 M HU or 0.015% MMS. Isogenic strains used in A and B were as follows: wild-type (GA-180); sgs1∷TRP1 (GA-734); orc2-1 (GA-1254); uba1-o1 (GA-1256); orc2-1 uba1-o1 (GA-463); orc2-1 sgs1∷TRP1 (GA-1387); uba1-o1 sgs1∷TRP1 (GA-1388); orc2-1 uba1-o1 sgs1∷TRP1 (GA-1389); rad53 (mec2-1; GA-1230); rad97∷LEU2 (GA-1148), orc2-1 rad9∷LEU2 (GA-1860); uba1-o1 rad9∷LEU2 (GA-1861). (C) Rad53p phosphorylation in response to HU is restored in an orc2-1 uba1-o1 double mutant (see Fig. 1B). Strains were as follows: wild-type (GA-1835); orc2-1 (GA-1836); uba1-o1 (GA-1837); orc2-1 uba1-o1 (GA-1838). (D) The uba1-o1 mutation restores the cell cycle arrest response to HU in orc2-1 cells. Cells synchronized in G1 as in A were released into YPAD + 0.2 M HU. Spindle elongation was scored as in Figure 1F. The drop in long spindles in the rad53 strain at 6 h reflects entry into the next cell cycle.
The single and double orc2-1 uba1-o1 mutant strains were analyzed for Rad53p activation and spindle elongation in the presence of HU. The double mutant fully restores the Rad53p response to 0.2 M HU and arrests spindle elongation, whereas entry into S phase was unaltered (Fig. 2C,D; data not shown). We conclude that a loss or alteration in ubiquitin-dependent protein degradation specifically suppresses the orc2-1 checkpoint defect at 23°C as well as suppressing orc2-1 lethality at 30°C.
orc2-1p is highly labile and is stabilized by the uba1-o1 mutation
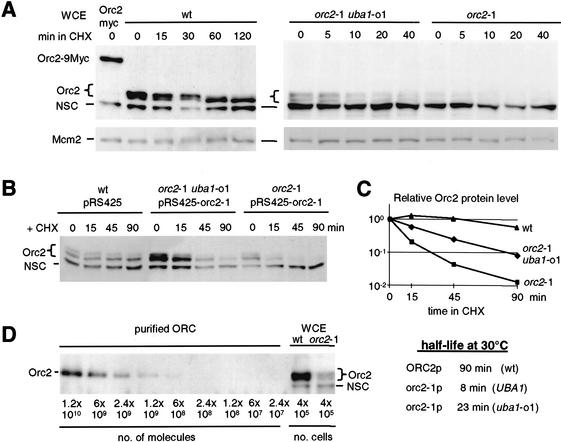
We entertained the thought that Orc2p is itself a target of degradation. Using an affinity-purified antibody that recognizes the N-terminal domain of Orc2p, we detect a doublet of 70 and 72 kD in whole-cell extracts of unsynchronized cultures, as well as a cross-reacting cytoplasmic protein of 60 kD (labeled NSC for nonspecific cross-reaction; see Fig. 3A). The identity of the upper doublet as Orc2p was confirmed by fusing nine c-Myc epitopes to its genomic coding sequence, which retard its electrophoretic mobility (Fig. 3A). The stabilities of wild-type Orc2p and mutant orc2-1p were determined by adding cycloheximide (CHX) to cultures and quantifying residual protein levels at subsequent time points on Western blots.
Figure 3.
The highly labile protein orc2-1p is partially stabilized by the uba1-o1 mutation. (A) Cycloheximide was added to exponentially growing cells and total protein extracts were prepared at indicated times. Western blots were sequentially probed with goat anti-MCM2 (Santa Cruz Antibodies) or affinity-purified anti-Orc2p antibody. The brace indicates to a doublet of Orc2p-specific bands (the upper band being phosphorylated); NSC indicates a nonspecific cross-reaction with a cytoplasmic protein that serves as an internal control for loading. Strains used were orc27∷ORC2-9Myc (GA-893); wild-type (GA-180); orc2-1 uba1-o1 (GA-463); orc2-1 (GA-1254). (B) Isogenic orc2-1 and orc2-1 uba1-o1 cells transformed with a multicopy plasmid expressing mutant orc2-1p (pRS425-orc2-1) and wild-type cells with the empty vector, were treated with cycloheximide (CHX) at 30°C, and whole-cell extracts were prepared at the indicated times. Orc2p protein was revealed as in A. (C) Orc2p protein signals in B were quantified by the AIDA program (Fuji PhosPhorimager) and normalized to the lower nonspecific band (NSC). Relative intensity is plotted as a function of time after CHX addition and the half-life of Orc2p (t1/2) was calculated. (D) Decreasing amounts of purified recombinant Baculovirus-expressed yeast ORC complex (gift of Dr. S. Bell), and total protein extracts equivalent to 4 × 105 cells from isogenic wild-type and orc2-1 cells grown at 23°C, were analyzed by Western blot using affinity-purified anti-Orc2 antibodies.
Wild-type Orc2p protein is extremely stable (t1/2 = 90 min at 30°C). Its level does not drop significantly during 2 h in CHX (Fig. 3A), yet as cycloheximide arrests cells in G1 phase, we do see the doublet collapse into the lower 70-kD band as it loses its S-phase specific phosphorylation (Vas et al. 2001). In the mutant, orc2-1p is nearly undetectable, despite an increased exposure time for the Western (compare signals from the cross-reacting bands versus Mcm2p, both used as controls for loading and transfer efficiency). Importantly, the uba1-o1 suppressor mutation increases the steady-state orc2-1p level, such that Orc2-specific bands are detected both before and after CHX addition (Fig. 3A).
To facilitate half-life calculations, we cloned the mutant orc2-1 allele and reintroduced it on a multicopy vector into both orc2-1 and orc2-1 uba1-o1 double mutant strains (pRS425-orc2-1; Fig. 3B). The presence of the uba1-o1 suppressor increases the half-life of orc2-1p from 8 to 23 min, or nearly threefold at 30°C (Fig. 3B,C). The rates of orc2-1p degradation in G1 and S phases were identical to those shown for random cultures (data not shown), and uba1-o1 slows degradation at all cell cycle stages (t1/2 = 23 min in orc2-1 uba1-o1 versus 90 min in wild type).
To estimate the copy number of wild-type and mutant Orc2 proteins, we have used a purified preparation of recombinant ORC (gift of S. Bell, Massachusetts Institute of Technology) to calibrate our Western blots. We calculate ∼3000 copies of orc2-1p per mutant cell, whereas copy number is >2 × 104 for the wild-type protein (Fig. 3D). Controls for purified protein concentration, transfer efficiency, and antibody specificity (to be published elsewhere) confirm that mutant orc2-1p levels are about 10-fold lower than those of wild-type Orc2p. Although these numbers are significantly higher than previous estimations (Rowley et al. 1995), the orc2-1p level is clearly insufficient to ensure normal initiation from all origins (see below).
Restoration of the S-phase checkpoint in an orc2-1 dose-dependent fashion
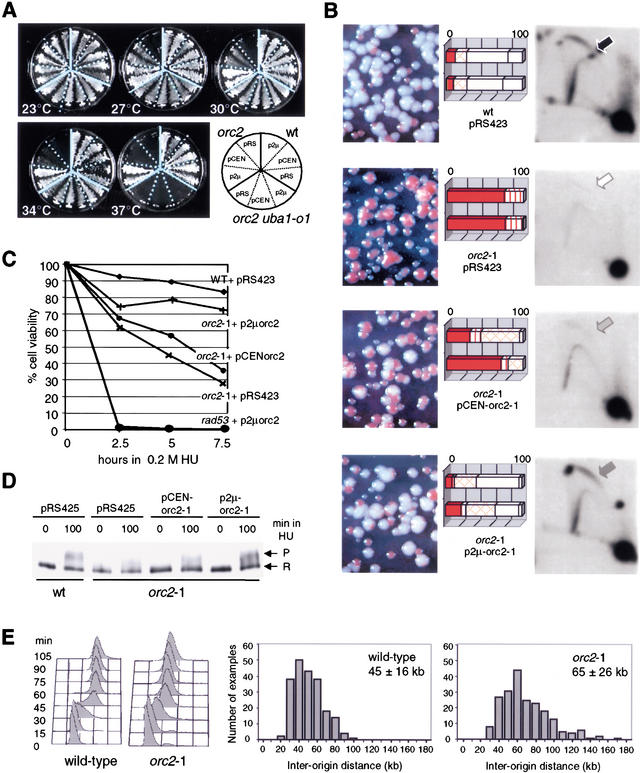
We note that even the partial restoration of orc2-1p levels by the uba1-o1 mutation fully restores the S-phase checkpoint function. To confirm that this exclusively reflects orc2-1p stabilization, we checked whether overexpression of the mutant protein from either low- (pCEN-orc2-1) or multi-copy (p2μ-orc2-1) vectors would have the same effects. Increased expression of orc2-1 restores viability to the mutant strain in a dose-dependent manner; the pCEN-orc2-1 plasmid works partially at intermediate temperatures, whereas the p2μ high-copy vector restores viability efficiently even at 37°C (Fig. 4A).
