Abstract
Large-scale gene expression profiling was performed on embryo-derived stem cell lines to identify molecular signatures of pluripotency and lineage specificity. Analysis of pluripotent embryonic stem (ES) cells, extraembryonic-restricted trophoblast stem (TS) cells, and terminally-differentiated mouse embryo fibroblast (MEF) cells identified expression profiles unique to each cell type, as well as genes common only to ES and TS cells. Whereas most of the MEF-specific genes had been characterized previously, the majority (67%) of the ES-specific genes were novel and did not include known differentiated cell markers. Comparison with microarray data from embryonic material demonstrated that ES-specific genes were underrepresented in all stages sampled, whereas TS-specific genes included known placental markers. Investigation of four novel TS-specific genes showed trophoblast-restricted expression in cell lines and in vivo, whereas one uncharacterized ES-specific gene, Esg-1, was found to be exclusively associated with pluripotency. We suggest that pluripotency requires a set of genes not expressed in other cell types, whereas lineage-restricted stem cells, like TS cells, express genes predictive of their differentiated lineage.
[Supplemental material is available online at www.genome.org and http://lgsun.grc.nia.nih.gov/microarray/data.html]
Fertilized eggs are the ultimate totipotent cells, giving rise to all cell types. Animal development can therefore be described as a progressive loss of totipotency, then of pluripotency, and ultimately differentiation into specific cell types. In mammals, the initial loss of totipotency occurs during preimplantation development, marked by the segregation of two distinct cell lineages in the blastocyst: the inner cell mass (ICM), which gives rise to the embryo proper, and the trophectoderm (TE), which contributes exclusively to the trophoblast portion of the placenta. Representative stem cell lines have been established from these two cell types: embryonic stem (ES) cells derived from the ICM (Evans and Kaufman 1981; Martin 1981) and trophoblast stem (TS) cells derived from the TE (Tanaka et al. 1998). ES cells are pluripotent and can differentiate into all embryonic cell lineages and some extraembryonic ones, whereas TS cells can differentiate into multiple cell types, but only of the trophoblast lineage.
The undifferentiated, pluripotent state of ES cells is maintained by Leukemia Inhibitory Factor (LIF) and JAK/STAT signaling (for review, see Smith 2001). On withdrawal of LIF, ES cells differentiate into various cell types (Shen and Leder 1992). In chimeric embryos, ES cells are able to contribute to various cell types, including the germ line (Nagy et al. 1993) but excluding trophoblast lineages (Beddington and Robertson 1989). In contrast, the undifferentiated, multipotent state of TS cells is maintained by Fibroblast Growth Factor 4 (FGF4) signaling (for review, see Kunath et al. 2001). On withdrawal of FGF4, TS cells differentiate into several trophoblast subtypes, including trophoblast giant cells. Therefore, although both ES and TS cells originate from the blastocyst and possess characteristics of stem cells, they differ in their differentiation potential, with ES cells being more pluripotent than TS cells.
Pluripotency has been defined experimentally by assessing whether cells can differentiate into multiple lineages. Several key genes, such as Eomes and Cdx2, have been identified to regulate the differentiation of TE (for review, see Rossant and Cross 2001) and the POU-domain transcription factor Oct3/4 (Scholer et al. 1989; Rosner et al. 1990; Okazawa et al. 1991) has been demonstrated to be essential for the maintenance of pluripotency of both the ICM (Nichols et al. 1998) and ES cells (Niwa et al. 2000). Expression profiling with cDNA microarray technology has been used to discover genes that are differentially expressed in developmental processes (Tanaka et al. 2000; Hemberger et al. 2001; Kim et al. 2001). In this study, we used the NIA mouse 15K cDNA microarray to profile ES cells, TS cells, and mouse embryo fibroblast (MEF) cells to identify genes specific to the earliest cell lineages (ICM and TE) that arise during development and to investigate the differences and similarities of a pluripotent stem cell and a lineage-restricted stem cell.
