Abstract
Methanogenesis, the biological production of methane, plays a pivotal role in the global carbon cycle and contributes significantly to global warming. The majority of methane in nature is derived from acetate. Here we report the complete genome sequence of an acetate-utilizing methanogen, Methanosarcina acetivorans C2A. Methanosarcineae are the most metabolically diverse methanogens, thrive in a broad range of environments, and are unique among the Archaea in forming complex multicellular structures. This diversity is reflected in the genome of M. acetivorans. At 5,751,492 base pairs it is by far the largest known archaeal genome. The 4524 open reading frames code for a strikingly wide and unanticipated variety of metabolic and cellular capabilities. The presence of novel methyltransferases indicates the likelihood of undiscovered natural energy sources for methanogenesis, whereas the presence of single-subunit carbon monoxide dehydrogenases raises the possibility of nonmethanogenic growth. Although motility has not been observed in any Methanosarcineae, a flagellin gene cluster and two complete chemotaxis gene clusters were identified. The availability of genetic methods, coupled with its physiological and metabolic diversity, makes M. acetivorans a powerful model organism for the study of archaeal biology.
[Sequence, data, annotations, and analyses are available at http://www-genome.wi.mit.edu/. The sequence data described in this paper have been submitted to the GenBank data library under accession no. AE010299.]
The Archaea remain the most poorly understood domain of life despite their importance to the biosphere. Methanogenesis, which plays a pivotal role in the global carbon cycle, is unique to the Archaea. Each year, an estimated 900 million metric tons of methane are biologically produced, representing the major global source for this greenhouse gas and contributing significantly to global warming (Schlesinger 1997). Methanogenesis is critical to the waste-treatment industry and biologically produced methane also represents an important alternative fuel source. At least two-thirds of the methane in nature is derived from acetate, although only two genera of methanogens are known to be capable of utilizing this substrate. We report here the first complete genome sequence of an acetate-utilizing (acetoclastic) methanogen, Methanosarcina acetivorans C2A.
The Methanosarcineae are metabolically and physiologically the most versatile methanogens. Only Methanosarcina species possess all three known pathways for methanogenesis (Fig. 1) and are capable of utilizing no less than nine methanogenic substrates, including acetate. In contrast, all other orders of methanogens possess a single pathway for methanogenesis, and many utilize no more than two substrates. Among methanogens, the Methanosarcineae also display extensive environmental diversity. Individual species of Methanosarcina have been found in freshwater and marine sediments, decaying leaves and garden soils, oil wells, sewage and animal waste digesters and lagoons, thermophilic digesters, feces of herbivorous animals, and the rumens of ungulates (Zinder 1993).
Figure 1.
Three pathways for methanogenesis. Methanogenesis is a form of anaerobic respiration using a variety of one-carbon (C-1) compounds or acetic acid as a terminal electron acceptor. All three pathways converge on the reduction of methyl-CoM to methane (CH4). Many methanogens can reduce CO2 to methane using electrons derived by oxidizing H2 (the hydrogenotrophic pathway, red arrows). Others can utilize C-1 compounds such as methanol or methylamines with one molecule of C-1 compound being oxidized to provide electrons for reducing three additional molecules to methane (the methylotrophic pathway, green arrows). Still other methanogens split acetate into a methyl group and an enzyme-bound CO, with the CO subsequently oxidized to provide electrons for the reduction of the methyl group to methane (the acetoclastic pathway, blue arrows). In all cases, an electrochemical gradient is generated for use in ATP synthesis. Most methanogens possess only one of the three methanogenic pathways. Methanosarcina species possess all three. CoM, coenzyme M; H4SPT, tetrahydrosarcinapterin; MF, methanofuran.
The Methanosarcineae are unique among the Archaea in forming complex multicellular structures during different phases of growth and in response to environmental change (Fig. 2). Within the Methanosarcineae, a number of distinct morphological forms have been characterized, including single cells with and without a cell envelope, as well as multicellular packets and lamina (Macario and Conway de Macario 2001). Packets and lamina display internal morphological heterogeneity, suggesting the possibility of cellular differentiation. Moreover, it has been suggested that cells within lamina may display differential production of extracellular material, a potential form of cellular specialization (Macario and Conway de Macario 2001). The formation of multicellular structures has been proposed to act as an adaptation to stress and likely plays a role in the ability of Methanosarcina species to colonize diverse environments.
Figure 2.
Different morphological forms of Methanosarcina acetivorans. Thin-section electron micrographs showing M. acetivorans growing as both single cells (center of micrograph) and within multicellular aggregates (top left, bottom right). Cells were harvested during late-exponential growth in medium containing sodium acetate and prepared for electron microscopy as described previously (Sowers and Ferry 1983). Electron micrographs were taken with a JOEL JEM 100B transmission electron microscope.
Significantly, powerful methods for genetic analysis exist for Methanosarcina species. These tools include plasmid shuttle vectors (Metcalf et al. 1997), very high efficiency transformation (Metcalf et al. 1997), random in vivo transposon mutagenesis (Zhang et al. 2000), directed mutagenesis of specific genes (Zhang et al. 2000), multiple selectable markers (Boccazzi et al. 2000), reporter gene fusions (M. Pritchett and W. Metcalf, unpubl.), integration vectors (Conway de Macario et al. 1996), and anaerobic incubators for large-scale growth of methanogens on solid media (Metcalf et al. 1998). Furthermore, and in contrast to other known methanogens, genetic analysis can be used to study the process of methanogenesis: Because Methanosarcina species are able to utilize each of the three known methanogenic pathways, mutants in a single pathway are viable (M. Pritchett and W. Metcalf, unpubl.). The availability of genetic methods allowing immediate exploitation of genomic sequence, coupled with the genetic, physiological, and environmental diversity of M. acetivorans make this species an outstanding model organism for the study of archaeal biology. For these reasons, we set out to study the genome of M. acetivorans.
RESULTS
Genome Structure and Content
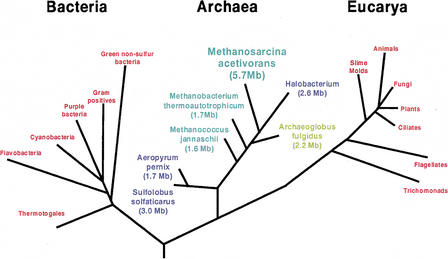
We sequenced M. acetivorans using a combination of whole-genome shotgun and directed finishing (see Methods). The genome consists of a single circular chromosome of 5,751,492 base pairs with 42.7% G + C content (Table 1). M. acetivorans has thus the largest genome known for an archaeon and the fourth largest among sequenced prokaryotes. The M. acetivorans genome is twice as large as those of its phylogenetically nearest sequenced relatives, Archaeoglobus fulgidus and Halobacterium, and over three times as large as the two previously sequenced methanogens, Methanobacterium thermoautotrophicum and Methanococcus jannaschii (Fig. 3).
Table 1.