Figure 4.
Ectopic expression of the orc2-1 mutant protein suppresses orc2 defects in a dosage-dependent manner. (A) Two independent transformants of orc2-1, orc2-1 uba1-o1, and wild-type cells carrying with p2μ (pRS425-orc2-1), pCEN (pRS415-orc2-1) or pRS425 (labeled pRS) were streaked on SD-leu and cultured at the indicated temperature (see diagram). (B) The orc2-1 strain was cotransformed with pRS415-ADE2 (carrying ADE2 and CEN6/H4ARS) and pRS423-orc2-1 (p2μ-orc2-1), or pRS413-orc2-1 (pCEN-orc2-1), and both wild-type and orc2-1 cells were cotransformed with pRS415-ADE2 and pRS423. Transformants were first selected on SD-leu-his, then plated on SD-his containing limiting amounts of adenine for 3–4 d at 23°C. The rate of pRS415-ADE2 loss scored by the level of red pigment reflects the initiation efficiency and was scored for 100–200 colonies from two independent transformants. Values are presented as percentage of total colony number. Solid box, all red colony; vertical-lined box, >80% red; cross-hatched box, 20%–80% red; diagonal-lined box, <20% red; open box, white. NcoI-digested genomic DNA from each transformant was probed for the genomic ARS1 after two-dimensional-gel electrophoresis (see Materials and Methods). Arrows identify bubble arcs (i.e., initiation events). (C) Isogenic wild-type, orc2-1 and rad53 strains were transformed with pRS423, whereas rad53 and orc2-1 strains were transformed with p2μ-orc2-1 (pRS423-orc2-1) or pCEN-orc2-1 (pRS413-orc2-1). Transformants cultured in SD-his at 23°C were scored for cell viability as in Figure 1A. (D) Isogenic wild-type and orc2-1 strains transformed with vector or orc2-1 expressing plasmids (A–C) were cultured in SD-leu at 23°C. Western analysis of Rad53-myc phosphorylation was performed (see Fig. 1) and Rad53p phosphorylation levels in orc2-1 cells after 100 min in HU were 49% (pRS425), 63% (pCEN-orc2-1), and 97% (p2μ-orc2-1) of the wild-type levels. (E) DNA combing and quantitation of the distances between stretches of BrdU incorporation were performed as described in Materials and Methods by use of wild-type (E-1000) and orc2-1 cells (E-1313) cultured at 24°C. The inter-origin distances (kilobase) for 208 origins are plotted, and the mean and S.D. indicated. Cell cycle progression through S phase was monitored by FACS analysis at 24°C.
We next confirmed that the replication function of ORC is restored when the mutant protein is overexpressed by scoring for ADE2-episome maintenance on nonselective medium (Fig. 4B). In wild-type cells, 95% of the colonies are white (indicative of pCEN-ADE2 replication and segregation), whereas >95% of the orc2-1 colonies are sectored or red, indicating high rates of plasmid loss. Upon mutant orc2-1p overexpression, orc2-1 cells can maintain the ADE2-containing plasmid at near wild-type levels. Again, this is dose dependent; pCEN-orc2-1 restores 25%–60%, and p2μ-orc2-1 restores 90% of wild-type maintenance rates. Consistent with the suppression phenotype of the uba1-o1 mutation, the double orc2-1 uba1-o1 mutant also improves pCEN-ADE2 stability (data not shown).
To demonstrate the restoration of genomic initiation events, we checked the efficiency of origin activation of the genomic ARS1 origin in these strains (Fig. 4B). The so-called bubble arc indicates efficient origin firing (see wild-type cells), and its appearance in orc2-1 mutant cells requires the presence of low- or high-copy number orc2-1 expression plasmids (Fig. 4B, arrows). Again, the increase in bubble arc intensity is dosage dependent. Similar dose-dependent increases in initiation rates were obtained for a second early-firing origin, ARS607 (data not shown).
Finally, to correlate restored origin efficiency with checkpoint function, we examined the S-phase checkpoint in these same transformed strains. The viability of cells was determined after exposure to HU and shows an orc2-1 dose-dependent restoration of survival (Fig. 4C). As expected, this effect is not dominant over other mutations; the rad53 defect is unaffected by orc2-1p levels (Fig. 4C). To confirm that Rad53p activation was restored by orc2-1 expression, the transformed strains were exposed to 0.2 M HU, and Rad53p phosphorylation was monitored. Again, we see a dose-dependent recovery of the checkpoint response, and full restoration requires the high-copy p2μ-orc2-1 plasmid (Fig. 4D).
In summary, the orc2-1 mutation destabilizes Orc2p, reducing its level 10-fold. Increased expression of the mutant protein, achieved either by increased gene dosage or the uba1-o1 mutation, restores replication efficiency, viability on HU, and the ability of the cells to activate the intra-S-phase checkpoint in the presence of either HU (Fig. 4C,D) or of MMS (data not shown). This correlation between the dose-dependent restoration of origin function and of the checkpoint response suggests that checkpoint activation in S phase requires a sufficient number of replication forks (or sufficient amount of signal from stalled forks).
orc2-1 mutants have 30% fewer replication forks
To determine the degree to which the orc2-1 mutation reduces fork number in vivo, a molecular combing method (Michalet et al. 1997) was applied to isogenic wild-type and orc2-1 cells grown at permissive temperature and synchronized in S phase (Fig. 4E). In this method, the frequency of genomic origin firing is estimated from the average distance scored between them on DNA isolated from cells that have incorporated a derivatized nucleotide (Lengronne et al. 2001). In the orc2-1 strain, the average origin-to-origin distance is 65 ± 26 kb, which implies ∼210 active origins, as compared with 45 ± 16 kb and ∼320 origins in wild-type cells. Surprisingly, orc2-1 cells progress into and through S phase with unperturbed kinetics despite this reduced level of active origins (FACS analysis; Fig. 4E).
Orc2p expression is not necessary for S phase after establishment of the preRC
The reduction in active origins in the orc2-1 strain, combined with the dose-dependent restoration of origin function and checkpoint response, suggests that ORC's execution point for the S-phase checkpoint corresponds to the establishment of the preRC in G1. The best-characterized function of ORC is to facilitate the origin-specific loading of Cdc6p, Cdt1p, and the MCM helicase, which in turn bind Cdc45 and help load pol α-primase for the initiation step (Diffley 2001). To rule out a direct role of Orc2p in checkpoint signaling, and to confirm that its unique role in the checkpoint is the licensing of a sufficient number of origins, we designed a strain in which we could eliminate Orc2p after preRC formation, but before S phase.
