Abstract
Pax6 is a critical transcription factor in the development of the eye, pancreas, and central nervous system. It is composed of two DNA-binding domains, the paired domain (PD), which has two helix–turn–helix (HTH) motifs, and the homeodomain (HD), made up from another HTH motif. Each HTH motif can bind to DNA separately or in combination with the others. We identified three novel binding sites that are specific for the PD and HD domains of human PAX6 from single-copy human genomic DNA libraries using cyclic amplification of protein binding sequences (CAPBS) and electrophoretic mobility shift assays (EMSAs). One of the binding sites was found within sequences of repetitive Alu elements. However, most of the Alu sequences were unable to bind to PAX6 because of a small number of mismatches (mostly in CpG dinucleotide hot spots) in the consensus Alu sequences. PAX6 binding Alu elements are found primarily in old and intermediate-aged Alu subfamilies. These data along with our previously identified B1-type Pax6 binding site showed that evolutionarily conserved Pax6 has target sites that are disparate in primates and rodents. This difference indicates that human and mouse Pax6-regulated gene networks may have evolved through these lineage-specific repeat elements.
[The sequence data from this study have been submitted to GenBank under accession no. AF451322. The following individual kindly provided reagents, samples, or unpublished information as indicated in this paper: L. Yu.]
Pax6 is an important regulator of transcription in the development of the eye and central nervous system in vertebrates and invertebrates (Quiring et al. 1994). The protein sequence and function of Pax6 are evolutionarily conserved so that murine and human Pax6 proteins are identical. Even human and zebrafish Pax6 proteins share a 97% homology. With few exceptions, Pax6 is expressed during central nervous system development and during fundamental sensory processes, particularly of the photoreceptive organ (Callaerts et al. 1997). The expression of Pax6 in adult mammals is restricted to the eye, brain, and pancreas (Mansouri et al. 1996; Sander et al. 1997). Mutations that reduce or abnormally increase functional Pax6 level lead to eye and brain malformations in humans and mice (Aalfs et al. 1997; Engelkamp et al. 1999; Prosser and van Heyningen 1999; Fukuda et al. 2000; Sisodiya et al. 2001).
Pax6 is composed of a C-terminal transactivation domain and two N-terminal DNA-binding domains, a paired domain (PD), and a paired-type of homeodomain (HD). The PD, which characterizes Pax protein, is a bipartite DNA-binding domain that contains two helix–turn–helix (HTH) motifs. Each of these two subdomains, PAI and RED, binds to DNA, and of them the PAI subdomain is the most essential (Xu et al. 1995; Jun and Desplan 1996). A spicing variant, Pax6–(5a), which is expressed at a lower level than Pax6, has an additional 14 amino acids in its PAI subdomain, resulting in DNA-specific binding involving the RED subdomain (Epstein et al. 1994a; Kozmik et al. 1997). It has been demonstrated that the PD and HD can function independently and together to regulate different genes (Sheng et al. 1997). In contrast to HDs, which recognize a short but closely related DNA sequence (TAAT), the PDs recognize 16- to 20-bp sequences, as determined through in vitro random selection (Epstein et al. 1994b). Several different Pax6 binding sites have been identified in murine and human genomes (Holst et al. 1997; Meech et al. 1999; Zhou et al. 2000), but none shares a marked similarity with the PD consensus binding site. Although to date these have not been identified, we surmise that there are additional, distinct Pax6 binding sites. This assumption is based on the ability of Pax6 to bind to many different DNA sequences and on its involvement in complex regulatory networks.
Using in vitro protein–DNA binding and polymerase chain reaction (PCR)-based cyclic amplification of protein binding sequences (CAPBS) from a human MboI-digested single-copy (sc) DNA library, we previously identified a novel PAX6 binding site, the B1 type (Zhou et al. 2000). Excluding high- and mid-level repetitive DNA fragments from the target DNA used for CAPBS successfully limited the isolated DNA fragments primarily to gene-rich DNA fractions in the sc-DNA library. Our CAPBS method is, however, most sensitive for identifying small DNA fragments (0.1–1 kb) that contain protein-binding sites. To clone other PAX6 binding sequences, we used CAPBS screening against genomic DNA digested with different 4-base-cutting restriction enzymes (Tsp509I and MseI). Here we report the identification of three novel PAX6 binding sites in the human genome. One site, the Alu-type, was similar to a region in the Alu repetitive element, of which more than 5000 copies have been found in the humangenome.
Alu repetitive elements are the major short interspersed nucleotide element (SINE) family in the primate genome. They are ∼300 bp long, occupy ∼10.7% of the human genome, and, according to a recent report by Li et al. (2001), are present in more than one million copies in the human genome. Of particular interest is that some of the PAX6 binding sites were found in SINEs, indicating a role in the evolution of gene regulation.