Methanosarcina acetivorans Genome Features
| General | ||||
| Size | 5,751,492 bp | |||
| % G + C | 42.7% | |||
| Protein-coding genes | 4524 | |||
| RNA genes | 68 | |||
| Pseudogenes | 117 | |||
| Percent coding | 74% | |||
| Average gene size | 936 bp (312 aa) | |||
| Average intergenic space size | 340 bp | |||
| Predicted protein-coding sequences | ||||
| Identified by similarity to known sequences | 2226 (49%) | |||
| Conserved hypothetical proteins | 908 (20%) | |||
| Predicted proteins (no similarity to known sequences) | 1390 (31%) | |||
| Multigene families | ||||
| Number of multigene families | 539 | |||
| Number of genes in multigene families | 2178 (48%) | |||
| Domain comparison | Genes with top BLASTX hit to domain | Genes with BLASTX hits only to domain | ||
| Archaea | 1852 (41%) | 759 (17%) | ||
| Bacteria | 945 (21%) | 387 (9%) | ||
| Eucarya | 98 (2%) | 47 (1%) | ||
| Organism comparison | Genes w/ BLAST hits | Genes w/ top BLASTX hits | Genes w/ BBH hitsa | Conserved clustersb |
| Archaeoglobus fulgidus | 1618 (36%) | 549 (12%) | 967 (21%) | 93 (256 genes) |
| Methanococcus thermoautotrophicum | 1485 (33%) | 377 (8%) | 897 (20%) | 90 (295 genes) |
| Methanococcus jannaschii | 1331 (29%) | 237 (5%) | 822 (18%) | 70 (194 genes) |
| Pyrococcus abyssi | 1193 (26%) | 116 (3%) | 730 (16%) | 58 (151 genes) |
| Pyrococcus horikoshii | 1141 (25%) | 96 (2%) | 690 (15%) | 47 (117 genes) |
| Halobacterium sp NRC-1 | 1067 (24%) | 125 (3%) | 731 (16%) | 65 (195 genes) |
Best bidirectional hit: Defined between two species as a pair of proteins, each of which has its top best BLASTP hit to the other when all proteins from both species are compared to one another.
A cluster in one species was defined as a set of genes with each member in the set no further than 10 genes from at least 1 other gene in the set. A conserved cluster was defined as a cluster in M. acetivorans with each member of the cluster having a best bidirectional hit (BBH) with a member of a single cluster in a second species. Thus, a conserved cluster consists of a set of genes “close” in the M. acetivorans genome whose BBH genes are also “close” in the genome of the second species. The number of genes indicates the number of genes in conserved clusters.
Figure 3.
Universal tree of life based on small-subunit ribosomal (SSU) RNA sequences. Selected sequenced Archaea are highlighted and have their genome sizes indicated. All known methane-producing organisms are members of the domain Archaea (shown in green). The Methanosarcineae are the most metabolically and environmentally diverse methanogens. They are also members of the lineage giving rise to the anaerobic sulfate-reducing Archaeoglobus fulgidus (shown in yellow) and the aerobic halophilic Halobacterium species (shown in blue).
Origin of Replication
A putative origin of replication (selected as base pair 1) was identified using the method of cumulative skew (Grigoriev 1998), as well as colocalization with features identified in archaeal replication origins (Kelman 2000).
Genes
A total of 4524 open reading frames (ORFs) longer than 200 bp were identified as putative protein-coding genes (see Methods). A summary of the characteristics of these genes is shown in Table 1. The average protein-coding region is 936 bp. Predicted coding sequences cover 74% of the genome. This gene density is low compared to other sequenced archaea (for example, it is 92% in A. fulgidus) and is consistent with a general negative correlation between gene density and genome size in sequenced prokaryotic genomes (data not shown). The average length of intergenic regions is 341 bp, nearly three times the average of 122 bp seen in A. fulgidus. Putative functions were assigned to 2226 (49%) of the predicted protein-coding regions based on similarity to identified protein sequences in public databases. A further 908 (20%) coding regions had sequence similarity to hypothetical coding sequences from other species, and 1390 (31%) had no similarity to known proteins. Comparisons of M. acetivorans proteins with those of other fully sequenced genomes (Table 1) indicate that the M. acetivorans proteome is most similar on average to that of A. fulgidus. This is in contrast with small subunit (SSU) rRNA data that places the halophiles phylogenetically closer to the Methanosarcineae. The physiological similarities between the Methanosarcineae and the sulfate-reducing, strictly anaerobic A. fulgidus, as compared to the facultative, phototrophic aerobe Halobacterium, as well as adaptations by Halobacterium to a high salt environment, may explain this observation.
Multigene Families
A considerable portion of the M. acetivorans genome can be accounted for by multigene families. Nearly half of all annotated genes belong to one of 539 multigene families (Table 2), with 98 families containing 315 predicted proteins that appear to be unique to M. acetivorans. Comparisons with related archaeal genomes (see Methods) indicate likely gene family expansion in M. acetivorans. Multigene families in M. acetivorans are 1.3–1.4 times larger on average than related families in A. fulgidus, Halobacterium spp., M. thermoautotrophicum, M. jannaschii, Pyrococcus abysii, and Pyrococcus horikoshii. Of course, family contraction in these related species cannot be ruled out with the available data. In comparison, the number of proteins in M. acetivorans without homology to other known proteins was two to three times larger than in these species. Thus both the differences in gene family size and the greater number of apparently unique genes appear to contribute to the larger genome of M. acetivorans relative to these related archaeal species.
Table 2.
Largest 30 Multigene Families
| Name | Size |
|---|---|
| ABC transporter ATP-binding protein | 85 |
| Surface protein | 62 |
| Sensory transduction histidine kinase | 49 |
| Methyltransferase | 46 |
| Predicted protein | 29 |
| Oligopeptide ABC transporter (permease) | 28 |
| Conserved hypothetical protein | 25 |
| Transposase (Archaeoglobus fulgidus IS0963 family) | 21 |
| Iron ABC transporter, solute-binding protein | 21 |
| Transposase | 20 |
| Oligopeptide ABC transporter, solute-binding protein | 19 |
| Iron-sulfure flavoprotein | 19 |
| Transposase (Bacillus atoarothermophilus ORF A family) | 18 |
| IstB-like ATP-binding protein | 18 |
| Conserved hypothetical protein | 16 |
| Conserved hypothetical protein | 15 |
| Conserved hypothetical protein | 15 |
| MFS transporter family protein | 14 |
| Polysaccharide biosynthesis protein/LPS biosynthesis protein/sugar transferase | 14 |
| Predicted protein | 14 |
| ABC transporter, permease protein | 13 |
| Fe (III) ABC transporter, permease protein | 13 |
| Nitrogenase, subunit | 13 |
| Transposase | 13 |
| Methylamine corrinoid protein | 12 |
| Metallo cofactor biosynthesis protein family | 12 |
| O-linked GlcNAc transferase | 11 |
| Response regulator | 11 |
| Sulfate/molybdate/phosphate ABC transporter, permease protein | 11 |
| Transcription repressor; MarR family protein | 10 |
The largest multigene families include several transport-related families. In all, at least 243 genes were members of transport-related families and ∼160 membrane transport systems were identified. A small number of large families account for a significant proportion of the membrane transporters. For example, 47 ABC transporter genes putatively involved in iron chelate uptake were identified including 17 different binding proteins, more than twice the number of iron ABC transporter genes than in any other sequenced prokaryote (http://www-biology.ucsd.edu/∼ipaulsen/transport/). Although originally isolated in an iron-rich environment, these predicted genes suggest the ability of M. acetivorans to adapt to iron-poor environments, as well as to outcompete other organisms for iron uptake. Both possibilities underscore the adaptability of M. acetivorans.
Examination of the multigene families also highlights the importance of surface structures to M. acetivorans. The second largest multigene family consists of 62 annotated surface proteins. The majority of these genes were predicted not to contain transmembrane domains, suggesting that these proteins are secreted and play a role in generating the cell envelope (or S-layer), as well as an extracellular matrix during the formation of multicellular structures (see below).
Finally, widespread genetic diversity was also observed at the level of cellular systems. For example, M. acetivorans is distinct among all prokaryotes in possessing all of the four major molecular chaperoning systems, including the GroES/L system, which has not been described in any other archaeon. Additional examples of the genetic diversity of M. acetivorans are described below.