RESULTS AND DISCUSSION
Expression Profiling of Stem Cell Lines by cDNA Microarrays
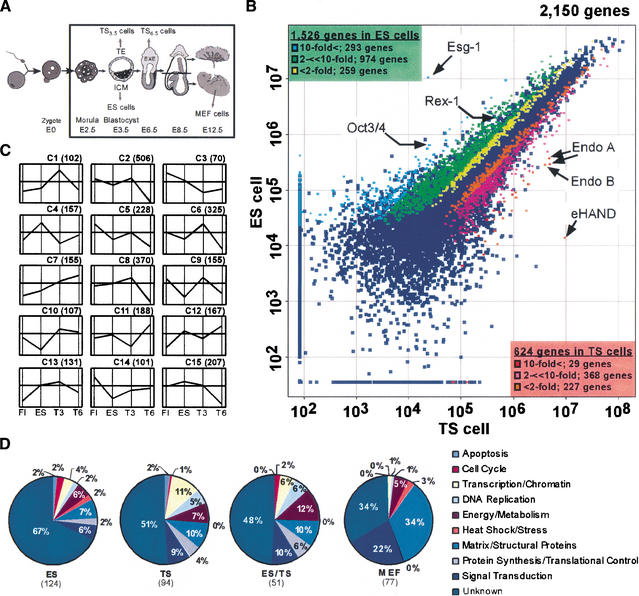
Gene expression profiles were obtained from ES cells, TS3.5 cells (derived from E3.5 blastocysts), TS6.5 cells (derived from E6.5 trophoblast tissue), and mouse embryo fibroblast (MEF) cells from E12.5 embryos (Fig. 1A) using the NIA mouse 15K cDNA microarray. This clone set consists of ∼12,000 unique mouse genes and is enriched for genes expressed in placental tissue and early mouse embryos, including preimplantation stages; it is therefore well suited to this study (Tanaka et al. 2000; Kargul et al. 2001). Approximately half of the genes are novel, and ∼90% are sequence-verified (Kargul et al. 2001). Each hybridization experiment was performed in triplicate, and the data were globally normalized after background subtraction. Both pair-wise and multiple comparisons were performed on the data sets to identify candidate stem cell-specific, lineage-specific, and differentiation-specific clusters. For example, a pair-wise comparison between ES and TS6.5 cells identified 2,150 differentially expressed genes, as determined by the Student's t-test (P < 0.05). Among these genes, 1,526 genes were more highly expressed in ES cells and 624 genes were more highly expressed in TS6.5 cells (Fig. 1B; Supplemental Table 1). Analysis of known genes within these lists agreed well with expected expression patterns. The TS cell profile showed characteristic features of the trophoblast lineage, expressing known placental-specific genes such as EndoA, EndoB, eHand/Hand1, Transglutaminase, Pem, Secretin, Tbx1, and Psx1 (Tanaka et al. 2000). Also, genes known to function in ES cells, such as Rex-1 (Hosler et al. 1989) and Oct3/4 were included in the ES cell list (Fig. 1B). In addition, many novel, uncharacterized genes were present in each list.
Figure 1.
(A) Schematic representation of tissues and cell lines used for microarray hybridization experiments and Northern analysis. Embryos, tissue types, and cell lines in the rectangle were used in this study. (B) Scatter-plot analysis of ES versus TS6.5 cells microarray experiment. Average expression levels (arbitrary units) of each gene were calculated from three independent hybridizations. Genes that show significantly different expression levels between ES and TS6.5 cells at the 5% significance level (P < 0.05) with expression levels above the background are displayed as colored spots, and the other genes are displayed as dark blue spots. For genes that are expressed higher in ES cells than in TS6.5 cells, blue indicates genes expressed greater than 10-fold, green indicates those expressed between two- and 10-fold, and yellow indicates those expressed less than twofold. For genes that are expressed higher in TS6.5 cells than in ES cells, red indicates genes expressed greater than 10-fold, pink indicates those expressed between two- and 10-fold, and orange indicates those expressed less than twofold. Several examples of genes are indicated. (C) k-means cluster-analysis of differentially expressed genes among ES, TS3.5, TS6.5, and MEF at a 5% significance level (P < 0.05). By using the k-means algorithm, 2,969 genes were grouped into 15 distinct clusters based on their similarities of expression patterns among ES, TS3.5, TS6.5, and MEF. The name of individual clusters is indicated, followed in parentheses with the number of genes in each cluster. Only statistically significant, sequence-validated genes are represented here. (D) Functional classifications of ES-, TS-, MEF-, and stem cell-specific genes. The number of genes identified as specific to the cell type is indicated within parentheses. Annotated genes were classified by their function (Kargul et al. 2001), and are indicated on the right with its color co-ordinate. The unknown category includes sequences that are in the database but have yet to be characterized, as well as completely novel sequences.