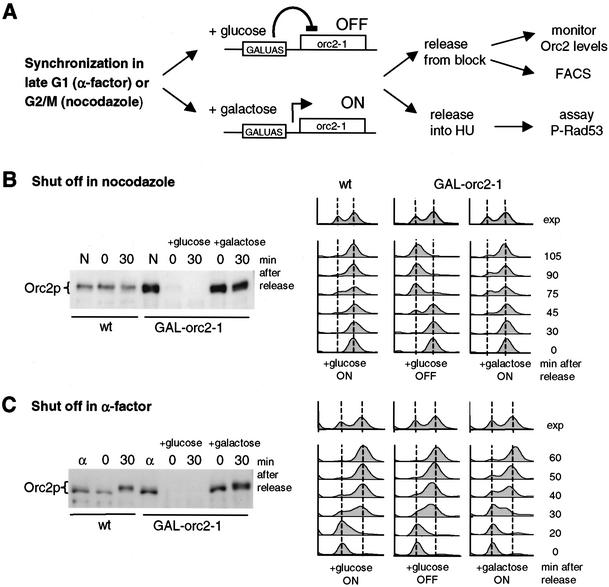
The integration of a galactose-inducible UAS upstream of the endogenous copy of orc2-1 (Fig. 5A) renders its expression repressible by glucose-containing medium. When repressed, the existing orc2-1p is rapidly depleted due to its inherent instability. Given its 8-min half-life (Fig. 3), and assuming a wild-type dosage of 2 × 104 copies per cell, the orc2-1p complement should be reduced to <100 copies/cell by 1 h. This was confirmed on Western blots after shutting off transcription by glucose addition to either a random or synchronized population of cells. Within an hour after the switch to glucose, orc2-1p levels drop from above-wild-type levels to undetectable amounts (Fig. 5, Supplementary Fig. 2).
Figure 5.
The integrated GAL1UAS∷orc2-1 construct allows rapid depletion of orc2-1p in synchronized cells. (A) After integration of Gal1UAS upstream of the mutant orc2-1 gene (GAL1UAS∷orc2-1), orc2-1p levels are slightly elevated on galactose and the cells exhibit wild-type growth rates (B,C). Replacing galactose with glucose suppresses expression and all detectable orc2-1p disappears within 60 min. (B,C) Wild-type (GA-180) and GAL1UAS∷orc2-1 cells (GA-1680) were cultured in YPA-2% galactose at 30°C, and cells were arrested at G2/M with nocodazole (7.5 μg/mL, B) for 70 min or at G1 with α-factor for 60 min (C). Thereafter, the wild-type and half of the GAL1UAS∷orc2-1 cells were shifted for 1 h to 2% glucose medium with nocodazole or pheromone to allow depletion of orc2-1p. Cells were then released from the nocodazole or α-factor block into either glucose or galactose, as indicated. Samples were taken for Western blots with anti-Orc2p at 0 or 30 min (minutes after release), or for FACS analysis at 30–105 min (B) and 20–60 min (C).
To confirm that the depletion was sufficient to confer a complete orc− phenotype, wild-type and GAL1UAS∷orc2-1 cells were grown on galactose and synchronized in mitosis with nocodazole. In the middle of the 2-h nocodazole arrest, cells were switched to glucose to repress orc2-1 transcription for 1 h, after which cells were released into glucose-containing medium lacking nocodazole. Samples were taken at 0 and 30 min after release for Western analysis, and at later time intervals to monitor cell cycle progression by FACS (Fig. 5B). As expected, orc2-1p is not detectable 1 h after the switch to glucose (time 0 after release), even though the initial GAL1UAS∷orc2 expression level is higher on galactose than from the endogenous promoter (Fig. 5B, cf. lanes labeled N in wild-type and GAL–orc2-1 cells). Importantly, FACS analysis confirms that the released GAL1UAS∷orc2-1 cells accumulate at the G1/S boundary, unable to initiate DNA replication in the absence of preRC formation (Fig. 5B).
In contrast, when the same protocol was applied to GAL1UAS∷orc2-1 cells in mid-G1 (α-factor arrest; Fig. 5C), progression into and through S phase proceeds with normal kinetics (Fig. 5C). Moreover, DNA replication in the absence of orc2-1p has no lethal side effects, for if orc2-1 expression is reinduced at G2/M following its depletion in G1, cells resume growth efficiently (data not shown). These controls demonstrate that ORC plays an essential role in the establishment of the preRC in early G1, but suggests that the preRC's formed prior to α-factor-induced arrest remain competent for initiation in the absence of detectable levels of Orc2p.
As an independent control for this, we scored for association of MCM complexes with an insoluble chromatin fraction (Pasero et al. 1999) using GAL1UAS∷orc2-1 cells depleted for orc2-1p during pheromone arrest, as described above. Roughly equal levels of Mcm2p are recovered in chromatin-bound fractions isolated from wild-type and orc2-1p-deleted cells (data not shown). Combined with FACS and two-dimensional-gel data (Figs. 5, 6B), this argues that a continued presence of Orc2p in late G1 is not necessary for maintenance of an initiation competent preRC, nor for progression into S phase. This agrees with observations made in Xenopus egg extracts, in which depletion of ORC following preRC formation had no effect on replication efficiency per se (Hua and Newport 1998; Rowles et al. 1999).
Figure 6.
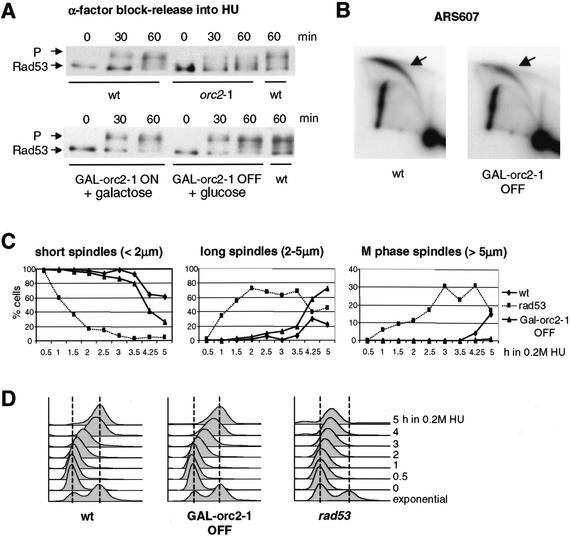
The S-phase checkpoint requires ORC2 function in G1, but not in S phase. (A) Congenic wild-type (GA-1682), GAL1UAS∷orc2-1 (GA-1681), and orc2-1 (GA-1836) strains were cultured in YPA-2% galactose (YPAG) at 23°C. After synchronization in α-factor, wild-type, orc2-1, and half of the GAL1UAS∷orc2-1 (GAL∷orc2-1 OFF) cells were switched to YPAD + α-factor, whereas the other half of the GAL1UAS∷orc2-1 culture was kept on YPAG + α-factor (GAL∷orc2-1 ON). After 60 min at 30°C, cultures were washed and released from pheromone into 0.2 M HU in YPAD or YPAG at 30°C. Rad53p phosphorylation was monitored at 0, 30, and 60 min after release. (B) Two-dimensionl gel analysis of replication intermediates at the genomic origin ARS607, based on genomic DNA prepared from wild-type and GAL∷orc2-1 OFF cells released from α-factor into HU for 60 min. PstI–ClaI-digested genomic DNA was probed as described in Materials and Methods. (C,D) Wild-type (GA-1535), rad53 (GA-1499), and GAL1UAS∷orc2-1 (GA-1780) cells were synchronized by α-factor in YPAG at 30°C. After depletion of orc2-1p during α-factor block (A), cells were released into YPAD + 0.2 M HU, and aliquots were taken for spindle elongation analysis and FACS (D) as in Figure 1C–E. At 4–5 h, rad53 cells enter the next cell cycle.
The intra-S checkpoint does not need the continued presence of Orc2p
We could now test whether the intra-S-phase checkpoint remains functional in the absence of detectable Orc2p. GAL1UAS∷orc2-1 cells were synchronized with pheromone on galactose, switched to glucose, and finally released after an hour to enter S phase in the presence of HU (Fig. 5A). The Rad53p phosphorylation state was then monitored at 0, 30, and 60 min after addition of HU. The observed response is fully intact in GALUAS∷orc2-1 cells on glucose (Fig. 6A; GAL–orc2-1 OFF, + glucose 60 min). Controls were performed on galactose and with normal orc2-1 cells (Fig. 6A), whereas FACS analyses confirm that GALUAS∷orc2-1 and wild-type cells respond to HU identically (Fig. 6D). Finally, the analysis of ARS607 by two-dimensional gels indicates that this origin fires with wild-type efficiency after the depletion of orc2-1p in mid-G1 (Fig. 6B). If similar manipulations are performed such that orc2-1 transcription is repressed prior to preRC formation (i.e., in a cdc15-2 arrest), then Rad53p fails to be activated in response to HU, as cells do not efficiently enter S phase (data not shown).