Methanogenesis
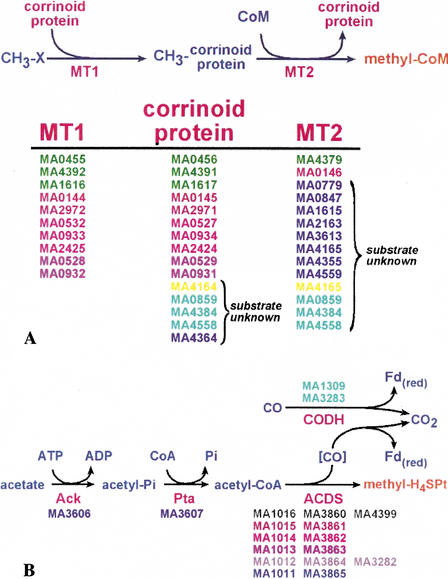
The only known energy-yielding process available to methanogenic organisms is methanogenesis (Fig. 1) and the importance of this metabolic process is reflected in the genome of M. acetivorans. Nearly 200 genes related to methanogenesis were identified. These include genes required for known steps in methanogenesis from methylated compounds such as methanol and methylated amines (the methylotrophic pathway; Fig. 4A) and those required for methanogenesis from acetate (the acetoclastic pathway; Fig. 4B). Many of these genes are present in multiple copies in the M. acetivorans genome. These include two or three copies each of genes encoding components of the methyl transferase systems for methanol, trimethylamine, dimethylamine and monomethylamine, and two copies of a six-gene operon encoding the acetyl-CoA decarbonylase/synthase (ACDS) complex essential for the acetoclastic pathway. The duplication of the ACDS complex in M. acetivorans is particularly striking; the two operons are 95% identical over a 6.6-kbp stretch, including a subsection of nearly 4 kbp with only 6 nucleotide changes.
Figure 4.
Redundant and novel genes involved in methylotrophic and acetoclastic methanogenesis. (A) The pathway for conversion of one-carbon (C-1) compounds to the central-pathway intermediate methyl-CoM (see Fig. 1). Multiple copies of substrate-specific methyltransferases (MT1 and MT2) and methylotrophic corrinoid proteins were identified for each substrate (CH3-X). The gene IDs (MA numbers) for proteins involved in the metabolism of methanol (green) and methylamine (pink); dimethylamine (red) and trimethylamine (dark red) are indicated. Five corrinoid and 12 MT2 homologs without known substrates were identified. Two shown in yellow form a putative operon; three shown in light blue comprise a paralogous family with fused corrinoid and MT2 domains. Additional MT1 homologs were not identified as these proteins do not constitute a homologous family. (B) The pathway for conversion of acetate to the central-pathway intermediate methyl-H4MPT. Two nearly identical copies of genes encoding the acetyl-CoA decarbonylase/synthase (ACDS) complex were identified (homologous genes indicated by identical color, a third lone copy of cdhA [MA4399] was also identified). Two genes (cyan) encoding single-subunit, bacterial-type CO2 dehydrogenase (CODH) proteins were also identified suggesting the possibility of exogenous CO metabolism (see text for details).
Methanogenic Specialization
The biochemical functions of the multiple methanogenic genes provides strong evidence that M. acetivorans specializes in the utilization of acetate and a variety of one-carbon compounds as growth substrates. It is possible that these multiple gene copies provide added flexibility in the utilization of these substrates. The individual copies of the duplicated genes could display differential regulation or encode enzymes with altered kinetic properties, allowing the organism to rapidly change between substrates and to cope with variable substrate concentrations. The emphasis on the utilization of multiple substrates is consistent with the hypothesis that Methanosarcina species succeed in nature by following a generalist strategy (Zinder 1993). This is in contrast to the previously sequenced genomes of M. thermoautotrophicum and M. jannaschii that specialize in the utilization of a single substrate, molecular hydrogen (H2).
Hydrogen Utilization
M. acetivorans differs from many other Methanosarcina species in being incapable of growing methanogenically using H2 to reduce CO2 (the hydrogenotrophic pathway; Sowers et al. 1984). The hydrogenotrophic pathway shares many steps in common with the oxidative branch of the methylotrophic pathway and utilizes the same enzymes for these steps, although the reactions are run in the reductive rather than the oxidative direction (Fig. 1). Electrons required only for the reduction reactions are provided by the oxidation of molecular hydrogen by enzymes known as hydrogenases. These hydrogenases are characteristic enzymes of the hydrogenotrophic pathway and would not be expected to play a role in the oxidative reactions.
Therefore, it was surprising that most of the hydrogenases identified in hydrogenotrophic Methanosarcina species were also present in the M. acetivorans genome. The genes include a homolog of the Methanosarcina barkeri frh operon encoding an F420-reducing hydrogenase (Vaupel and Thauer 1998) and homologs of two Methanosarcina mazei operons encoding F420-nonreducing hydrogenases (in which F420 is a redox coenzyme unique to methanogenesis). The identification of these hydrogenases in M. acetivorans strongly implies that it is capable of metabolizing hydrogen for purposes other than CO2 reduction. Thus, a reassessment of the role of hydrogen metabolism in so-called nonhydrogenotrophic organisms is warranted.
The inability of M. acetivorans to grow methanogenically using H2/CO2 may be explained by the absence of two critical hydrogenases. Neither the F420-reducing hydrogenase encoded by the fre operon (Vaupel and Thauer 1998) nor the energy-conserving ferredoxin-dependent hydrogenase encoded by the ech operon (Kunkel et al. 1998; Meuer et al. 1999) are present in M. acetivorans, whereas both are present in M. barkeri, which is capable of methanogenesis using H2/CO2. The presence of these two genes in hydrogen-utilizing methanogens, and their absence in M. acetivorans, provides compelling evidence for their roles in hydrogenotrophic methanogenesis.
Carbon Monoxide Metabolism
Certain Methanosarcina species have been shown to grow using carbon monoxide (CO) as a methanogenic substrate. It has been hypothesized that oxidation of CO is mediated by the CO dehydrogenase (CODH), now known to be part of the multisubunit ACDS complex (O'Brien et al. 1984), during methanogenic growth on this substrate. Therefore, it was unexpected that M. acetivorans was found to contain two homologs of the single-subunit enzyme utilized by Rhodospirillum rubrum for growth on CO (Fig. 4B). The presence of this single-subunit enzyme indicates a possible alternative mechanism for methanogenic growth on CO. Furthermore, the fact that this enzyme is used by R. rubrum for nonmethanogenic growth on CO raises the surprising possibility that M. acetivorans may grow nonmethanogenically, using CO as a substrate. This would require a major reassessment of the metabolism of methanogens, which heretofore have been thought to rely solely on methanogenesis for energy production.
Undiscovered Growth Substrates
A number of genes were identified indicating the possibility that other, as yet unidentified, growth substrates may be utilized by M. acetivorans. As shown in Figure 4A, methylotrophic methanogenesis begins with the transfer of the methyl group from a variety of substrates to coenzyme M, via two methyl-transferases, designated MT1 and MT2, and a substrate-specific methylotrophic corrinoid protein (containing a modified B12 derivative). Fourteen MT2 and 15 corrinoid protein-encoding genes were identified in the M. acetivorans genome. These include three proteins with both MT2 and corrinoid-binding domains. Two of the MT2 homologs and 10 of the corrinoid proteins are known to be involved in the utilization of either methanol or methylated amines (see Fig. 4A). The remaining homologs have not been attributed to known substrates. Some of these are likely to be involved in the use of methyl-sulfides, substrates known to be used by M. acetivorans. However, no close homologs of the known M. barkeri methyl-sulfide methyltransferase genes (Paul and Krzycki 1996) were identified in M. acetivorans. It is also possible that additional MT1 homologs exist but were not identified, as these proteins do not constitute a homologous family that would allow identification by sequence comparison. Therefore, 12 unaccounted MT2 homologs and five unaccounted corrinoid proteins appear to be encoded by M. acetivorans genome (Fig. 4A), suggesting that additional one-carbon compounds may act as energy-yielding substrates for this organism.