RESULTS
PD of PAX6 (PD6)-Specific Binding Site Mse180B1s
After three rounds of CAPBS with in vitro-synthesized Flag-tagged PAX6 protein, we obtained a single DNA band from screening the MseI-digested sc-DNA library. This 362-bp DNA fragment was cloned and subjected to direct EMSA to confirm binding to PAX6. Southern hybridization using the 362-bp DNA fragment as a probe against human genomic DNA digested with eight 4-base-cutting restriction enzymes revealed two bands in each digestion (data not shown), indicating that the probe sequence contained DNA fragments from two different locations in the genome. Sequence data from genomic clones isolated from a human placental genomic library later confirmed that the probe was composed of two MseI fragments from two different locations in the genome and showed that the PAX6 binding site was located in a 180-bp MseI fragment (Mse180, GenBank accession no. AF451322; data not shown). This chimeric DNA fragment was likely generated during adaptor ligation by T4 DNA ligase during the construction of the MseI sc-DNA library. A GenBank search located Mse180 in intron 6 of Cyp39A1.
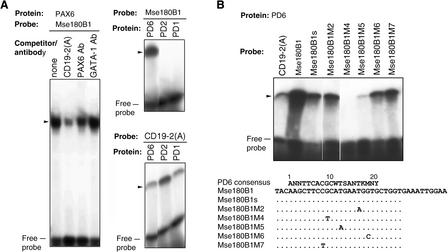
Within Mse180, we identified several regions that were 70% to 75% similar to the human PD6 consensus binding sequence. We synthesized five oligomers (31–44 bp) that entirely covered Mse180, with the putative PD6 binding sites centered in the middle of each oligomer. EMSA revealed that only the 38-bp oligomer Mse180B1 bound PAX6 (data not shown). Mse180B1 binding was competed using a Pax6 binding oligomer CD19-2(A) (Fig. 1A). Adding PAX6 antibody decreased the amount of the complex, but adding the control antibody GATA-1 did not. This result revealed that Mse180B1 bound specifically to PAX6. Figure 1A shows that only PD6, but not the murine PD of Pax1 (PD1) or Pax2 (PD2), bound Mse180B1, indicating that Mse180B1 is a PD6-specific binding site. Figure 1B shows the sequence similarity (75%) between the 5′ end of Mse180B1 (probe Mse180B1s) and the PAX6 consensus binding site. The binding affinity of Mse180B1s was less than that of Mse180B1 but similar to that of CD19-2(A) (Table 1).
Figure 1.
(A)EMSAs of the PD6 binding site Mse180B1. CD19-2(A) was used as a positive control. PAX6 binding competitor CD19-2(A) and rabbit serum (PAX6 and GATA-1) were added to the EMSA reactions. (B) EMSA of Mse180B1 mutants and the probe sequences, aligned with the PAX6 consensus binding sequence; (W) A or T, (S) C or G, (K) G or T, (M) A or C, (R) A or G, (Y) T or C, and (N) G, T, A, or C. Dots represent nucleotides identical to those in Mse180B1. The proteins and peptides used for binding were in vitro synthesized mouse PD1 and PD2, human PD6, the N-terminal half of PAX6, including the PD and HD domains (PD + HD), and the entire PAX6 protein (PAX6).
Table 1.
PAX6-Binding Sequences in the Human Genome
| Name | sc-DNA library | Sequence (5′–3′) | Homology to SINE | Location | Binding specificity | Binding affinityb |
|---|---|---|---|---|---|---|
| GCα1BLs5a | MboI | TTAGTTCCAGGTCAG | Rodent B1 | Intron 6 of GCα1 | PAX6 | 0.60 |
| Tsp11Bs | Tsp509I | AATTAGCCAGGCATGGTAGCACA | Primate Alu | Multiple | PAX6 | 0.56 |
| Mse180B1s | MseI | TACAAGCTTCCGCATGAATGGTGCTGGT | None | Intron 6 of Cyp39A1 | PD6 | 0.65 |
| Tsp2As1 | Tsp509I | GGCCAGTCACGCGTGTAATT | None | AL157938 | PAX6 | 1.5 |
Data from Zhou et al. (2000).
Binding affinity to PAX6 relative to CD19-2(A).
To identify residues critical for Mse180B1s binding, we searched GenBank for primate and rodent homologs with one mismatch in the 15-bp core region of Mse180B1s (CTTCCG CATGAATGG). Positions with frequent mismatches in the Mse180B1s homologs were assessed using EMSA for their ability to bind PAX6. Figure 1B shows that sequences with single nucleotide changes at positions 8, 16, and 18 in the PD6 consensus binding site (probes Mse180B1M7, Mse180B1M2, and Mse180B1M6, respectively) bound to PAX6 with similar affinities to the original sequences, indicating that these three sites are not critical for PAX6 binding. In contrast, sequences with single nucleotide changes at positions 9 and 12 (probes Mse180B1M4 and Mse180B1M5, respectively) did not bind to or bound weakly to PAX6, indicating that these two sites are critical for binding in the Mse180B1-type of PAX6 binding site. A PATSCAN search of the human genome for the 15-bp core region of Mse183B1s, allowing one mismatch at positions 8, 16, or 18, found 16 locations with putative PD6 binding sequences, including the Cyp39A1 gene (http://odin.mdacc.tmc.edu/zhou).