Using the same depletion protocol during α-factor arrest, we monitored spindle length and DNA content in the presence of HU, comparing the isogenic GAL1UAS∷orc2-1 cells with wild-type and rad53-deficient strains. Consistent with the Rad53p phosphorylation data, cells depleted for orc2-1p retain short S-phase spindles and progress slowly through S phase in HU like the parental strain, whereas the rad53 mutant fails to repress spindle growth despite incomplete DNA replication (Fig. 6C,D). These experiments argue strongly that the execution point of Orc2p for intra-S-phase checkpoint competence is in G1, and suggest that the impaired checkpoint response stems exclusively from insufficient preRC formation.
Damage and fork arrest signals are additive for intra-S-phase Rad53p activation
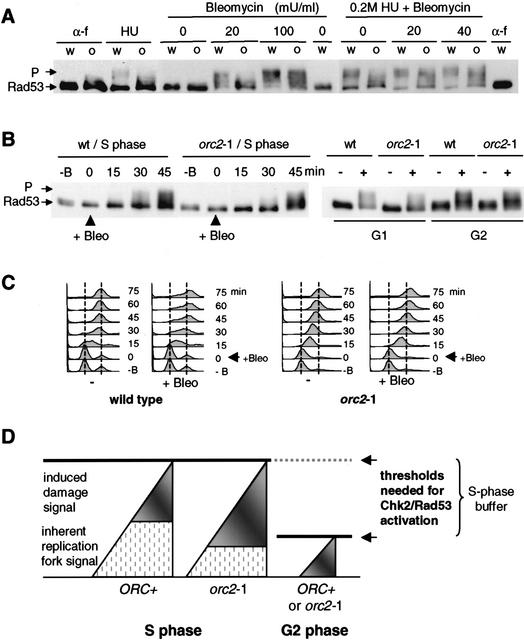
The striking, dose-dependent restoration of the intra-S-phase checkpoint (Fig. 4) implies that a certain number of stalled forks are necessary to provoke Rad53p activation on HU. On the other hand, stalled forks are not the only type of damage that provokes Rad53p activation in mid-S phase. To see whether the threshold applies to other types of DNA damage, and to test whether these are additive with HU-induced signals, we treated orc2-1 cells synchronized in S phase, with HU alone, with a titration of the radiomimetic drug bleomycin, or both. Figure 7A shows that neither HU alone nor 20 mU of bleomycin alone can activate Rad53p in the orc2-1 strain in S phase, whereas wild-type cells show a strong checkpoint response 30 min after drug addition. On the other hand, very high levels of bleomycin (100 mU/m), or a combined treatment of HU and 20 or 40 mU/mL bleomycin activates Rad53p fully even in the orc2-1 strain, suggesting that the signals are additive and integrated at the level of Rad53p. The combined effects of fork arrest and bleomycin-induced DNA damage on Rad53p kinase activation indicates that unlike G2 phase, in which a single DSB can impose a prolonged G2/M delay (Sandell and Zakian 1993), S-phase cells have a threshold that requires a pronounced accumulation of signal to provoke the Rad53p-dependent response.
Figure 7.
Combined damage reaches the threshold for intra-S-phase checkpoint activation. (A) Wild-type (w, GA-1829) and orc2-1 (o, GA-1831) strains were cultured at 23°C and synchronized in G1 with α-factor (α-f). After release into YPAD without HU, early S-phase cells were exposed to 0, 20, or 100 mU/mL bleomycin for 30 min, and then analyzed for Rad53p phosphorylation. Samples to the right were identical, except that cells were first released into YPAD + 0.2 M HU for 50 min (HU), prior to bleomycin addition (0, 20, or 40 mU/ml for 30 min). (B) For the analysis of Rad53p activation in S-phase, wild-type (wt, GA-1835) and orc2-1 (GA-1836) cells were blocked with α-factor at 23°C and released into YPAD. The functional equivalent of 20 mU/mL bleomycin was added after 20 min, and Rad53-myc phosphorylation (P) was monitored at the indicated times. For G1- or G2-phase cells, Rad53 activation was analyzed in α-factor-arrested or nocodazole-arrested cells, either with (+) or without (−) exposure to the same level of bleomycin for 30 min (right). Batch-specific variations in milliunits of bleomycin are normalized by functional assays. (C) S-phase progression with (+Bleo) or without (−) 20 mU/mL bleomycin was determined by FACS on samples from B, left. (D) We propose that the Chk2 (Rad53p)-dependent checkpoint in S phase, unlike that in G2 phase, has a buffer zone that tolerates a certain level of activating signal to allow normal fork progression. The signal arising from natural replication fork pausing is a likely source of background signal (hatched region) that can be complemented by signals arising from fork arrest (DNA pol ɛ and/or Sgs1-dependent signals) or DNA damage (Rad9- or Rad24-dependent signals), to reach an activation threshold. In orc2-1 cells, replication fork number is lower than in wild-type cells, more damage-induced signal is necessary to achieve Rad53p activation.
To demonstrate that this high tolerance of damage, or threshold for Rad53p activation, is unique to S-phase cells, we compared the response of synchronized populations of wild-type and orc2-1 cells with bleomycin in G1, G2, and S phases of the cell cycle. The response of G1 and G2 phase orc2-1 cells is nearly identical to the wild-type control after 30 min (Fig. 7B). On the other hand, when bleomycin is added as cells enter S phase (i.e., time 0 here indicates 15 min after release from pheromone), there is both a temporal lag in the response and a reduction in the level of Rad53p phosphorylation (Fig. 7B). If these same cells are analyzed by FACS, we see that unlike wild-type cells that progress slowly through S phase, the orc2-1 population moves rapidly through S phase and accumulates in G2 by 45 min after drug addition, accounting for the Rad53p response at this late time point (Fig. 7C). We note that even in wild-type cells, the S-phase response to bleomycin is significantly reduced in comparison with that provoked by the same amount of reagent in G2 phase. Again, this reinforces the notion that the threshold of damage needed to activate Rad53p changes in S phase (see Fig. 7).
Discussion
The molecular response of a eukaryotic cell to stalled replication forks and DNA damage in mid-S phase is poorly understood. Here, we have focused specifically on the intra-S-phase checkpoint, which requires the activation of the yeast homolog of the CHK2 kinase, Rad53p. The checkpoint response includes the stabilization of DNA polymerases at stalled forks, down-regulation of late-firing origins, and slowed progression through S phase as monitored by the length of the budding yeast intranuclear spindle (Santocanale et al. 1998; Shirahige et al. 1998; Lopes et al. 2001; Tercero and Diffley 2001). This response is distinct from the G2-specific arrest that prevents entry into mitosis in the presence of unreplicated DNA or damage in G2, which depends on the Chk1 kinase response (for review, see Rhind and Russell 2000; Zhou and Elledge 2000; Melo and Toczyski 2002).
The instability of a mutant form of Orc2p and a serendipitous suppressor mutation have revealed the dose-dependent character of the intra-S checkpoint response to HU and MMS. We find that the orc2-1 mutant is impaired in its ability to activate Rad53p, which accounts for its hypersensitivity to HU and DNA-damaging agents (Figs. 1, 7; see also Weinberger et al. 1999), as well as its inability to down-regulate late firing origins in the presence of MMS (Shirahige et al. 1998). Not only does Orc2p act upstream of Rad53p, but the execution point of its checkpoint function coincides with preRC formation in early G1 phase. The depletion of all detectable Orc2p in late G1 phase after origin licencing does not impair the intra-S-phase response to either MMS-induced DNA damage (data not shown) or dNTP depletion (Fig. 6). Using plasmids that express the unstable orc2-1p at different levels, we can demonstrate a strict correlation between the dose-dependent restoration of ORC's replication function and its checkpoint function, suggesting that the intra-S checkpoint can be activated by HU or MMS only when a sufficient number of replication forks are affected (Fig. 7D). Our ability to complement this reduced fork signal with low doses of the radiomimetic drug bleomycin, which alone fail to activate the checkpoint, indicates that the S-phase threshold for activation can be overcome through accumulated genetic insult, even in orc2-1 cells.