Central Metabolism
Oxygen Metabolism
Methanogenesis is a form of anaerobic respiration using compounds such as CO2 or methanol as terminal electron acceptors (Fig. 1). This is analogous to the use of oxygen as the terminal electron acceptor by mitochondria and many bacteria, or the use of nitrate, sulfate, or iron by various microbes. Despite decades of search, no methanogen has ever been observed to utilize nonmethanogenic electron acceptors to support growth. Moreover, methanogens are exquisitely sensitive to oxygen under laboratory conditions and are rapidly killed by even very low concentrations of O2.
It thus is interesting that a putative operon was identified, designated cydAB, encoding an M. acetivorans homolog of Escherichia coli cytochrome d oxidase. Cytochrome d oxidase is a key step in O2-dependent respiration in E. coli under low O2 conditions (Cotter et al. 1997). It transfers electrons from quinones to O2. In E. coli, the reaction contributes to the production of a membrane electrochemical gradient that can be used for ATP synthesis.
The primary role of cytochrome d oxidase in M. acetivorans is unclear. Cytochrome d oxidase has been suggested to play a role in protection from oxygen toxicity in numerous organisms, presumably by consuming O2 and lowering its concentration (Cotter et al. 1997; Duwat et al. 2001; Endley et al. 2001). Such a protective role thus is possible in M. acetivorans. In addition to cydAB, two superoxide dismutases, one superoxide reductase, and one catalase were identified. Each of these proteins normally functions in protection from reactive oxygen species. Alternatively, cytochrome d oxidase may play a role in energy conservation. In particular, it has been shown that Lactococcus lactis, often considered a strict anaerobe, is capable of O2-dependent respiration using a cytochrome d oxidase homolog (Duwat et al. 2001). The finding of cytochrome d oxidase encoding genes thus suggests the possibility that M. acetivorans may be capable of O2-dependent respiration under appropriate circumstances. This possibility will require careful study to evaluate. In this regard, it is important to note that Methanosarcina species are not known to contain quinones, although genes homologous to those known to be involved in ubiquinone and menaquinone biosynthesis were identified.
Nitrogen Fixation
Methanogens are the only archaea known to fix nitrogen. Nitrogen fixation is mediated by a nitrogenase complex consisting of two metalloproteins, a dinitrogenase, and a dinitrogenase reductase. Three types of nitrogenases are known: a molybdenum- and iron-containing nitrogenase, a vanadium- and iron-containing nitrogenase, and an iron-only-containing nitrogenase. The latter two nitrogenases are termed alternative nitrogenases and have been hypothesized to serve as mechanisms of nitrogen fixation in molybdenum-limiting environments.
Unique among the archaea, M. acetivorans appears to possess all three types of nitrogenases. The molybdenum nitrogenase gene cluster is most similar to that of the gram-positive bacterium Clostridium pasteurianum, whereas the vanadium alternative nitrogenase cluster is similar to that of the cyanobacterium Anabaena variabilis (Thiel 1993). The iron-only nitrogenase cluster lacks a homolog encoding the dinitrogenase reductase; however, a gene just upstream encodes a protein with 91% identity to the dinitrogenase reductase of the vanadium nitrogenase cluster and may function as the dinitrogenase reductase for the iron-only complex. No archaeon outside the genus Methanosarcina is known to contain more than one nitrogenase system. Some bacteria are known to possess three sets of nitrogenase genes, including Azotobacter vinlandii (Bishop and Premakumar 1992), an organism considered highly adapted to nitrogen fixation. The presence of all three nitrogenases in M. acetivorans suggests the importance of nitrogen fixation to M. acetivorans and again underscores the adaptability of this organism.
Information Processing
Transcription and Gene Regulation
Transcriptional regulation within archaea in general, and within the methanogens in particular, is not fully understood. The minimal transcriptional apparatus for most archaeal promoters consists of a TATA-box-binding protein (TBP) and an archaeal transcription factor B (TFB), complexed with an archaeal RNA polymerase (RNAP; Bell and Jackson 2001). Interestingly, M. acetivorans appears to code for three substantially different TBPs, in addition to a single TFB. Although multiple TBP and TFB have been described in several archaea (Kawarabayasi et al. 1998; Thompson et al. 1999; Ng et al. 2000), M. acetivorans (together with M. barkeri; E.Conway de Macario, unpubl.) is the only known methanogen with multiple identified TBPs. The presence of multiple TBPs raises the possibility of differential gene expression through the formation of alternative TBP–TFB–RNA polymerase complexes as previously suggested for Halobacterium species (Baliga et al. 2000). However, all other archaea with multiple TBPs also contain multiple TFBs. The presence of only one TFB in M. acetivorans, along with available genetic methods, offers the capability to investigate the role of multiple TBPs independently of the potential role of multiple TFBs.
Several additional transcription factors were identified, including a single archaeal transcription factor E (TFE), a single archaeal transcription factor S (TFS), and numerous bacterial-like regulators. Comparisons with related sequenced archaea indicate a strikingly different profile for bacterial-like regulators in M. acetivorans. For example, M. acetivorans possesses a family of 15 tetR regulator proteins and a family of 11 marR repressor proteins, whereas no other sequenced archaeon possesses more than 3 members of either family, and A. fulgidus contains only one of each. In contrast, only a single Lrp regulatory protein was identified in M. acetivorans, whereas 14 were identified in A. fulgidus.
Translation
M. acetivorans was found to contain both a class I and a class II lysyl-tRNA synthetase. The class I lysyl-tRNA synthetase is found in many archaea and some bacteria, and the class II synthetase is found in most bacteria and all eukarya (Soll et al. 2000). M. acetivorans (together with M. barkeri; G. Srinivasa, G. Paul, and J. Krzycki, unpubl.) represents the first instance of any organism in any domain possessing both classes.
We speculate that the possession of both classes of lysyl-tRNA synthetase may relate to the presence of read-through amber codons (UAG codons not read as a translation stop). It has been noted in methanogens that all known methylamine methyltransferase genes contain a single conserved in-frame read-through amber codon (Paul et al. 2000), and studies have shown that this codon corresponds to a lysine residue in the product of one of these genes (James et al. 2001). Recent studies have also identified a putative amber decoding tRNA (J. Krzycki, unpubl.). This raises the possibility of conserved amber read-through codons in other gene families. In fact, a preliminary analysis identified a family of four putative transposase genes each containing an in-frame amber codon at an identical position. In addition, a methlycobamide:CoM methylase of unknown function was also identified with an amber codon. Furthermore, use of UAG as a stop codon appears depressed in M. acetivorans as compared with related species, supporting the hypothesis of an alternative role for this codon. For example, only 5% of coding regions in M. acetivorans end with UAG in comparison to 19% in A. fulgidus. Further analysis is required to determine the extent of the read-through amber-codon phenomenon.