PAX6-Specific Binding Site Tsp2As1
Flag-tagged and His-tagged PAX6 were used to screen PAX6 binding sequences from the Tsp509I sc-DNA library. For this library, using two disparately tagged PAX6s for CAPBS yields a greater number of PAX6-specific binding DNA fragments than using either alone. After two rounds of CAPBS with Flag-tagged PAX6 and two with His-tagged PAX6, we cloned all PCR products. Direct EMSA of 15 clones showed that 13 of them contained sequences that formed complexes with PAX6 and could be competed with the PAX6 binding oligomer CD19-2(A) (data not shown). Sequencing the PAX6 binding clones showed inserts flanked by TspAR and TspBR primers, which were used to construct the library. However, these inserts were composed of two or three blocks of 24- to 26-bp genomic DNA fragments (Tsp2A, Tsp2B, Tsp11B, and Tsp12A), each flanked by the PCR primers TspAR or TspBR or by both. These artificial DNA fragments were likely produced during adaptor ligation. The patterns of the shifted bands showed that one, two, or three PAX6/DNA complexes were formed, and the sequences of these bands indicated that only Tsp2A and Tsp11B bound PAX6 strongly (data not shown).
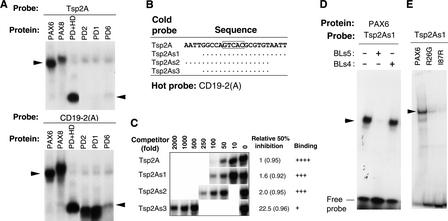
Figure 2A shows an EMSA of Tsp2A binding to in vitro synthesized PAX6, PAX8, PD6, PD1, PD2, and PD + HD of PAX6. Tsp2A did not bind to PD6 but bound to PAX6 and PD + HD. Tsp2A bound to PAX8 weakly, perhaps because PAX8 does not contain a complete HD (Dahl et al. 1997). A search of GenBank revealed one copy of the last 22-bp sequence of Tsp2A in the human genome (Fig. 2B), and the first two nucleotides in Tsp2A did not match the genomic DNA. The first Tsp509 site (AATT) in Tsp2A may thus originate from the adaptor.
Figure 2.
(A)EMSA of PAX6-specific binding site Tsp2A with whole or partial Pax proteins (the abbreviation for proteins are defined in the legend for Fig. 1). CD19-2(A) was used as a positive control. (B) Probe sequences. Additional random sequences (GAAGAGCTC at the 5′ ends and GAGCTCGAC at the 3′ ends) were added to ends of the probe sequences shown here as possible docking sites. The relative binding affinities were deduced based on the amount of competitor inhibiting protein binding by 50% from EMSA in panel C. The core sequence GTCAC for binding by the N-terminal subdomain of the PD (Jun and Desplan 1996) is boxed. Dots represent nucleotides identical to those in Tsp2A. The numbers in parentheses are the correlation coefficients (R) between the amount of cold probe and the amount of the complex formed. (D) Sequence-specific binding of PAX6 to Tsp2As1. BLs5 is a PAX6-specific binding probe and BLs4 is a non-PAX6-specific binding probe (Zhou et al. 2000). (E) EMSA of PAX6, and mutant PAX6 R26G and I87R to Tsp2As1.
Because AATT is a binding site for HD and there were two such sites in Tsp2A, we deleted them singly and together to examine whether they were necessary for PAX6 binding. As shown in Figure 2, B and C, PAX6 binding was not significantly affected when one AATT copy was deleted (probes Tsp2As1 and Tsp2As2). When both copies of AATT were deleted (probe Tsp2As3), however, binding to PAX6 decreased by 22-fold. Because only Tsp2As1 exists in the human genome, this probe was used to detect binding specificity for PAX6. As shown in Figure 2D, Tsp2As1 bound PAX6 in a sequence-dependent manner, as the complex could be abolished by the competitor oligomer BLs5 but not by the non-PAX6 binding oligomer BLs4, which differs from BLs5 by only one base (Zhou et al. 2000). To clarify the specific PAX6 binding domain required for binding to this site, EMSA analysis was performed using in vitro synthesized PAX6 and two PAX6 mutants identified in aniridia patients, R26G and I87R (Tang et al. 1997). R26G was mutated in the N-terminal HTH motif of PD, and the I87R mutation occurred in the C-terminal HTH motif of PD. Western hybridization with PAX6 antibody showed that the amount of PAX6 protein used in EMSA is about twofold higher than the amount of R26G and I87R (data not shown). As shown in Figure 2E, R26G and I87R binding to Tsp2As1 is markedly less than binding from wild-type PAX6. Because PAX8 (the paired-box protein with a half HD) did not bind to Tsp2As1, it indicates that both HTH domains of PD and HD are required for binding to Tsp2As1.
Alu-Type PAX6-Specific Binding Sites
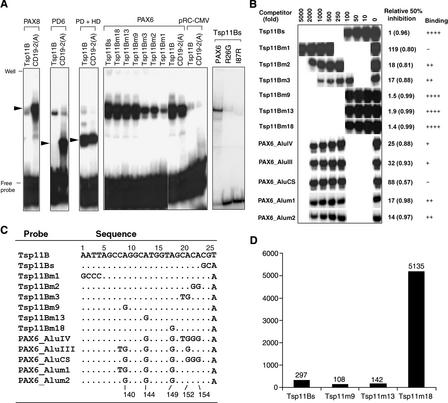
Using CAPBS screening of the Tsp509I sc-DNA library, we also isolated a 26-bp PAX6 binding sequence, Tsp11B (Fig. 3). A GenBank search (release 121, December 26, 2000) revealed two locations in the human genome that were a complete match for Tsp11B. When three mismatches were allowed in the Tsp11B search, however, 4622 unique locations were found in the primate database, but only 6 were found in the rodent database. It was, therefore, not surprising that Tsp11B shared a high degree of sequence similarity to a region found within primate Alu repetitive elements. As shown in Figure 3A, Tsp11B bound to PAX6 but not to PD6, mutant PAX6 R26G and I87R, or PAX8. Similar to binding to Tsp2As1, PAX6 binding to Tsp11B required both HTH domains of PD and HD.