A cellular mechanism sensitive to replication fork number
Although the temporal progression of the orc2-1 mutant into and through S phase at permissive temperature is indistinguishable from that of a wild-type cell, the average distance between replication forks is ∼20 kb larger (Fig. 4E). This translates into ∼30% fewer forks per genome, a reduction that is apparently sufficient to compromise Rad53p activation on HU, MMS, or the radiomimetic drug, bleomycin. Even in a wild-type strain, levels of bleomycin that are sufficient to activate the G2-phase checkpoint, fail to activate Rad53p in S phase (Fig. 7B). This S-phase-specific dampening of the DNA damage response appears to occur in mammalian cells as well. CHO cells undergoing DNA replication do not respond to levels of ionizing radiation that are able to block cells in G1 (Larner et al. 1994; Lee et al. 1997). Moreover, it was shown that p53 is stabilized, but not active, when S-phase cells are treated with ionizing radiation, possibly suggesting that mammalian cells also have a suppressed checkpoint response during DNA replication arrest (Gottifredi et al. 2001).
We have demonstrated that different types of damage in S phase are additive for Rad53p-dependent checkpoint activation, despite the fact that different damage-specific sensors and adaptors are functioning upstream of Rad53p. This emphasizes the role of the Rad53p (Chk2) kinase as an integrator for diverse signals, and supports genetic data that argue for parallel pathways within the intra-S-phase damage and replication checkpoint that merge at Rad53p (Foiani et al. 2000; Frei and Gasser 2000; Falck et al. 2002; Myung and Kolodner 2002). We argue that the high threshold for Rad53p activation is an essential characteristic of the response to genetic insult during DNA replication. The Chk2-activation pathway in S phase may be designed to tolerate a certain level of damage-like signals from replication forks, to enable efficient fork progression.
It is clear that replication forks pause frequently during a normal S phase (Deshpande and Newlon 1996; Wang et al. 2001), often leading to parental strand breakage. Because these can generally be repaired by recombination with the adjacent sister chromatid (Myung et al. 2001; Schär 2001), it would not, in most situations, be in the interest of the cell to provoke a cell-cycle arrest in response to these fork-associated events. The noise of damage arising from normal fork progression should fall, therefore, below the threshold necessary for the intra-S-phase checkpoint activation (Fig. 7). Although such tolerance may be essential to ensure S-phase progression, it would not only be unnecessary, but dangerous, in G2-phase cells.
A rationale for the delayed initiation of origins
If normal replication-fork pausing can create signals similar to those evoked by HU or MMS, then a cell may need to limit the number of replication forks active at any given point in S phase to stay below the intra-S checkpoint response threshold. This may provide a rationale for the temporal patterns of eukaryotic origin activation, as the distribution of origin firing throughout S phase also temporally distributes the background damage signal arising from naturally paused forks. In support of this, we note that wild-type, but not rad53 cells delay S phase when forced to replicate very high-copy number plasmids (>200 copies per cell of a leu2d-plasmid; data not shown).
Embryonic nuclei, which have frequent initiation events (one per 5 kb) and no clear demarcation of early-, mid-, and late-S-phase replication events (Blow et al. 2001), may suppress this aspect of the dosage-dependent intra-S-phase checkpoint pathway. Although Xenopus egg extracts do support a Chk1-dependent arrest prior to mitosis in response to the presence of aphidicolin (Hekmat-Nejad et al. 2000; Kumagai and Dunphy 2000), this S/M checkpoint differs fundamentally from the intra-S-phase Chk2p (Rad53p) activation described here. We also predict that the S/M checkpoint may not require the threshold of activation shown here for the intra-S arrest.
S/M regulation is distinct from the threshold dependent intra-S-phase response
It is important to note that the checkpoint defects ascribed to the orc2-1 mutant are not equivalent to those of the Saccharomyces cerevisiae cdc6 or the Schizosaccharomyces pombe cdc18 mutations (Kelly et al. 1993; Piatti et al. 1995), nor to those provoked by the depletion of MCM components in late G1- or S-phase cells (Labib et al. 2001). First of all, the checkpoint defect of cdc6-1sc cells is observed under conditions that block the initiation of DNA replication, and lead to a reductional mitosis, due to the absence of the S/M checkpoint that would normally prevent the metaphase–anaphase transition. In contrast, the orc2-1 defect in S-phase checkpoint activation is manifest under conditions that support rates of elongation similar to those in wild-type cells. Moreover, orc2-1 cells incur a low-level activation of Rad53p at the G2/M boundary accumulate prior to metaphase even in the absence of HU (data not shown). Thus, although the orc2-1 mutation compromises Rad53p activation during the intra-S-phase response, these same cells do activate Rad53p to block G2/M progression, unlike cdc6sc or cdc18sp mutants. Finally, the degron-induced degradation of MCM proteins in S phase itself induces fork stalling (Labib et al. 2001), which is not observed in the absence of Orc2p (Fig. 5).
On the basis of orc2-1p depletion in late G1 (Figs. 6, 7), we can argue that ORC plays no direct role in sensing stalled forks in budding yeast. Our data are inconsistent with the interpretation of Murakami et al. (2002), who suggest that S. pombe ORC is directly involved in maintaining S-phase checkpoint activation, on the basis of analysis of progression into mitosis in the orp1tscdc18ts mutant. Although mutation of the CDC6 and MCM genes discussed above do not act like orc2-1, it is possible that other mutants in the replication machinery that enhance HU and MMS sensitivity, do so by reducing the number of functional replication forks and thus compromising Rad53p activation. This requires further investigation.
Mechanisms for attenuating S-phase checkpoint kinase activation
We envision two potential mechanisms for the S-phase buffering of Rad53p activation. First, the sensitivity of Rad53p itself may be modulated in S phase, that is, more activating signal may be needed to activate the kinase due to an S-phase-specific modification or ligand of Rad53p. For example, Mrc1p could buffer Rad53p activation, as well as being required for its full activation in response to damage signals arising either from a stalled fork or from damage detected by the Rad17/Mec3/Ddc1 complex (Alcasabas et al. 2001; Tanaka and Russell 2001). Alternatively Asf1p, which appears to form a complex with Rad53p in an inactive form, could sequester Rad53p from stimulatory signal (Emili et al. 2001; Hu et al. 2001).
An alternative threshold mechanism may involve modulation of the signal itself. It has been argued that the creation of single-strand DNA and the binding of Replication factor A (RFA), is part of a dose-dependent signal for Rad53p activation in response to either a targeted DSB or to subtelomeric damage in a cdc13-1 allele (Garvik et al. 1995; Lee et al. 1998; Pellicioli et al. 2001). Other studies have implicated RFA in an S-phase response (Longhese et al. 1996; Kim and Brill 2001). Thus, the S-phase-specific phosphorylation of Rfa2p (Din et al. 1990; Dutta and Stillman 1992) may quench its ability to signal to Rad53p. Consistent with a role of ssDNA as an intra-S-phase checkpoint-activating signal (Longhese et al. 1996), it has been shown that the ssDNA associated with a replication fork increases by ∼100 nucleotides in HU (Sogo et al. 2002). This increase of 200 nucleotides per bidirectional fork could serve to count stalled forks in a dose-dependent manner.
In summary, our analysis of the role of Orc2p has revealed the unique and hitherto uncharacterized ability of the DNA damage checkpoint to adapt to S phase by imposing a threshold on checkpoint activation. This may allow the cell to tolerate replication-associated structures that look like damage, while preserving the cell's ability to respond to damage above a certain level.