Multicellular Structures
Methanosarcineae are the only archaea known to undergo major morphological changes involving the formation of distinct multicellular structures (Fig. 2; Macario and Conway de Macario 2001). Thus, M. acetivorans provides a model for dissecting the mechanisms regulating multicellularity in this domain of life. M. acetivorans synthesizes a cell envelope, termed an S-layer, which consists of protein subunits adjacent to the cell membrane (Sowers et al. 1984a ; Kandler and Konig 1993). Under certain conditions, the organism also grows as multiple cells, each with an S-layer, contained within a common sacculus-like structure. Furthermore, M. acetivorans is capable of growing as multicellular bodies embedded within a complex extracellular matrix. The importance of the formation of multicellular structures is underscored by the number and diversity of proteins potentially associated with this process.
Polysaccharide Biosynthesis
Over 30 predicted gene products were identified having sequence similarity to polysaccharide biosynthetic enzymes. Many occurred in clusters of adjacent genes. One cluster, characteristic of a typical bacterial capsular biosynthetic operon, includes a dolichyl-phosphate glycosyl transferase, several additional glycosyltransferases, and a gene predicted to belong to a family of polysaccharide extrusion systems. Several other genes have high sequence identity to other hypothetical biosynthetic genes in both S-layer-producing species and pseudo-murein-producing species. This suggests that these genes code for synthesis of extracellular polysaccharides such as lipopolysaccharides, S-layer glycoproteins, and methanochondroitin because pathways for the biosynthesis of all three involve sequential addition of UDP-PP-monosaccharides to a glycan chain (Hartmann and Konig 1989; Kandler and Konig 1998).
Surface Proteins
M. acetivorans contains a large number of surface proteins with a diverse array of domains and architectures (Fig. 5). For instance, the second largest multigene family (Table 2) consists of 62 proteins, the majority of which lack transmembrane domains and are homologous to surface antigens in M. mazei and thus are likely components of the S-layer (Mayerhofer et al. 1995). In addition, many genes in this family are large multidomain proteins containing domains homologous to metazoan cell adhesion surface receptor protein domains, including polycystic kidney disease (PKD) domains (or C repeats in M. mazei [Mayerhofer et al. 1995]) and YVTN β-propeller domains (or AB repeats in M. mazei [Mayerhofer et al. 1995]; Fig 5; H. Jing, in prep.). This suggests an additional regulatory or structural role in the formation of M. acetivorans multicellular structures. Consistent with the unique ability to form multicellular structures, the fraction of total proteins that are S-layer proteins and the fraction of proteins that contain PKD domains or YVTN β-propeller domains are strikingly larger in M. acetivorans than in other sequenced archaea. For example, A. fulgidus possesses only two proteins containing PKD domains. Furthermore, the number and diversity of surface proteins provides a mechanism for generating variations in surface structures, consistent with the antigenic variation observed in M. mazei (Yao et al. 1992).
Figure 5.
Diversity of surface proteins. Consistent with the unique ability of M. acetivorans among the archaea to form complex multicellular structures, a large number and diverse array of surface proteins were identified. Illustrated are examples of domain architectures that contain β-propeller, polycystic kidney disease (PKD), and β-helix domains. The PKD and YVTN β-domains share sequence similarity to metazoan cell-adhesion molecules, suggesting a role for these proteins in the formation of multicellular structures. The domains were identified by similarity to structurally defined domains in M. mazei surface antigens (H. Jing, in prep.). The helical transmembrane regions (HTM) were predicted using PHDhtm (Rost et al. 1996). The Ca2+-binding β-hairpin motifs were predicted based on sequence similarity with a previously identified Ca2+-binding motif (Springer et al. 2000). One β-propeller domain appears intermediate between YVTN and WD40 β-propellers.
Environmental Sensing and Interaction
Signal Transduction
The genome of M. acetivorans indicates a wide repertoire of behavioral responses to external stimuli via two-component regulatory systems. The basic two-component system, as characterized in bacteria, consists of a sensory transduction histidine kinase and a cognate response regulator protein. Typically these two components are paired in a one-to-one ratio. Activation of the sensory transduction histidine kinase causes autophosphorylation and subsequent phosphoryl transfer to the receiver domain of the response regulator. Most bacterial response regulators include an additional effector domain responsible for the final cellular output.
In contrast, the genome of M. acetivorans indicates a different approach to two-component signal transduction. The sensory transduction histidine kinases comprise the third largest multigene family in M. acetivorans with 50 representatives (not including 2 cheA genes involved in chemotaxis signaling). The majority of these contain one or more PAS domains (Ponting and Aravind 1997; Pellequer et al. 1998), predicted to be small-molecule-binding domains, although the exact nature of the small molecules they may bind cannot be determined by similarity alone. In contrast, the genome contains only 18 response regulator receiver domains (excluding 2 cheY and 2 cheB genes). Seven of these occurrences are as domains within histidine kinases themselves. Of the remaining 11 occurrences, 10 are in small single-domain proteins containing only the response regulator receiver domain itself, whereas only a single protein is of sufficient length to contain a bacterial-like effector domain.
The differences in the nature of two-component signaling in M. acetivorans, relative to bacteria, led us to examine the genomes of other archaea. A departure from the one-to-one relationship between kinases and regulators has been observed in other archaea, and this appears to be the case for all archaea with annotated two-component systems. In addition, most annotated response regulators in archaea appear to be single-domain proteins. These observations imply a different mode of action for two-component signal-transduction systems in archaea in comparison to well-characterized bacterial species such as E. coli. This implication, to our knowledge, has not been previously noted. One possibility is a very high degree of signal integration at the response regulator level with, on average, five histidine kinases per autonomous receiver domain. Alternatively, the kinases may phosphorylate other substrates in addition to recognizable receiver domains. In addition, receiver-domain peptides may interact with as yet uncharacterized effector domains on separate peptides.
Flagella and Motility
Although motility has not been observed in any Methanosarcineae, genes encoding products for both flagella and chemotaxis are present. M. acetivorans contains a single flagellin cluster organized in a manner similar to that in A. fulgidus. This cluster includes two flagellin genes followed by three genes with conserved sequences and appearing in the same order as in A. fulgidus, followed by flagella accessory genes H, I, and J. The flagellin genes appear more similar to those of A. fulgidus than those from methanococci. Intergenic spacing suggests that all of these genes could be cotranscribed, in keeping with observations in methanococci (Kalmokoff and Jarrell 1991), extreme halophiles (Patenge et al. 2001), and Pyrococcus (Nagahisa et al. 1999).
M. acetivorans also possesses two very similar clusters of chemotaxis genes, che-1 and che-2, both of which comprise an apparently complete chemotactic system. The che-1 cluster lies adjacent to the flagellin cluster and consists of homologs of cheR, cheD, cheC, cheA, cheB, cheY, cheW, and a methyl-accepting chemotaxis protein. The che-2 cluster contains homologs of cheD, cheC, cheR, cheA, cheB, cheY, a methyl-accepting chemotaxis protein, and cheW. This cluster also contains a single-domain response regulator.
Despite the presence of flagellin and chemotaxis genes, we were unable to detect processing of preflagellin in a preflagellin peptidase assay using Methanococcus voltae preflagellin and membranes obtained from M. acetivorans growing as single cells (Correia and Jarrell 2000). Thus, it seems likely that flagella and chemotaxis genes are expressed in specific environmental conditions not previously created in culture (as has been seen for other organisms [Faguy et al. 1993; Mukhopadhyay et al. 2000]), possibly in conjunction with changes in morphology.