Identification of Signature Genes
The pair-wise comparisons revealed many differentially-expressed genes, but multiple comparisons can reveal clusters of genes that are unique to a particular cell type or cell state, which are of particular interest here. To identify such genes, we first analyzed the four triplicate data sets by a statistical analysis of variance (ANOVA) at a 5% level of significance, and identified 3,910 genes differentially expressed among the four cell types. Of these genes, 2,969 sequence-verified genes were analyzed further. First, the number of gene clusters was estimated to be 15 by Partek program. We then performed k-means clustering to group the 2,969 genes into 15 distinctive clusters based on the similarities of their expression patterns (Chen et al. 2002). The average expression levels for each cluster of genes were plotted (Fig. 1C). We identified clusters representative of particular cell types, and further focused on genes that showed greater than twofold expression difference among ES, TS3.5, TS6.5, and MEF cells. We obtained 124 ES-specific genes in cluster 4, 94 TS-specific genes in cluster 7, and 77 MEF-specific genes in cluster 14. The 51 genes in clusters 12 and 13 were expressed in both ES and TS cells, but not in MEF cells, and are therefore designated as potential “stem cell-specific” genes. A total of 346 genes were therefore identified as signature genes that are characteristic of one or another cell type and investigated further below (lists of genes are available in Supplemental Tables 2 and 3). Although the ES/TS cluster is small, and half of the genes are novel, we did observe two genes involved in the inositol phospholipid signaling pathway, myo-inositol 1-phosphate synthase A1 and diacylglycerol kinase a (Szebenyi and Fallon 1999). Furthermore, an in vivo comparison clustered these two genes into a preimplantation embryo-specific cluster (Fig. 2, cluster D). These results indicate that a common signal transduction pathway, working in both ES and TS cells (and the preimplantation embryo), may have a common function such as self-renewal.
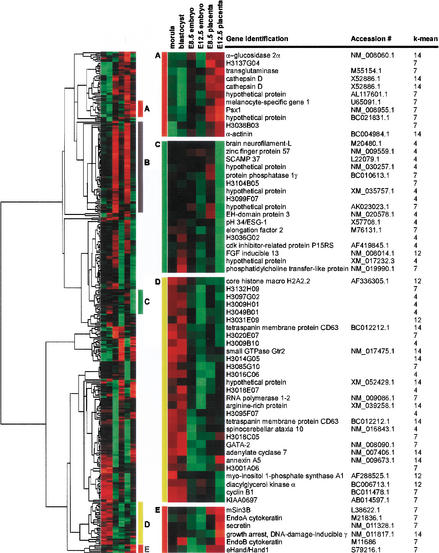
Figure 2.
List of genes showing unique cluster-patterns. Based on hierarchical clustering of 346 genes expressed specifically in either ES, TS, MEF, or in stem cells, we could identify five distinct clusters: Clusters A and E are for extraembryonic cell-lineage-specific clusters; B is for an E8.5 stage-specific cluster; C is a low expression cluster; D is for preimplantation embryo-specific cluster. Lists of genes in these clusters are shown. Cluster 4 in k-means analysis indicates that the gene is ES cell-specific, number 7 is for TS cell-specific, 14 is for MEF-specific, and 12 is for genes commonly expressed in both ES and TS cells.
Based on gene annotation (Kargul et al. 2001), these signature genes were classified into functional categories. Surprisingly, ES cells expressed many uncharacterized genes (67%), whereas the TS and ES/TS clusters exhibited fewer uncharacterized genes (51% and 48%, respectively) and differentiated MEF cells showed the lowest representation of novel genes (34%; Fig. 1D). Well-characterized differentiation markers for mesoderm and endoderm were not found in the ES cell signature gene list (Supplemental Table 3) and many genes uniquely expressed in ES cells have not been studied thus far and remain uncharacterized. In contrast, TS cell signature genes included transcription factors and other genes with well-defined roles in placental differentiation, such as eHand/Hand1, Pem, and Psx1. This indicates that one of the conspicuous features of lineage-committed TS cells is the expression of marker genes for differentiated trophoblast lineages.
Cell Culture and In Vivo cDNA Microarray Comparisons
To determine whether there is any relationship between clusters identified from the cell culture lines and clusters identified from their in vivo counterparts, we compared the present results with microarray data obtained from the following in vivo tissues: morulae, blastocysts, E8.5 embryo, E8.5 placenta, E12.5 embryo, and E12.5 placenta (Fig. 1A) (Tanaka et al. 2000). The comparison was performed by selecting the 346 signature genes and hierarchically clustering them based on their expression in vivo (Eisen et al. 1998).