Materials and methods
Strains and plasmids
All strains used are listed in Table 1. We found that our original orc2-1 strain (YJL861 or JRY4125, from J. Li, University of California, San Francisco) was viable at 30°C but not at 37°C, whereas orc2-1 spores recovered after backcrossing to the strain's parental background W303, died at 30°C. Suspecting that the original strain had an extragenic suppressor mutation, we backcrossed repeatedly and scored for temperature-sensitive lethality at various temperatures, identifying two temperature-sensitive alleles, orc2-1 (dead at ≥30°C; GA-1254) and uba1-o1 (dead at 37°C; GA-1256). A second orc2-1 strain from another source (CFY 240, Dr. C.A. Fox, University of Wisconsin) was also viable at 30°C, and from this strain we isolated a different extragenic suppressor, suggesting that such mutations occur frequently. To avoid spontaneous suppressors, all experiments using orc2-1 are performed with freshly defrosted cells checked regularly for slow growth at 23°C and lethality at 30°C. The sgs1∷TRP1 and sgs1∷LEU2 both eliminate Sgs1p and are collectively called sgs1Δ (Frei and Gasser 2000). The rad53 allele used is mec2-1 or rad53-11 (Weinert et al. 1994). Cells carrying a GFP–TUB1 fusion at the URA3 locus were created by the transformation with pAFS125 (gift of Dr. K. Bloom, University of North Carolina). The GAL1UAS∷orc2-1 strain (GA-1680) was created by integration of a PCR-based GAL–UAS cassette (Longtine et al. 1998) upstream of orc2-1 in GA-1254.
Table 1.
Yeast strains used in this study
| Strain
|
Genotype
|
Source
|
|---|---|---|
| GA-180 | MATα ade2-1 trp1-1 his3-11,15 ura3-1 leu2-3,112 can1-100 | R. Rothstein (W303-1A) |
| GA-435 | MATα ade2-1 trp1-1 his3-11,15 ura3-1 leu2-3,112 can1-100 orc2-1 uba1-o1 | J. Li (YJL861) formerly JRY4125 |
| GA-463 | GA-180 with orc2-1 uba1-o1 | This study |
| GA-734 | GA-180 with sgs1-3::TRP1 | R. Sternglanz (RS1077) |
| GA-893 | GA-180 with orc2::ORC2-9Myc-LEU2 | K Nasmyth (K6447) |
| GA-1048 | MATα his3 ura3 leu2 trp1 mec1-1 sml1 rad53::RAD53-13Myc-KanMX6 | Frei and Gasser (2000) |
| GA-1148 | GA-180 with rad9::LEU2 | D. Shore (S114) |
| GA-1230 | GA-180 with mec2-1(rad53-11) bar1::hisG ssd1-d2 | T. Weinert (DLY259) |
| GA-1254 | GA-180 with orc2-1 | This study |
| GA-1256 | GA-180 with uba1-o1 | This study |
| GA-1387 | GA-180 with orc2-1 sgs1-3::TRP1 | This study |
| GA-1388 | GA-180 with uba1-o1 sgs1-3::TRP1 | This study |
| GA-1389 | GA-180 with orc2-1 uba1-o1 sgs1-3::TRP1 | This study |
| GA-1499 | MATα ade2-1 leu2 his3 mec2-1(rad53-11) URA3::GFP-TUB1 | This study |
| GA-1533 | GA-180 with orc2-1 URA3::GFP-TUB1 | This study |
| GA-1535 | GA-180 with URA3::GFP-TUB1 | This study |
| GA-1680 | GA-180 with His3MX6-UASGal-orc2-1 | This study |
| GA-1681 | GA-1682 with His3MX6-UASGal-orc2-1 | This study |
| GA-1682 | MATα ura3 his3 trp1 leu2 rad53::RAD53-13Myc-KanMX6 | This study |
| GA-1829 | MATα his3 ura3 leu2 trp1 rad53::RAD53-13Myc-KanMX6 (A364a background) | This study |
| GA-1780 | GA-180 with His3MX6-UASGal-orc2-1 URA3::GFP-TUB1 | This study |
| GA-1830 | GA-1829 with sgs1::LEU2 | This study |
| GA-1831 | GA-1829 with orc2-1 | This study |
| GA-1832 | GA-1829 with orc2-1 sgs1::LEU2 | This study |
| GA-1835 | MATα ade2-1 his3 leu2 ura3 trp1 rad53::RAD53-13Myc-KanMX6 | This study |
| GA-1836 | MATα ade2-1 his3 leu2 ura3 trp1 orc2-1 rad53::RAD53-13Myc-KanMX6 | This study |
| GA-1837 | MATα his3 leu2 ura3 trp1 uba1-o1 rad53::RAD53-13Myc-KanMX6 | This study |
| GA-1838 | MATα his3 leu2 ura3 trp1 orc2-1 uba1-o1 rad53::RAD53-13Myc-KanMX6 | This study |
| GA-1860 | GA-180 with orc2-1 rad9::LEU2 | This study |
| GA-1861 | GA-180 with uba1-o1 rad9::LEU2 | This study |
| E-1000 | GA-180 with URA3::GPD-TK7x | Lengronne et al. (2001) |
| E-1313 | GA-180 with orc2-1 URA3::GPD-TK7x | This study |
The plasmid pRS415-ADE2 contains a 2.5-kb BglII fragment containing the ADE2 gene, whereas the pCEN-orc2-1 and p2μ-orc2-1 vectors contain the orc2-1 gene on a 2.8-kb SacI fragment from pCF226 (gift of Dr. C.A. Fox, University of Wisconsin) in the appropriate pRS vectors. Medium preparation, HU, bleomycin and MMS sensitivity assays, preparation of protein extracts, FACS, and microscopic analyses of spindle length were carried out as described in Frei and Gasser (2000). Bleomycin units are normalized by a growth curve of wild-type yeast.
Replication intermediate analysis
For two-dimensional gels analysis, yeast genomic DNA was isolated from 7 × 108 cells by using G-20 column (QIAGEN) as described in Wu and Gilbert (1995). After the digestion with restriction enzymes (PstI–ClaI for the early ARS607, and NcoI for ARS1), replication intermediates were enriched by BND-cellulose and applied to two-dimensional gel (neutral/neutral) analysis as described (Huberman et al. 1987). DNA combing was performed as described in Michalet et al. (1997) and Lengronne et al. (2001). In brief, strains E1000 or E1313 were arrested with pheromone and released synchronously into S phase in the presence of 0.4 mg/mL bromodeoxyuridine. To pause forks, 0.2 M HU was added, and genomic DNA was isolated and combed on silanized coverslips. BrdU epitopes were detected with appropriate antibodies (Sera Labs; Molecular Probes). BrdU tracks were measured on CCD-recorded images with Metamorph (Universal Imaging Corp.), and converted to base pairs using λ DNA as a molecular standard.
Acknowledgments
We thank J. Li, R. Sternglanz, D. Shore, B. Tye, S. Bell, C.A. Fox, K. Bloom, J.M. Galan, K. Nasmyth, and T. Weinert for strains, plasmids, and/or protein. We thank Drs. A. Taddei, J. Cobb, K. Dubrana, and L. Bjergbaek for critical comments on the manuscript. K.S. acknowledges support from the IARC, Cancer and Solidarity Foundation, and ISREC. The Gasser laboratory thanks the Swiss National Science Foundation and the Swiss Cancer League for continued support.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL susan.gasser@molbio.unige.ch; FAX 41-22-702-6868.
Supplemental material is available at http://www.genesdev.org.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.239802.