DISCUSSION
Methanosarcineae are unsurpassed among methanogens in terms of metabolic, physiological, and environmental versatility and are unique among archaea in forming complex multicellular structures. These capabilities are reflected in the genome of M. acetivorans, the largest known for an archaeon. The genome reveals extensive genetic diversity and redundancy underlying the ability of M. acetivorans to adapt to varied environmental conditions. In addition, the genome sequence indicates the potential for entirely unexpected metabolic capabilities. These include the possibilities of methanogenesis from as yet undiscovered substrates, nonmethanogenic growth-utilizing CO, and perhaps even O2-dependent respiration.
The powerful genetic methods available for M. acetivorans allow the immediate exploitation of the complete genome sequence for understanding the organism's biochemical and environmental diversity and the regulatory mechanisms that underlie its adaptive abilities. In addition, these methods permit functional studies of the over 1300 predicted proteins lacking homology to known proteins. Moreover, the existence of ongoing sequencing efforts for M. mazei (Göttingen Genomics Laboratory at http://www.g2l.bio.uni-goettingen.de/ ), Methanosarcina thermophila, and Methanosarcina barkeri (DOE Joint Genome Institute [JGI] at http://www.jgi.doe.gov/JGI_microbial/html/ ) promises unparalleled opportunity for comparative studies. Such studies offer the potential for elucidating the evolutionary basis of the extraordinary genetic diversity of the Methanosarcineae, leading in turn to a greater understanding of the archaeal domain of life.
METHODS
Growth Conditions
Methanosarcina strains were grown in single-cell morphology (Sowers et al. 1993) at 35°C in HS broth medium containing 125 mM methanol plus 40 mM sodium acetate (HS-MA medium) (Metcalf et al. 1996).
Sequencing, Assembly, and Finishing
The genome was sequenced by the whole-genome shotgun method. Genomic DNA was isolated from M. acetivorans as described by Boccazzi et al. (2000). m13 (1.5-kb inserts) and plasmid (4-kb inserts) libraries were generated as described at http://www-genome.wi.mit.edu/. Methanosarcina Fosmid (40-kb inserts) library construction is described elsewhere (Zhang et al., 2002).
Sequencing methods are described at http://www-genome.wi.mit.edu/. Plasmid and Fosmid inserts were sequenced from both ends to generate paired-reads. We generated sequence coverage of 7× from plasmids, 1× from M13 and 0.076× from Fosmids and assembled it with Phrap. Initial analysis of the assembly was done with the Mapper software (M.C. Zody, pers. comm.) to select gap-spanning clones for finishing. Two hundred gaps spanned by plasmid clones were closed by transposon-based sequencing using the EZ::TN 〈KAN-2〉 kit from Epicentre. Forty-eight gaps spanned only by Fosmids were closed by sequencing Fosmid-derived PCR products. Sequence from 28 unspanned gaps was obtained from fragments generated by combinatorial PCR using genomic DNA as template and pooled primers (Tettelin et al. 1999). One unspanned gap was closed by sequencing a small-insert library (McMurray et al. 1998) produced from an 8.5-kb PCR product. Regions of low sequence quality were resolved by (1) use of ABI dGTP Big Dye Terminator sequencing mix, (2) transposon-primed sequencing of plasmid clones, or (3) sequencing PCR products obtained from plasmid or genomic template. Paired-reads within the assembly were visualized with the Mapper software and used in assembly validation. Regions of the assembly spanned by paired-reads occurring with the appropriate orientation and spacing were considered valid. Validation of 99.99% of the genome was performed in this manner, whereas only 6270 bases of the finished assembly were spanned soley by sequenced PCR products. Regions of the assembly containing nonsensical paired-reads were analyzed further. Eighteen of these regions proved to have been misassembled by Phrap and were resolved manually. The finished genome sequence was manually inspected for quality and edited using the Staden package viewer Gap4. During annotation 480 possible sequence errors (based on breaks in ORFs) were identified. These were manually reviewed and one was shown to be an editing error and was corrected. Five of the possible errors were not unambiguous by quality but after resequencing were shown to have been correct.
Annotation and Analysis
The M. acetivorans genome was annotated using the Calhoun annotation system (J. Galagan, in prep.). ORFs likely to encode proteins were predicted using GLIMMER (Delcher et al. 1999), and all ORFs were searched against two sets of protein family Hidden Markov Models (HMM), Pfam (Bateman et al. 2000) and TIGRFAM (Haft et al. 2001), using the HMMER program (Durbin et al. 1998). The entire genome was also searched against the public protein databases using BLASTX with threshold E ≤ 1e-5. ORFs identified by GLIMMER >200 bp in length and all ORFS with similarity to a protein family HMM or known proteins were annotated as genes. Genes were assigned identities by a team of seven annotators and three annotation reviewers. For genes with similarities to known proteins, the literature was consulted to identify those proteins experimentally characterized, and this information was captured by the Calhoun system. To supplement and verify the Calhoun automated annotation process, the genome sequence was applied to the annotation pipeline associated with the Comprehensive Microbial Resource (Peterson et al. 2001). Comparisons of results from both systems displayed no substantial inconsistencies.
Multigene families were constructed by searching each annotated gene against all other genes using BLASTP, requiring matches with E ≤ 1e-5 over 60% of the longer gene length, and subsequently clustering genes with matches. An identical analysis was performed on 14 other archaeal and bacterial species. Corresponding families in each of these species were based on using best bidirectional BLASTP hits (E ≤ 1e-5, score > 10, hit length ≧ 60%) between family members. Transfer RNAs were identified using the tRNAScan-SE program (version X). The origin of replication was identified as the point of maximum cumulative AT skew (defined as the cumulative sum of A – T/A + T on one strand).
Additional annotation and extensive genome analysis were performed as part of the Methanosarcina acetivorans Community Annotation Project. Over 30 members of the scientific community participated in the project. Gene annotations were submitted via a web interface and detailed analyses of the genome annotation performed. The project culminated in a two-day Genome Analysis Meeting at the Whitehead Institute Center for Genome Research.
Additional details, analysis results, and the genome sequence are available at http://www-genome.wi.mit.edu/ and at http://www.tigr.org/CMR.
WEB SITE REFERENCES
http://www-biology.ucsd.edu/∼ipaulsen/transport/
http://www.bioscience.org/2001/v6/d/macario/fulltext.htm.
http://www.g2l.bio.uni-goettingen.de/; Genomics laboratory site.
http://www-genome.wi.mit.edu/; Sequence, data, annotations, and analyses are available at this site.
http://www.jgi.doe.gov/JGI_microbial/html/; DOE Joint Genome Institute [JGI].
Acknowledgments
We gratefully acknowledge B. Whitman, R. Barrett, S. Talon, D. Gregoire, H. Harris, J.P. Leger, C. Davis, S. Diaz, E. Tsao, C. Urbytes, M. Modeski, C. Lopez, K.M. Peterson, R. Milbert, J. Klein, C. Neilson, S. Diaz-Perez, S. Maloy, S. Kasif, C. Woese, M.C. Zody, and all members of the sequencing group at the Whitehead Institute Center for Genome Research for their contributions to the project. Preliminary sequence data for M. barkeri was obtained from the DOE Joint Genome Institute (JGI) at http://www.jgi.doe.gov/JGI_microbial/html/. This work was supported in part by grant MCB-987459 from the National Science Foundation and by a Searle Scholars Award from the Chicago Community Trust to W.W.M. and with support from the Whitehead Institute for Biomedical Research and the NIH (E.S.L.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL bwb@genome.wi.mit.edu; FAX (617) 258-0903.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.223902. Article published online before print in March 2002.