Gene clusters with related expression patterns were easily discernable. First, genes with relatively high expression in E8.5 and E12.5 placentas were grouped in clusters A and E (Fig. 2; lists of genes available at http://lgsun.grc.nia.nih.gov/microarray/data.html). Most of these genes fell into the TS-specific k-means cluster (7), and therefore support the notion that TS cells already express marker genes for differentiated trophoblast lineages, including eHand/Hand1, EndoA, EndoB, Transglutaminase, and Psx1. Second, two stage-specific clusters were apparent. Genes with relatively high expression levels in morula and blastocyst and lower expression levels in other tissue types were grouped in cluster D and genes with high expression in both the E8.5 embryo and placenta were grouped in cluster B (Fig. 2). Mapping the cell line-derived k-means clusters onto these stage-specific clusters revealed a random distribution. This indicated that the cell lines were not representative of a particular stage of development. Third, genes with relatively low expression levels in essentially all tissues examined were grouped into cluster C. This group included many genes that were identified as ES-specific genes (k-means cluster 4), indicating that this cluster contains genes that are truly unique to ES cells and may be critically involved in the pluripotency of ES cells. This cluster included genes such as zinc finger protein 57, secretory carrier membrane protein (SCAMP) 37, EH-domain containing protein 2, Cdk inhibitor-related protein P15RS, and five uncharacterized genes, including embryonal stem cell specific gene 1 (Esg-1) (Fig. 2). It is notable that analysis of in vivo embryonic tissues alone would not have focused on many of these pluripotency-associated genes; the use of purified, established stem cells identified genes, such as Esg-1, that are apparently associated with pluripotency.
Examples of TS-Specific Genes
The expression and localization of several uncharacterized genes that are TS-specific by pair-wise comparisons were analyzed further by Northern blotting and in situ hybridization. Three clones, H3005B12, H3052E12, and H3138C09, all exhibited expression in TS6.5 cells and were undetectable in ES cells, in agreement with the microarray data. H3138C09 was found to be expressed in E9.5 placenta (data not shown) and E12.5 placenta, but not in the embryo at these stages (Fig. 3A). This novel, trophoblast-restricted gene encodes a putative 428-amino-acid protein with a RING zinc finger domain, a protease-associating (PA) domain, a signal peptide, and a single transmembrane domain. A fourth TS-specific gene, H3001A06, was highly expressed in the E9.5 placenta and decreased in later stage placental tissues. Similar to H3138C09, it was undetectable in embryos at the same stages. In situ hybridization at E9.5 revealed its expression in all trophoblast lineages, but not in allantoic mesoderm or decidua. By E11.5, its expression was weak, though still detectable in the labyrinth and spongiotrophoblast (Fig. 3B).
Figure 3.
(A, top) Three previously uncharacterized clones showing statistically significant high expression in TS6.5 cells were used as probes for Northern blots. The probe for β-actin was used as a control. (Middle) DNA probe for H3138C09 showed E12.5 placenta-specific signal by Northern hybridization. The probe for β-actin was used as a control. (Bottom) Schematic representation of one novel TS-specific gene, TC527107, from the TIGR Gene Index (Quackenbush et al. 2001). The cDNA clone H3138C09 is localized to the 3′UTR of this gene. See text for details. (B, top) Total RNA was extracted from extraembryonic and embryonic parts at the different stages indicated and transferred onto nylon membranes, which were probed with either H3001A06 or β-actin and exposed for 8 d to a Phosphorscreen. (Middle) DIG-labeled RNA probe for H3001A06 was hybridized in situ to a section of E9.5 conceptus. (Bottom left) HE-staining shows three different layers of the placenta at E11.5. Bottom right: DIG-labeled RNA probe for H3001A06 was hybridized in situ to a section of E11.5 placenta. The signals were weak, but detectable in the spongiotrophoblast and labyrinthine trophoblast layers, but undetectable in surrounding decidua. (TG2) Secondary trophoblast giant cells; (De) decidua; (P1) placenta; (A1) allantois; (Ex) exocoelom; (vYS) visceral yolk sac; (Sp) spongiotrophoblast layer; (La) labyrinthine trophoblast layer.
Although only four genes from the TS cell signature genes were examined, these results already indicate a trend—lineage-committed TS cells already express genes of the later placenta. This again contrasts with the ES cell signature genes. There may still be genes, however, that are unique to TS cells but are not expressed in mature trophoblast cells. An FGF-inducible 13 gene is such an example, as it was expressed in a TS-specific manner and grouped in cluster 12 (Fig. 1C), but was not expressed in E8.5 and E12.5 placentas (Fig. 2). Further analysis of the microarray data may provide candidates for such genes.