Figure 3.
(A)PAX6-specific binding to the Alu-type PAX6 binding sequences (Tsp11Bs, Tsp11Bm9, Tsp11Bm13, and Tsp11Bm18). CD19-2(A) was used as a positive control. For protein abbreviations, see the legend for Figures 1 and 2. (B) Binding affinities of the Alu-type PAX6 binding sites and their mutants were determined through competitive EMSA. The hot probe is Tsp11Bs. (C) The sequences of the probes. The numbers at the bottom of the alignments correspond to locations in Alu consensus sequences (Jurka and Smith 1988). (D) Proportion of different Alu-type PAX6 binding sites in the human genome (http://www.ncbi.nlm.nih.gov/hs_phase3.fna, release on May 8, 2002). All probes contained the additional random nucleotides (described in the legend to Fig. 2) at their ends, which served as possible docking sites.
Because the 26-bp Tsp11B sequence was very similar to a region found within Alu repetitive elements, we wanted to determine if the consensus Alu sequences contained the PAX6 binding site. We first identified the minimal PAX6 binding site in Tsp11B. As shown in Figure 3A, changing AATT at positions 1–4 to GCCC (probe Tsp11Bm1) almost abolished binding, indicating that the 5′ AATT residues are important for PAX6 binding. Changing the last three nucleotides (positions 24–26) from CGT to GCA (probe Tsp11Bs) did not affect binding. However, when nucleotides 22 and 23 were changed from CA to GG (probe Tsp11Bm2) and nucleotides 20 and 21 were changed from CA to TG (probe Tsp11Bm3), the binding affinity was dramatically decreased. These experiments showed that the Alu-type PAX6 binding site was 23 bp long (probe Tsp11Bs), occupying positions 1–23. This location corresponded to positions 132–154 in the Alu consensus sequences, which match the very beginning of the second monomer (Jurka and Smith 1988).
Because Tsp11Bs nucleotides 9, 13, and 18 differed from the Alu consensus sequences, we examined the effect of each individual change on PAX6 binding using normal (Fig. 3A) and competitive (Fig. 3B) EMSAs. Changing nucleotide A to G at positions 9 (probe Tsp11Bm9), 13 (probe Tsp11Bm13), or 18 (probe Tsp11Bm18) did not affect binding affinity. Tsp11Bs and its functional homologs (Tsp11Bm9, Tsp11Bm13, and Tsp11Bm18) were revealed as Alu-type PAX6 binding sites because they all bound PAX6 in vitro.
When nucleotides 9, 13, and 18 were each changed from A to G (in probe PAX6_Alum2) so that they were the same as in the Alu consensus sequences, PAX6 binding affinity to the probe was dramatically reduced. However, changing nucleotide 8 from C in probe PAX6_Alum2 to T in probe PAX6_Alum1 had no significant effect. In addition, minor changes at the 3′ region of Tsp11Bs (probes Tsp11Bm2 and Tsp11Bm3), which occurred in all consensus Alu sequences and were represented by PAX6_AluIV, PAX6_AluIII, and PAX6_AluCS probes, led to a significant or complete loss of binding to PAX6. Because these three Alu consensus probes are known to recognize almost all of the consensus sequences in the various Alu subfamilies (Britten et al. 1988; Batzer et al. 1996), we concluded that most Alu members do not contain Alu-type PAX6 binding sites.
The PAX6 Binding Alu Elements
Characterization of the Alu-type PAX6 binding site revealed the relevant binding nucleotides, which allowed us to estimate the abundance of these sites in the human genome, as well as their locations relative to genes. Using PATSCAN in the GCG program, a total of 5682 Alu-type PAX6 binding sites were found by searching the GenBank hs_phase 3 released on May 8, 2002. Based on the estimated Alu copy reported by Li et al. (2001), these 5682 sites (assumed to be present in Alu repeats) represent ∼0.2% of all of the Alu repeats in the human genome.
Using the computer software DNANavider (developed by Liping Yu, liping_yu@yahoo.com), we searched all 5682 Alu-type PAX6 binding sites against GenBank sequences for annotated information on the location of these sites. The detailed results can be found at http://odin.mdacc.tmc.edu/zhou/. The relative abundance of the four Alu-type PAX6 binding sites is shown in Figure 3D. Tsp11m18 was the most common type (90%) of Alu-type PAX6 binding motif found in the human genome. Comparing the Alu-type PAX6 binding sequences with their corresponding Alu sequences, changing four of the five G nucleotides to A in Alu consensus sequences at positions 140, 144, 149, 152, and 154 (Jurka and Smith 1988) produced Alu-type PAX6 binding sites. Except for the G nucleotide at position 149, which is in position 18 of Tsp11m18, all other G nucleotides are CpG dinucleotides. Because most mutations of Alu repeats occur repeatedly at CpG dinucleotide hot spots (Li 1997), the high proportion of Tsp11m18 sequences found in the Alu-type PAX6 binding motif is clearly caused by a high mutation efficiency of G nucleotides in the CpG dinucleotide. This could also explain why most Alu-type PAX6 binding sites are in older subfamilies that had a longer time to mutate. The presence of PAX6 binding sites in a series of older Alu subfamilies, but not in their consensus sequences, indicates that the sites are not distributed by these elements, but are instead created in these elements after their insertion at locations as a result of an accumulation of mutations.