REFERENCES
- Baliga NS, Goo YA, Ng WV, Hood L, Daniels CJ, DasSarma S. Is gene expression in Halobacterium NRC-1 regulated by multiple TBP and TFB transcription factors? Mol Microbiol. 2000;36:1184–1185. doi: 10.1046/j.1365-2958.2000.01916.x. [DOI] [PubMed] [Google Scholar]
- Bateman A, Birney E, Durbin R, Eddy SR, Howe KL, Sonnhammer EL. The Pfam protein families database. Nucleic Acids Res. 2000;28:263–266. doi: 10.1093/nar/28.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell SD, Jackson SP. Mechanism and regulation of transcription in archaea. Curr Opin Microbiol. 2001;4:208–213. doi: 10.1016/s1369-5274(00)00190-9. [DOI] [PubMed] [Google Scholar]
- Bishop PE, Premakumar R. Biochemical genetics of nitrogenase. In: Stacey G, et al., editors. Biological Nitrogen Fixation. New York: Chapman and Hall; 1992. pp. 736–762. [Google Scholar]
- Boccazzi P, Zhang JK, Metcalf WW. Generation of dominant selectable markers for resistance to pseudomonic acid by cloning and mutagenesis of the ileS gene from the archaeon Methanosarcina barkeri fusaro. J Bacteriol. 2000;182:2611–2618. doi: 10.1128/jb.182.9.2611-2618.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway de Macario E, Guerrini M, Dugan CB, Macario AJL. Integration of foreign DNA in an intergenic region of the archaeon Methanosarcina mazei without effect on transcription of adjacent genes. J Mol Biol. 1996;262:12–20. doi: 10.1006/jmbi.1996.0494. [DOI] [PubMed] [Google Scholar]
- Correia JD, Jarrell KF. Posttranslational processing of Methanococcus voltae preflagellin by preflagellin peptidases of M. voltae and other methanogens. J Bacteriol. 2000;182:855–858. doi: 10.1128/jb.182.3.855-858.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter PA, Melville SB, Albrecht JA, Gunsalus RP. Aerobic regulation of cytochrome d oxidase (cydAB) operon expression in Escherichia coli: Roles of Fnr and ArcA in repression and activation. Mol Microbiol. 1997;25:605–615. doi: 10.1046/j.1365-2958.1997.5031860.x. [DOI] [PubMed] [Google Scholar]
- Delcher AL, Harmon D, Kasif S, White O, Salzberg SL. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 1999;27:4636–4641. doi: 10.1093/nar/27.23.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DOE Joint Genome Institute at http://www.jgi.doe.gov/JGI_microbial/html/.
- Durbin R, Eddy S, Krogh A, Mitchison G. Biological sequence analysis: Probabilistic models of proteins and nucleic acids. Cambridge, UK: Cambridge University Press; 1998. [Google Scholar]
- Duwat P, Sourice S, Cesselin B, Lamberet G, Vido K, Gaudu P, Le Loir Y, Violet F, Loubiere P, Gruss A. Respiration capacity of the fermenting bacterium Lactococcus lactis and its positive effects on growth and survival. J Bacteriol. 2001;183:4509–4516. doi: 10.1128/JB.183.15.4509-4516.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endley S, McMurray D, Ficht TA. Interruption of the cydB locus in Brucella abortus attenuates intracellular survival and virulence in the mouse model of infection. J Bacteriol. 2001;183:2454–2462. doi: 10.1128/JB.183.8.2454-2462.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faguy D, Koval SF, Jarrell KF. Effect of changes in mineral composition and growth temperature on filament length and flagellation in the archaeon Methanospirillum hungatei. Arch Microbiol. 1993;159:512–520. [Google Scholar]
- Göttingen Genomics Laboratory at http://www.g2l.bio.uni-goettingen.de/.
- Grigoriev A. Analyzing genomes with cumulative skew diagrams. Nucleic Acids Res. 1998;26:2286–2290. doi: 10.1093/nar/26.10.2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haft DH, Loftus BJ, Richardson DL, Yang F, Eisen JA, Paulsen IT, White O. TIGRFAMs: A protein family resource for the functional identification of proteins. Nucleic Acids Res. 2001;29:41–43. doi: 10.1093/nar/29.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann E, Konig H. Uridine and dolichyl diphosphate activated oligosaccharides are intermediates in the biosynthesis of the S-layer glycoprotein of Metanothermus fervidus. Arch Microbiol. 1989;151:274. [Google Scholar]
- James CM, Ferguson TK, Leykam JF, Krzycki JA. The amber codon in the gene encoding the monomethylamine methyltransferase isolated from Methanosarcina barkeri is translated as a sense codon. J Biol Chem. 2001;276:34252–34258. doi: 10.1074/jbc.M102929200. [DOI] [PubMed] [Google Scholar]
- Kalmokoff ML, Jarrell KF. Cloning and sequencing of a multigene family encoding the flagellins of Methanococcus voltae. J Bacteriol. 1991;173:7113–7125. doi: 10.1128/jb.173.22.7113-7125.1991. . Cell envelopes of Archaea: structure and chemistry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandler O, Konig H. Cell envelopes of Archaea: Structure and chemistry. In: Kates M, et al., editors. The biochemistry of Archaea. Amsterdam: Elsevier; 1993. pp. 223–259. [Google Scholar]
- ————— Cell wall polymers in Archaea (Archaebacteria) Cell Mol Life Sci. 1998;54:305–308. doi: 10.1007/s000180050156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawarabayasi Y, Sawada M, Horikawa H, Haikawa Y, Hino Y, Yamamoto S, Sekine M, Baba S, Kosugi H, Hosoyama A, et al. Complete sequence and gene organization of the genome of a hyper-thermophilic archaebacterium, Pyrococcus horikoshii OT3. DNA Res. 1998;5:55–76. doi: 10.1093/dnares/5.2.55. [DOI] [PubMed] [Google Scholar]
- Kelman Z. The replication origin of archaea is finally revealed. Trends Biochem Sci. 2000;25:521–523. doi: 10.1016/s0968-0004(00)01687-x. [DOI] [PubMed] [Google Scholar]
- Kunkel A, Vorholt JA, Thauer RK, Hedderich R. An Escherichia coli hydrogenase-3-type hydrogenase in methanogenic archaea. Eur J Biochem. 1998;252:467–476. doi: 10.1046/j.1432-1327.1998.2520467.x. [DOI] [PubMed] [Google Scholar]
- Macario AJL, Conway de Macario E. The molecular chaperone system and other anti-stress mechanisms in archaea. Frontiers Biosc. 2001;6:d262–283. doi: 10.2741/macario. http://www.bioscience.org/2001/v6/d/macario/fulltext.htm ; http://www.bioscience.org/2001/v6/d/macario/fulltext.htm. . [DOI] [PubMed] [Google Scholar]
- Mayerhofer LE, Conway de Macario E, Macario AJL. Conservation and variability in Archaea: Protein antigens with tandem repeats encoded by a cluster of genes with common motifs in Methanosarcina mazei S-6. Gene. 1995;165:87–91. doi: 10.1016/0378-1119(95)00524-a. [DOI] [PubMed] [Google Scholar]