References
- Alcasabas AA, Osborn AJ, Bachant J, Hu F, Werler PJ, Bousset K, Furuya K, Diffley JF, Carr AM, Elledge SJ. Mrc1 transduces signals of DNA replication stress to activate Rad53. Nat Cell Biol. 2001;3:958–965. doi: 10.1038/ncb1101-958. [DOI] [PubMed] [Google Scholar]
- Bell SP. The origin recognition complex: From simple origins to complex functions. Genes & Dev. 2002;16:659–672. doi: 10.1101/gad.969602. [DOI] [PubMed] [Google Scholar]
- Blow JJ, Gillespie PJ, Francis D, Jackson DA. Replication origins in Xenopus egg extract are 5–15 kilobases apart and are activated in clusters that fire at different times. J Cell Biol. 2001;152:15–25. doi: 10.1083/jcb.152.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande AM, Newlon CS. DNA replication fork pause sites dependent on transcription. Science. 1996;272:1030–1033. doi: 10.1126/science.272.5264.1030. [DOI] [PubMed] [Google Scholar]
- Diffley JF. DNA replication: building the perfect switch. Curr Biol. 2001;11:R367–R370. doi: 10.1016/s0960-9822(01)00196-8. [DOI] [PubMed] [Google Scholar]
- Din S, Brill SJ, Fairman MP, Stillman B. Cell-cycle-regulated phosphorylation of DNA replication factor A from human and yeast cells. Genes & Dev. 1990;4:968–977. doi: 10.1101/gad.4.6.968. [DOI] [PubMed] [Google Scholar]
- Dutta A, Stillman B. Cdc2 family kinases phosphorylate a human cell DNA replication factor, Rpa, and activate DNA replication. EMBO J. 1992;11:2189–2199. doi: 10.1002/j.1460-2075.1992.tb05278.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emili A, Schieltz DM, Yates JR, III, Hartwell LH. Dynamic interaction of DNA damage checkpoint protein Rad53 with chromatin assembly factor Asf1. Mol Cell. 2001;7:13–20. doi: 10.1016/s1097-2765(01)00150-2. [DOI] [PubMed] [Google Scholar]
- Falck J, Petrini JH, Williams BR, Lukas J, Bartek J. The DNA damage-dependent intra-S phase checkpoint is regulated by parallel pathways. Nat Genet. 2002;30:290–294. doi: 10.1038/ng845. [DOI] [PubMed] [Google Scholar]
- Foiani M, Pellicioli A, Lopes M, Lucca C, Ferrari M, Liberi G, Muzi Falconi M, Plevani P. DNA damage checkpoints and DNA replication controls in S. cerevisiae. Mutat Res. 2000;451:187–196. doi: 10.1016/s0027-5107(00)00049-x. [DOI] [PubMed] [Google Scholar]
- Foss M, McNally FJ, Laurenson P, Rine J. Origin recognition complex (ORC) in transcriptional silencing and DNA replication in S. cerevisiae. Science. 1993;262:1838–1844. doi: 10.1126/science.8266071. [DOI] [PubMed] [Google Scholar]
- Frei C, Gasser SM. The yeast Sgs1p helicase acts upstream of Rad53p in the DNA replication checkpoint and colocalizes with Rad53p in S-phase-specific foci. Genes & Dev. 2000;14:81–96. [PMC free article] [PubMed] [Google Scholar]
- Garvik B, Carson M, Hartwell L. Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol Cell Biol. 1995;11:6128–6138. doi: 10.1128/mcb.15.11.6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottifredi V, Shieh S, Taya Y, Prives C. p53 accumulates but is functionally impaired when DNA synthesis is blocked. Proc Natl Acad Sci. 2001;98:1036–1041. doi: 10.1073/pnas.021282898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hekmat-Nejad M, You Z, Yee MC, Newport JW, Cimprich KA. Xenopus ATR is a replication-dependent chromatin-binding protein required for the DNA replication checkpoint. Curr Biol. 2000;10:1565–1573. doi: 10.1016/s0960-9822(00)00855-1. [DOI] [PubMed] [Google Scholar]
- Hilt W, Enenkel C, Gruhler A, Singer T, Wolf DH. The PRE4 gene codes for a subunit of the yeast proteasome necessary for peptidylglutamyl-peptide-hydrolyzing activity. Mutations link the proteasome to stress- and ubiquitin-dependent proteolysis. J Biol Chem. 1993;268:3479–3486. [PubMed] [Google Scholar]
- Hu F, Alcasabas AA, Elledge SJ. Asf1 links Rad53 to control of chromatin assembly. Genes & Dev. 2001;15:1061–1066. doi: 10.1101/gad.873201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua XH, Newport J. Identification of a preinitiation step in DNA replication that is independent of origin recognition complex and cdc6, but dependent on cdk2. J Cell Biol. 1998;140:271–281. doi: 10.1083/jcb.140.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huberman JA, Spotila LD, Nawotka KA, el-Assouli SM, Davis LR. The in vivo replication origin of the yeast 2 microns plasmid. Cell. 1987;51:473–481. doi: 10.1016/0092-8674(87)90643-x. [DOI] [PubMed] [Google Scholar]
- Kastan MB. Checkpoint controls and cancer. Introduction. Cancer Surv. 1997;29:1–6. [PubMed] [Google Scholar]
- Kelly TJ, Martin GS, Forsburg SL, Stephen RJ, Russo A, Nurse P. The fission yeast cdc18+ gene product couples S phase to START and mitosis. Cell. 1993;74:371–382. doi: 10.1016/0092-8674(93)90427-r. [DOI] [PubMed] [Google Scholar]
- Kim HS, Brill SJ. Rfc4 interacts with Rpa1 and is required for both DNA replication and DNA damage checkpoints in S. cerevisiae. Mol Cell Biol. 2001;21:3725–3737. doi: 10.1128/MCB.21.11.3725-3737.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai A, Dunphy WG. Claspin, a novel protein required for the activation of Chk1 during a DNA replication checkpoint response in Xenopus egg extracts. Mol Cell. 2000;6:839–849. doi: 10.1016/s1097-2765(05)00092-4. [DOI] [PubMed] [Google Scholar]
- Labib K, Tercero JA, Diffley JF. Uninterrupted MCM2-7 function required for DNA replication fork progression. Science. 2000;288:1643–1647. doi: 10.1126/science.288.5471.1643. [DOI] [PubMed] [Google Scholar]
- Labib K, Kearsey SE, Diffley JF. MCM2-7 proteins are essential components of prereplicative complexes that accumulate cooperatively in the nucleus during G1-phase and are required to establish, but not maintain, the S-phase checkpoint. Mol Biol Cell. 2001;12:3658–3667. doi: 10.1091/mbc.12.11.3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larner JM, Lee J, Hamlin JL. Radiation effects on DNA synthesis in a defined chromosomal replicon. Mol Cell Biol. 1994;14:1901–1008. doi: 10.1128/mcb.14.3.1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Larner JM, Hamlin JL. A p53-independent damage sensing mechanism that functions as a checkpoint at the G1/S transition in Chinese Hamster Ovary cells. Proc Natl Acad Sci. 1997;94:526–531. doi: 10.1073/pnas.94.2.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SE, Moore JK, Holmes A, Umezu K, Kolodner RD, Haber JE. Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell. 1998;94:399–409. doi: 10.1016/s0092-8674(00)81482-8. [DOI] [PubMed] [Google Scholar]
- Lengronne A, Pasero P, Bensimon A, Schwob E. Monitoring S phase progression globally and locally using BrdU incorporation in TK(+) yeast strains. Nucleic Acids Res. 2001;29:1433–1442. doi: 10.1093/nar/29.7.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longhese MP, Neecke H, Paciotti V, Lucchini G, Plevani P. The 70 kDa subunit of replication protein A is required for the G1/S and intra-S DNA damage checkpoints in budding yeast. Nucleic Acids Res. 1996;24:3533–3537. doi: 10.1093/nar/24.18.3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in S. cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Lopes M, Cotta-Ramusino C, Pellicioli A, Liberi G, Plevani P, Muzi-Falconi M, Newlon CS, Foiani M. The DNA replication checkpoint response stabilizes stalled replication forks. Nature. 2001;412:557–561. doi: 10.1038/35087613. [DOI] [PubMed] [Google Scholar]
- Lydall D, Weinert T. G2/M checkpoint genes of Saccharomyces cerevisiae: Further evidence for roles in DNA replication and/or repair. Mol Gen Genet. 1997;256:638–651. doi: 10.1007/s004380050612. [DOI] [PubMed] [Google Scholar]