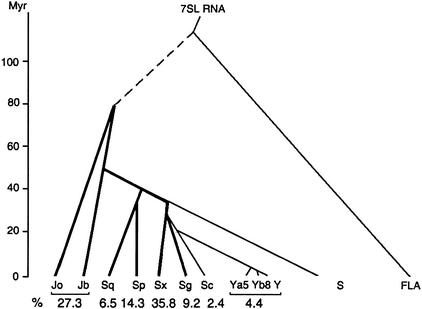
From 4317 Alu-type PAX6 binding sites processed by DNANavider, 293 locations were found in 5 or 6 major Alu subfamilies that are annotated according to the standard nomenclature for Alu repeats (Fig. 4; Batzer et al. 1996). More than one-third (36%) of these PAX6 binding Alu elements belonged to the AluSx subfamily. The second largest group was from the AluJ subfamily (14% AluJo and 13% AluJb). To generalize, the J Alu subfamily is the oldest; the S subfamily is intermediate in age; and the Y subfamily is the youngest (Batzer et al. 1996). Thus, >68.3% of the PAX6 binding Alu elements were in intermediate-age Alu families; 27.3% were in old Alu families; and only 4.4% were in young Alu families. This trend can be explicitly demonstrated by mapping the distribution of Alu-type PAX6 binding sites onto the phylogeny of Alu major subfamilies (Fig. 4). The explosive increase in AluSx PAX6 binding sites indicates that Alu-mediated amplification of PAX6 binding sites occurred after the split between AluSx and AluSg, which was ∼35 million years ago.
Figure 4.
PAX6 binding Alu elements. Branching order of Alu subfamilies (reprinted with permission from the J. Mol. Evol., The Age of Alu Subfamilies, by Kapitonov and Jurka, 42: 59–65, 1996 © Springer-Verlag GmbH & Co.). The branches with PAX6 binding Alu members are indicated by heavy lines, and the percentages in each Alu subfamily are shown.
DISCUSSION
Through an in vitro protein–DNA binding approach, we identified three new types of PAX6 binding sequences in the human genome (Table 1). Two exist in a single copy in genome, and one within Alu repetitive elements. Another binding sequence (GCa1BLs1), reported by us in an earlier study, is found in a gene in the human genome but is present in multiple copies in the rodent genome. This sequence is similar to the B1 repetitive element sequence (Zhou et al. 2000). All four types of elements bound PAX6 in vitro, but each had a different sequence. Only one (Mse180B1s and its variants), which is able to bind PD6, was similar (75%) to the consensus binding site for PD6. We also determined the minimal length of these binding sites and several residues that are critical for binding. This process facilitated the identification of putative PAX6 target genes (i.e., genes containing PAX6 binding sites) in the human genome. One of these putative PAX6 target genes encodes PDGFRA, in which an Alu-type PAX6 binding site was located in intron 7 (data shown in web site http://odin.mdacc.tmc.edu/zhou/). PDGFRA expression in the U251HF glioma cell line was down-regulated by stable transfection of a PAX6 expression construct (Zhou et al. 2001). Whether the PAX6 binding site in PDGFRA is involved in regulating gene expression will be investigated.
The Pax6 protein sequence is identical in human and mouse. Xenopus Pax6, which is >97% similar to human and murine Pax6, bound the Alu-type PAX6 binding site in vitro with an affinity similar to that of human PAX6 (data not shown). It is striking that such an evolutionarily conserved transcription factor can bind to diverse sequences in a sequence-specific manner. These sites were relatively long (15–22 bp), which would hardly be found in multiple copies in a randomized genome of 4 billion base pairs.
The Alu-type PAX6 binding sites were ubiquitous in the human genome. This fact supports our previous findings that B1-type PAX6 binding sites were widely distributed throughout the rodent genome (Zhou et al. 2000). Although Alu and B1 elements originated from a common 7SL RNA-derived retroposon monomeric element and are the major SINE families in the primate and rodent genomes, respectively (Quentin 1994; Zietkiewicz and Labuda 1996), the PAX6 binding sites identified in B1 and Alu elements have no clear sequence similarity. This is consistent with their different locations in each repeat: the very beginning of the second Alu monomer for the Alu-type and the internal 29-bp duplicate region of the B1 quasidimers for the B-type PAX6 binding sites.