- McMurray AA, Sulston JE, Quail MA. Short-insert libraries as a method of problem solving in genome sequencing. Genome Res. 1998;8:562–566. doi: 10.1101/gr.8.5.562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalf WW, Zhang JK, Shi X, Wolfe RS. Molecular, genetic, and biochemical characterization of the serC gene of Methanosarcina barkeri Fusaro. J Bacteriol. 1996;178:5797–5802. doi: 10.1128/jb.178.19.5797-5802.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalf WW, Zhang JK, Apolinario E, Sowers KR, Wolfe RS. A genetic system for Archaea of the genus Methanosarcina: Liposome-mediated transformation and construction of shuttle vectors. Proc Natl Acad Sci. 1997;94:2626–2631. doi: 10.1073/pnas.94.6.2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalf WW, Zhang JK, Wolfe RS. An anaerobic, intrachamber incubator for growth of Methanosarcina spp. on methanol-containing solid media. Appl Environ Microbiol. 1998;64:768–770. doi: 10.1128/aem.64.2.768-770.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuer J, Bartoschek S, Koch J, Kunkel A, Hedderich R. Purification and catalytic properties of Ech hydrogenase from Methanosarcina barkeri. Eur J Biochem. 1999;265:325–335. doi: 10.1046/j.1432-1327.1999.00738.x. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay B, Johnson EF, Wolfe RS. A novel pH2 control on the expression of flagella in the hyperthermophilic strictly hydrogenotrophic methanarchaeaon Methanococcus jannaschii. Proc Natl Acad Sci. 2000;97:11522–11527. doi: 10.1073/pnas.97.21.11522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagahisa K, Ezaki S, Fujiwara S, Imanaka T, Takagi M. Sequence and transcriptional studies of five clustered flagellin genes from hyperthermophilic archaeon Pyrococcus kodakaraensis KOD1. FEMS Microbiol Lett. 1999;178:183–190. doi: 10.1111/j.1574-6968.1999.tb13776.x. [DOI] [PubMed] [Google Scholar]
- Ng WV, Kennedy SP, Mahairas GG, Berquist B, Pan M, Shukla HD, Lasky SR, Baliga NS, Thorsson V, Sbrogna J, et al. Genome sequence of Halobacterium species NRC-1. Proc Natl Acad Sci. 2000;97:12176–12181. doi: 10.1073/pnas.190337797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien JM, Wolkin RH, Moench TT, Morgan JB, Zeikus JG. Association of hydrogen metabolism with unitrophic or mixotrophic growth of Methanosarcina barkeri on carbon monoxide. J Bacteriol. 1984;158:373–375. doi: 10.1128/jb.158.1.373-375.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patenge N, Berendes A, Englehardt SC, Schuster SC, Oesterhelt D. The fla gene cluster is involved in biogenesis of flagella in Halobacterium salinarum. Mol Microbiol. 2001;41:653–663. doi: 10.1046/j.1365-2958.2001.02542.x. [DOI] [PubMed] [Google Scholar]
- Paul L, Krzycki JA. Sequence and transcript analysis of a novel Methanosarcina barkeri methyltransferase II homolog and its associated corrinoid protein homologous to methionine synthase. J Bacteriol. 1996;178:6599–6607. doi: 10.1128/jb.178.22.6599-6607.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul L, Ferguson DJ, Jr, Krzycki JA. The trimethylamine methyltransferase gene and multiple dimethylamine methyltransferase genes of Methanosarcina barkeri contain in-frame and read-through amber codons. J Bacteriol. 2000;182:2520–2529. doi: 10.1128/jb.182.9.2520-2529.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellequer JL, Wager-Smith KA, Kay SA, Getzoff ED. Photoactive yellow protein: A structural prototype for the three-dimensional fold of the PAS domain superfamily. Proc Natl Acad Sci. 1998;95:5884–5890. doi: 10.1073/pnas.95.11.5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson JD, Umayam LA, Dickinson T, Hickey EK, White O. The comprehensive microbial resource. Nucleic Acids Res. 2001;29:123–125. doi: 10.1093/nar/29.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting CP, Aravind L. PAS: A multifunctional domain family comes to light. Curr Biol. 1997;7:R674–677. doi: 10.1016/s0960-9822(06)00352-6. [DOI] [PubMed] [Google Scholar]
- Rost B, Fariselli P, Casadio R. Topology prediction for helical transmembrane proteins at 86% accuracy. Protein Sci. 1996;5:1704–1718. doi: 10.1002/pro.5560050824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlesinger WH. Biogeochemistry: An analysis of global change. San Diego, CA: Academic Press; 1997. [Google Scholar]
- Soll D, Becker HD, Plateau P, Blanquet S, Ibba M. Context-dependent anticodon recognition by class I lysyl-tRNA synthetases. Proc Natl Acad Sci. 2000;97:14224–14228. doi: 10.1073/pnas.97.26.14224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowers KR, Ferry JG. Isolation and characterization of a methylotrophic marine methanogen, Methanococcoides methylutens. Appl Environ Microbiol. 1983;45:684–690. doi: 10.1128/aem.45.2.684-690.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowers KR, Baron SF, Ferry JG. Methanosarcina acetivorans sp. nov., an acetotrophic methane-producing bacerium isolated from marine sediments. Appl Environ Microbiol. 1984;47:971–978. doi: 10.1128/aem.47.5.971-978.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowers KR, Boone J, Gunsalus RP. Disaggregation of Methanosarcina spp. and growth as single cells at elevated osmolarity. Appl Environ Microbiol. 1993;59:3832–3839. doi: 10.1128/aem.59.11.3832-3839.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer TA, Jing H, Takagi J. A novel Ca2+ binding β hairpin loop better resembles integrin sequence motifs than the EF hand. Cell. 2000;102:275–277. doi: 10.1016/s0092-8674(00)00033-7. [DOI] [PubMed] [Google Scholar]
- Tettelin H, Radune D, Kasif S, Khouri H, Salzberg SL. Optimized multiplex PCR: Efficiently closing a whole-genome shotgun sequencing project. Genomics. 1999;62:500–507. doi: 10.1006/geno.1999.6048. [DOI] [PubMed] [Google Scholar]
- Thiel T. Characterization of genes for an alternative nitrogenase in the cyanobacterium Anabaena variabilis. J Bacteriol. 1993;175:6276–6286. doi: 10.1128/jb.175.19.6276-6286.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson DK, Palmer JR, Daniels CJ. Expression and heat-responsive regulation of a TFIIB homologue from the archaeon Haloferax volcanii. Mol Microbiol. 1999;33:1081–1092. doi: 10.1046/j.1365-2958.1999.01551.x. [DOI] [PubMed] [Google Scholar]
- Vaupel M, Thauer RK. Two F420-reducing hydrogenases in methanosarcina barkeri. Arch Microbiol. 1998;169:201–205. doi: 10.1007/s002030050561. [DOI] [PubMed] [Google Scholar]
- Yao R, Macario AJL, Conway de Macario E. Immunochemical differences among Methanosarcina mazei S-6 morphologic forms. J Bacteriol. 1992;174:4683–4688. doi: 10.1128/jb.174.14.4683-4688.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JK, Pritchett MA, Lampe DJ, Robertson HM, Metcalf WW. In vivo transposon mutagenesis of the methanogenic archaeon Methanosarcina acetivorans C2A using a modified version of the insect mariner-family transposable element Himar1. Proc Natl Acad Sci. 2000;97:9665–9670. doi: 10.1073/pnas.160272597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JK, White AK, Kuettner HC, Boccazzi P, Metcalf WW. Directed mutagenesis and plasmid-based complementation in the methanogenic archaeon Methanosarcina acetivorans C2A demonstrated by genetic analysis of proline biosynthesis. J Bacteriol. 2002;184:1449–1454. doi: 10.1128/JB.184.5.1449-1454.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinder SH. Physiological ecology of methanogens. In: Ferry JG, editor. Methanogenesis. Routledge, New York: Chapman Hall; 1993. pp. 128–206. [Google Scholar]