- Marini F, Pellicioli A, Paciotti V, Lucchini G, Plevani P, Stern DF, Foiani M. A role for DNA primase in coupling DNA replication to DNA damage response. EMBO J. 1997;16:639–650. doi: 10.1093/emboj/16.3.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath JP, Jentsch S, Varshavsky A. UBA 1: An essential yeast gene encoding ubiquitin-activating enzyme. EMBO J. 1991;10:227–236. doi: 10.1002/j.1460-2075.1991.tb07940.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo J, Toczyski D. A unified view of the DNA-damage checkpoint. Curr Opin Cell Biol. 2002;14:237–245. doi: 10.1016/s0955-0674(02)00312-5. [DOI] [PubMed] [Google Scholar]
- Michalet X, Ekong R, Fougerousse F, Rousseaux S, Schurra C, Hornigold N, van Slegtenhorst M, Wolfe J, Povey S, Beckmann JS, et al. Dynamic molecular combing: stretching the whole human genome for high-resolution studies. Science. 1997;277:1518–1523. doi: 10.1126/science.277.5331.1518. [DOI] [PubMed] [Google Scholar]
- Murakami H, Yanow SK, Griffiths D, Nakanishi M, Nurse P. Maintenance of replication forks and the S-phase checkpoint by Cdc18p and Orp1p. Nat Cell Biol. 2002;4:384–388. doi: 10.1038/ncb789. [DOI] [PubMed] [Google Scholar]
- Myung K, Kolodner RD. Suppression of genome instability by redundant S-phase checkpoint pathways in S. cerevisiae. Proc Natl Acad Sci. 2002;99:4500–4507. doi: 10.1073/pnas.062702199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myung K, Datta A, Kolodner RD. Suppression of spontaneous chromosomal rearrangements by S phase checkpoint functions in S. cerevisiae. Cell. 2001;104:397–408. doi: 10.1016/s0092-8674(01)00227-6. [DOI] [PubMed] [Google Scholar]
- Pasero P, Duncker BP, Schwob E, Gasser SM. A role for the Cdc7 kinase regulatory subunit Dbf4p in the formation of initiation-competent origins of replication. Genes & Dev. 1999;13:2159–2176. doi: 10.1101/gad.13.16.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulovich AG, Hartwell LH. A checkpoint regulates the rate of progression through S phase in S. cerevisiae in response to DNA damage. Cell. 1995;82:841–847. doi: 10.1016/0092-8674(95)90481-6. [DOI] [PubMed] [Google Scholar]
- Paulovich AG, Margulies RU, Garvik BM, Hartwell LH. RAD9, RAD17, and RAD24 are required for S phase regulation in S. cerevisiae in response to DNA damage. Genetics. 1997;145:45–62. doi: 10.1093/genetics/145.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellicioli A, Lee SE, Lucca C, Foiani M, Haber JE. Regulation of Saccharomyces Rad53 checkpoint kinase during adaptation from DNA damage-induced G2/M arrest. Mol Cell. 2001;7:293–300. doi: 10.1016/s1097-2765(01)00177-0. [DOI] [PubMed] [Google Scholar]
- Piatti S, Lengauer C, Nasmyth K. Cdc6 is an unstable protein whose de novo synthesis in G1 is important for the onset of S phase and for preventing a “reductional” anaphase in the budding yeast S. cerevisiae. EMBO J. 1995;14:3788–3799. doi: 10.1002/j.1460-2075.1995.tb00048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhind N, Russell P. Chk1 and Cds1: Linchpins of the DNA damage and replication checkpoint pathways. J Cell Sci. 2000;113:3889–3896. doi: 10.1242/jcs.113.22.3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowles A, Tada S, Blow JJ. Changes in association of the Xenopus origin recognition complex with chromatin on licensing of replication origins. J Cell Sci. 1999;112:2011–2018. doi: 10.1242/jcs.112.12.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley A, Cocker JH, Harwood J, Diffley JF. Initiation complex assembly at budding yeast replication origins begins with the recognition of a bipartite sequence by limiting amounts of the initiator, ORC. EMBO J. 1995;14:2631–2641. doi: 10.1002/j.1460-2075.1995.tb07261.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez Y, Desany BA, Jones WJ, Liu Q, Wang B, Elledge SJ. Regulation of RAD53 by the ATM-like kinases MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science. 1996;271:357–360. doi: 10.1126/science.271.5247.357. [DOI] [PubMed] [Google Scholar]
- Sanchez Y, Bachant J, Wang H, Hu F, Liu D, Tetzlaff M, Elledge SJ. Control of the DNA damage checkpoint by chk1 and rad53 protein kinases through distinct mechanisms. Science. 1999;286:1166–1171. doi: 10.1126/science.286.5442.1166. [DOI] [PubMed] [Google Scholar]
- Sandell LL, Zakian VA. Loss of a yeast telomere: Arrest, recovery, and chromosome loss. Cell. 1993;75:729–739. doi: 10.1016/0092-8674(93)90493-a. [DOI] [PubMed] [Google Scholar]
- Santocanale C, Diffley JF. A Mec1- and Rad53-dependent checkpoint controls late-firing origins of DNA replication. Nature. 1998;395:615–618. doi: 10.1038/27001. [DOI] [PubMed] [Google Scholar]
- Schär P. Spontaneous DNA damage, genome instability, and cancer – when DNA replication escapes control. Cell. 2001;104:329–332. doi: 10.1016/s0092-8674(01)00220-3. [DOI] [PubMed] [Google Scholar]
- Shirahige K, Hori Y, Shiraishi K, Yamashita M, Takahashi K, Obuse C, Tsurimoto T, Yoshikawa H. Regulation of DNA-replication origins during cell-cycle progression. Nature. 1998;395:618–621. doi: 10.1038/27007. [DOI] [PubMed] [Google Scholar]
- Sogo JM, Lopes M, Foiani M. Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science. 2002;297:599–602. doi: 10.1126/science.1074023. [DOI] [PubMed] [Google Scholar]
- Sun Z, Fay DS, Marini F, Foiani M, Stern DF. Spk1/Rad53 is regulated by Mec1-dependent protein phosphorylation in DNA replication and damage checkpoint pathways. Genes & Dev. 1996;10:395–406. doi: 10.1101/gad.10.4.395. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Russell P. Mrc1 channels the DNA replication arrest signal to checkpoint kinase Cds1. Nat Cell Biol. 2001;3:966–972. doi: 10.1038/ncb1101-966. [DOI] [PubMed] [Google Scholar]
- Tercero JA, Diffley JF. Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature. 2001;412:553–557. doi: 10.1038/35087607. [DOI] [PubMed] [Google Scholar]
- Vas A, Mok W, Leatherwood J. Control of DNA rereplication via Cdc2 phosphorylation sites in the origin recognition complex. Mol Cell Biol. 2001;21:5767–5777. doi: 10.1128/MCB.21.17.5767-5777.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Vujcic M, Kowalski D. DNA replication forks pause at silent origins near the HML locus in budding yeast. Mol Cell Biol. 2001;21:4938–4948. doi: 10.1128/MCB.21.15.4938-4948.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberger M, Trabold PA, Lu M, Sharma K, Huberman JA, Burhans WC. Induction by adozelesin and hydroxyurea of origin recognition complex-dependent DNA damage and DNA replication checkpoints in S. cerevisiae. J Biol Chem. 1999;274:35975–35984. doi: 10.1074/jbc.274.50.35975. [DOI] [PubMed] [Google Scholar]
- Weinert T. DNA damage and checkpoint pathways: Molecular anatomy and interactions with repair. Cell. 1998;94:555–558. doi: 10.1016/s0092-8674(00)81597-4. [DOI] [PubMed] [Google Scholar]
- Weinert TA, Kiser GL, Hartwell LH. Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes & Dev. 1994;8:652–665. doi: 10.1101/gad.8.6.652. [DOI] [PubMed] [Google Scholar]
- Wu JR, Gilbert DM. Rapid DNA preparation for 2D gel analysis of replication intermediates. Nucleic Acids Res. 1995;23:3997–3998. doi: 10.1093/nar/23.19.3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou BBS, Elledge SJ. The DNA damage response: Putting checkpoints in perspective. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]