Based on their sequences, the PAX6 binding Alu elements did not form a unique subfamily. Instead, they were scattered through all the major Alu subfamilies, from the oldest to the youngest. However, >68% were in intermediate-age subfamilies (the AluS family), and few were in young Alu families. This is similar to the distribution of B1-type sites in B1 elements of the rodent genome. These observations led to a putative evolutionary scenario that describes how a transcription factor (Pax6) recruits new target genes in the genome. Mediated by repetitive elements and mutations in CpG dinucleotide hot spots (e.g., Alu or B1), several types of Pax6 binding sites have been generated and were spread over the entire genome. Although most of the B1- and Alu-type PAX6 binding sites probably do not function as Pax6 regulatory sites, they could be the origin of new regulation pathways. Consequently, genes that are regulated in rodents by Pax6 though B1-type Pax6 binding sites and genes that are regulated in primates by PAX6 through Alu-type PAX6 binding sites may exert different effects in rodents than in primates. Increasing experimental evidence supporting the involvement of the retroposon (especially examples from Alu elements) in gene regulation has already drawn attention to how retroposons function in the control of gene expression (Britten 1996; Tomilin 1999). Our data strengthen the concept that exploring regulatory sequences within repetitive elements could provide important information on the evolution of gene regulation, on the diversity of gene regulation between closely related species, and on the impact of these events on biomedical research, particularly in a murine model.
METHODS
Construction of Human Tsp509I and MseI sc-DNA Libraries
Human genomic DNA was isolated from the human leukemia cell line K562. First, DNA (0.2 mg) was digested completely with Tsp509I or MseI at 37°C for 20 h. The sc-DNA was separated from the medium and high-copy DNA based on the DNA reassociation rate using the method of Zhou et al. (2000). An aliquot of the recovered sc-DNA (0.5 μg) was ligated into two double-stranded adaptors (adaptor A, 5′-CTCGAGCTAGCGGCCGCTAT-3′; adaptor B, 5′-GAGCTCT TCTCTGGATCCAGA-3′), both having protruding 5′ ends (AATT in the Tsp509I-digested DNA and TA in the MseI-digested DNA). An aliquot of the sc-DNA digested with each enzyme was amplified for 35 cycles at 94°C for 1 min, 52°C for 1 min, and 72°C for 1 min to construct sc-DNA libraries. For the Tsp509I sc-DNA library, the PCR primers used were TspAR (5′-GCCGCTAGCTCGAGAATT-3′) and TspBR (5′-AT CCAGAGAAGAGCTCAATT-3′). For the MseI sc-DNA library, the PCR primers used were MseAR (5′-GGCCGCTAGCT CGAGTA-3′) and MseBR (5′-GATCCAGAGAAGAGCTCTA-3′). Aliquots of the PCR products were screened for PAX6 binding sequences using the CAPBS method.
In Vitro Synthesis of Two PAX6-Tagged Proteins
The PAX6–Flag/pRC-CMV clone was described by Zhou et al. (2000). The PAX6–His/pCITE-4b(+) clone was generated by cloning the full-length of PAX6 cDNA into SacI and SalI sites of pCITE-4b(+) (Novagen) with a six-histidine peptide in a frame at the 3′ end of the PAX6 cDNA. The PAX6–Flag and PAX6–His proteins were translated in vitro from PAX6–Flag/pRC-CMV and PAX6–His/pCITE-4b(+) using the TnT T7 Quick Coupled Transcription/Translation System (Promega) and the manufacturer's protocol.
Modified CAPBS Screening for Target Sequences
A modified CAPBS method (Zhou et al. 2000) was used to isolate PAX6 binding DNA fragments from each sc-DNA library. The same protein with different tags (PAX6–His and PAX6–Flag) was used to screen the target DNA fragments. PAX6–His/DNA complexes were isolated with Ni-NTA agarose (QIAGEN) and PAX6–Flag/DNA complexes with anti-Flag M2 agarose (Sigma). In two rounds of PAX6–His-mediated CAPBS followed by two rounds of PAX6–Flag-mediated CAPBS, the DNA fragments in the last PCR were ligated directly into pCR4-TOPO and transformed into TOP10 Escherichia coli competent cells (Invitrogen). Sixteen clones were picked randomly and subjected to PCR screening with M13F and M13R primers.
Direct Electrophoretic Mobility Shift Assay (EMSA) of PAX6 Binding Clones
About 40 ng of the putative pCR4-TOPO clone of the PAX6 binding sequence was digested with EcoRI at 37°C for 1 h. Both the insert and vector DNA were labeled directly with 5 μCi of [32α]dATP (3000 Ci/mmole), 1 unit of Klenow polymerase (Roche), and 0.5 nmole each of dCTP, dGTP, and dTTP at 37°C for 30 min. After heat inactivation and purification by chromatography through G-50 columns (Pharmacia), the labeled fragments were subjected to binding reactions with in vitro synthesized PAX6 protein. EMSA was performed as described by Zhou et al. (2000). The PAX6 binding specificity was estimated by adding excess amounts of the unlabeled competitor oligonucleotide CD19-2(A) (Epstein et al. 1994b). The cDNA constructs for PAX6, mutant PAX6 R26G and I87R (Tang et al. 1997), PAX8 (Fraizer et al. 1997), and for the human PD6 and the mouse PD1 and PD2 (Sun et al. 2001) were described previously. Xenopus Pax6 cDNA construct was provided by Dr. Ali Hemmati-Brivanlou.
Competitive EMSA
To estimate the binding affinity of different PAX6 binding sequences, increasing amounts of unlabeled double-stranded oligonucleotides (oligomers) were added to the EMSA reaction before adding a fixed amount (7 fmole) of labeled probe. The densitometric value of the PAX6 complex was measured with ImageQuanNT (Molecular Dynamics), and a graph was drawn with Excel 5.0 (Microsoft) to show the amount of cold probe versus the densitometric value of the complex. An equation was determined from the curve and calculated from the amount of competitor that inhibited protein binding by 50%.
Computer Analyses
FINDPATTERNS in the GCG program (Oxford Molecular) and PATSCAN (Dsouza et al. 1997) were used to search GenBank release 121 (December 26, 2000), hs_phase3 (November 1, 2001) for sequences containing the PAX6 binding sequences identified in this study. DNANavider, which was developed by Liping Yu (liping_yu@yahoo.com), was used to locate the gene and Alu annotations in GenBank.
WEB SITE REFERENCES
http://odin.mdacc.tmc.edu/zhou; 16 locations with putative PD6 binding sequences, including the Cyp39A1 gene.
http://www.ncbi.nlm.nih.gov/;hs_phase3.fna, release on November 1, 2001; proportion of different Alu-type PAX6 binding sites in the human genome.
http://www.ncbi.nlm.nih.gov/; release 121, December 26, 2000; two locations in the human genome that were a complete match for Tsp11B.
Acknowledgments
We thank Liping Yu for providing us with the computer software DNANavider. We thank Sheng Zhao for helping us use GCG. We thank Joann Aaron for editing this manuscript. This work was supported by National Research Service Award grant 5 F32 EY06949–02.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL sa51001@odin.mdacc.tmc.edu; FAX (713) 745-1183.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.188302.
REFERENCES
- Aalfs CM, Fantes JA, Wenniger-Prick LJ, Sluijter S, Hennekam RC, van Heyningen V, Hoovers JM. Tandem duplication of 11p12–p13 in a child with borderline development delay and eye abnormalities: Dose effect of the PAX6 gene product? Am J Med Genet. 1997;73:267–271. doi: 10.1002/(sici)1096-8628(19971219)73:3<267::aid-ajmg7>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- Batzer MA, Deininger PL, Hellmann-Blumberg U, Jurka J, Labuda D, Rubin CM, Schmid CW, Zietkiewicz E, Zuckerkandl E. Standardized nomenclature for Alu repeats. J Mol Evol. 1996;42:3–6. doi: 10.1007/BF00163204. [DOI] [PubMed] [Google Scholar]
- Britten RJ. DNA sequence insertion and evolutionary variation in gene regulation. Proc Natl Acad Sci. 1996;93:9374–9377. doi: 10.1073/pnas.93.18.9374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britten RJ, Baron WF, Stout DB, Davidson EH. Sources and evolution of human Alu repeated sequences. Proc Natl Acad Sci. 1988;85:4770–4774. doi: 10.1073/pnas.85.13.4770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaerts P, Halder G, Gehring WJ. Pax6 in development and evolution. Annu Rev Neurosci. 1997;20:483–532. doi: 10.1146/annurev.neuro.20.1.483. [DOI] [PubMed] [Google Scholar]
- Dahl E, Koseki H, Balling R. Pax genes and organogenesis. Bioessays. 1997;19:755–765. doi: 10.1002/bies.950190905. [DOI] [PubMed] [Google Scholar]
- Dsouza M, Larsen N, Overbeek R. Searching for patterns in genomic data. Trends Genet. 1997;13:497–498. doi: 10.1016/s0168-9525(97)01347-4. [DOI] [PubMed] [Google Scholar]
- Engelkamp D, Rashbass P, Seawright A, van Heyningen V. Role of Pax6 in development of the cerebellar system. Development. 1999;126:3585–3596. doi: 10.1242/dev.126.16.3585. [DOI] [PubMed] [Google Scholar]
- Epstein J, Glaser T, Cai J, Jepeal L, Walton D, Maas R. Two independent and interactive DNA-binding subdomains of the Pax6 paired domain are regulated by alternative splicing. Genes & Dev. 1994a;8:2022–2034. doi: 10.1101/gad.8.17.2022. [DOI] [PubMed] [Google Scholar]
- Epstein J, Cai J, Glaser T, Jepeal L, Maas R. Identification of a Pax paired domain recognition sequence and evidence for DNA-dependent conformational changes. J Biol Chem. 1994b;269:8355–8361. [PubMed] [Google Scholar]
- Fraizer GC, Shimamura R, Zhang X, Saunders GF. PAX8 regulates human WT1 transcription through a novel DNA binding site. J Biol Chem. 1997;272:30678–30687. doi: 10.1074/jbc.272.49.30678. [DOI] [PubMed] [Google Scholar]
- Fukuda T, Kawano H, Osumi N, Eto K, Kawamura K. Histogenesis of the cerebral cortex in rat fetuses with a mutation in the Pax6 gene. Brain Res Dev Brain Res. 2000;120:65–75. doi: 10.1016/s0165-3806(99)00187-x. [DOI] [PubMed] [Google Scholar]
- Holst BD, Wang Y, Jones FS, Edelman GM. A binding site for Pax proteins regulates expression of the gene for the neural cell adhesion molecule in the embryonic spinal cord. Proc Natl Acad Sci. 1997;94:1465–1470. doi: 10.1073/pnas.94.4.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun S, Desplan C. Cooperative interactions between paired domain and homeodomain. Development. 1996;122:2639–2650. doi: 10.1242/dev.122.9.2639. [DOI] [PubMed] [Google Scholar]
- Jurka J, Smith T. A fundamental division in the Alu family of repeated sequences. Proc Natl Acad Sci. 1988;85:4775–4778. doi: 10.1073/pnas.85.13.4775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitonov V, Jurka J. The age of Alu subfamilies. J Mol Evol. 1996;42:59–65. doi: 10.1007/BF00163212. [DOI] [PubMed] [Google Scholar]
- Kozmik Z, Czerny T, Busslinger M. Alternatively spliced insertions in the paired domain restrict the DNA sequence specificity of Pax6 and Pax8. EMBO J. 1997;16:6793–6803. doi: 10.1093/emboj/16.22.6793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W-H. Molecular evolution. Sunderlander, MA: Sinauer; 1997. [Google Scholar]
- Li W-H, Gu Z, Wang H, Nekrutenko A. Evolutionary analyses of the human genome. Nature. 2001;409:847–849. doi: 10.1038/35057039. [DOI] [PubMed] [Google Scholar]
- Mansouri A, Hallonet M, Gruss P. Pax genes and their roles in cell differentiation and development. Curr Opin Cell Biol. 1996;8:851–857. doi: 10.1016/s0955-0674(96)80087-1. [DOI] [PubMed] [Google Scholar]
- Meech R, Kallunki P, Edelman GM, Jones ES. A binding site for homeodomain and Pax proteins is necessary for L1 cell adhesion molecule gene expression by Pax6 and bone morphogenetic proteins. Proc Natl Acad Sci. 1999;96:2420–2425. doi: 10.1073/pnas.96.5.2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosser J, van Heyningen V. PAX6 mutations reviewed. Hum Mutat. 1999;11:93–108. doi: 10.1002/(SICI)1098-1004(1998)11:2<93::AID-HUMU1>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Quentin Y. A master sequence related to a free left Alu monomer (FLAM) at the origin of the B1 family in rodent genomes. Nucleic Acids Res. 1994;22:2222–2227. doi: 10.1093/nar/22.12.2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiring R, Walldorf U, Kloter U, Gehring WJ. Homology of the eyeless gene of Drosophila to the Small eye gene in mice and Aniridia in humans. Science. 1994;265:785–789. doi: 10.1126/science.7914031. [DOI] [PubMed] [Google Scholar]
- Sander M, Neubuser A, Kalamaras J, Ee HC, Martin GR, German MS. Genetic analysis reveals that PAX6 is required for normal transcription of pancreatic hormone genes and islet development. Genes & Dev. 1997;11:1662–1673. doi: 10.1101/gad.11.13.1662. [DOI] [PubMed] [Google Scholar]
- Sheng G, Harris E, Bertuccioli C, Desplan C. Modular organization of Pax/homeodomain proteins in transcriptional regulation. Biol Chem. 1997;378:863–872. doi: 10.1515/bchm.1997.378.8.863. [DOI] [PubMed] [Google Scholar]
- Sisodiya SM, Free SL, Williamson KA, Mitchell TN, Willis C, Stevens JM, Kendall BE, Shorvon SD, Hanson IM, Moore AT, et al. PAX6 haploinsufficiency causes cerebral malformation and olfactory dysfunction in humans. Nat Genet. 2001;28:214–216. doi: 10.1038/90042. [DOI] [PubMed] [Google Scholar]
- Sun H, Dickinson DP, Costello J, Li WH. Isolation of Cladonema Pax-B genes and studies of the DNA-binding properties of cnidarian Pax paired domains. Mol Biol Evol. 2001;18:1905–1918. doi: 10.1093/oxfordjournals.molbev.a003731. [DOI] [PubMed] [Google Scholar]
- Tang HK, Chao LY, Saunders GF. Functional analysis of paired box missense mutations in the PAX6 gene. Hum Mol Genet. 1997;6:381–386. doi: 10.1093/hmg/6.3.381. [DOI] [PubMed] [Google Scholar]
- Tomilin NV. Control of genes by mammalian retroposons. Intl Rev Cyto. 1999;186:1–48. doi: 10.1016/s0074-7696(08)61050-5. [DOI] [PubMed] [Google Scholar]
- Xu W, Rould MA, Jun S, Desplan C, Pabo CO. Crystal structure of a paired domain–DNA complex at 2.5 A resolution reveals structural basis for Pax developmental mutations. Cell. 1995;80:639–650. doi: 10.1016/0092-8674(95)90518-9. [DOI] [PubMed] [Google Scholar]
- Zhou YH, Zheng JB, Gu X, Li WH, Saunders GF. A novel Pax6 binding site in rodent B1 repetitive elements: Co-evolution between developmental regulation and repeat elements. Gene. 2000;245:319–328. doi: 10.1016/s0378-1119(00)00019-6. [DOI] [PubMed] [Google Scholar]
- Zhou YH, Glass T, Yung AWK. PAX6 down regulates the expression of PDGFRA and MCP-1 genes in glioblastoma cells. Neuro-Oncology. 2001;3:312. [Google Scholar]
- Zietkiewicz E, Labuda D. Mosaic evolution of rodent B1 elements. J Mol Evol. 1996;42:66–72. doi: 10.1007/BF00163213. [DOI] [PubMed] [Google Scholar]