Coexpression of the ES-Specific Gene Esg-1 and Oct3/4
Among the many novel ES-specific genes, we selected Esg-1 for further study, because the gene showed the greatest expression difference in ES:TS comparisons (Fig. 1B). It was originally described as a gene down-regulated during retinoic acid-induced differentiation of embryonic carcinoma (EC) cells (Astigiano et al. 1991), and later as a gene expressed uniquely in ES cells and preimplantation embryos (Bierbaum et al. 1994). We confirmed the previous report by showing specific expression in ES and EC cells with no detectable expression in a variety of other cell lines (Fig. 4A). We also found no expression in adult tissues such as heart, brain, spleen, lung, liver, skeletal muscle, kidney, and testis, and in postimplantation embryos at E7, E11, E15, and E17 (data not shown). Semi-quantitative RT-PCR analysis also showed that Esg-1 transcripts were present at low levels in the unfertilized egg, but increased dramatically by the zygotic genome activation at the two-cell stage, and continued throughout preimplantation stages to the blastocyst (Fig. 4B). This behavior is slightly different from that of Oct3/4, which is present in oocytes but is not zygotically activated until the eight-cell stage (for review, see Pesce et al. 1998). The inference of early zygotic formation of Esg-1 was supported further by showing that its expression was moderately sensitive to α-amanitin, rather like the Hsp70.1 gene, the earliest known marker gene for zygotic genome activation (Christians et al. 1995), and unlike the Oct3/4 gene, which is insensitive (Pesce et al. 1998) (Fig. 4C).
Figure 4.
(A) Northern hybridization of Esg-1. (Left) Total RNA from ES, TS3.5, TS6.5, and MEF cells were blotted onto a nylon membrane. The image of ribosomal RNA is shown as a loading control. (Right) poly(A)+ RNA from multiple cell-lines were blotted onto a nylon membrane (Clontech). Cell lines used for this blot are (1) neuroblastoma NB41A3; (2) mastocytoma P815; (3) lymphoma P388D1; (4) lymphocytic leukemia L1210; (5) lymphoma R1.1; (6) hepatoma Hepa1–6; (7) embryonic carcinoma P19; (8) subcutaneous connective tissue-type L-M; (9) fibroblast M-MSVBALB/3T3; (10) fibroblast k-BALB; (11) Abelson murine leukemia virus-induced tumor RAW264.7; (12) lymphoid tumor PU5–1.8. (B–D) Semi-quantitative RT-PCR analysis in preimplantation embryos. Primer pair for EF1α was used as a loading control. (B) Expression patterns of Esg-1 and Oct3/4 were examined in unfertilized eggs (UN; 278 eggs), fertilized eggs (1; 248 zygotes), and preimplantation embryos at stages of 2-, 4-, and 8-cells (289, 312, and 181 embryos, respectively), morula (M; 144 embryos), and blastocyst (B; 175 embryos). It was confirmed that RT negative controls of each sample gave no PCR products. Only RT negative control of blastocyst (−) is shown. Each cDNA was first adjusted to one egg or one embryo-equivalent and then serially diluted as indicated to the right. (C) Early zygotic transcription-dependent expression of Esg-1 was confirmed by α-amanitin treatment of fertilized eggs. Fertilized eggs with a polar body were collected at 27 h post-hCG injection and either uncultured (1; 313 zygotes), or cultured for 18 h in M16 with (+; 207 embryos) or without (−; 230 embryos) 100 μg/mL α-amanitin. Two-cell stage embryos (vivo; 226 embryos) were also collected directly from oviducts in parallel. Serially-diluted cDNAs were used as indicated to the right. Embryos were pooled from four series of experiments. Hsp70.1 was used as a positive control. On average, 75% and 67% of the fertilized eggs developed to two-cell stage embryos in the absence and presence of α-amanitin, respectively. (D) Expression level of Esg-1 in the ICM and TE was investigated by direct isolation of the ICM and TE by immunosurgery and FITC-ConA labeling followed by trypsinization, respectively. A combination of serial dilution of cDNAs and different PCR-cycles were indicated. Fgf4 and Oct3/4 were known to be expressed in the ICM, whereas EndoA is a marker of the TE (Brulet and Jacob 1982). (E) Real-time RT-PCR analysis of Esg-1 and Oct3/4 in manipulated ES cells. Two manipulated ES cell-lines either expressing active Stat3 conditionally, or suppressing Oct3/4 conditionally were used as sources of RNA. MEF cells were used as a negative control. The expression level of Oct3/4 was also measured as a marker of pluripotency. The expression levels of Esg-1 and Oct3/4 were normalized to those of EF1α, whose constant expression levels over different conditions were verified (data not shown). The levels of Esg-1 and Oct3/4 in the manipulated cell lines were compared with those of nonmanipulated ES cells grown under standard conditions.
Several lines of evidence further support the notion that Esg-1 expression is restricted to pluripotent cell types (Pesce et al. 1998). A BLAST search against dbEST (Boguski et al. 1993) detected Esg-1 only in cDNA libraries made from preimplantation embryos, ES cells, and EC cells (though one EST each was recovered from bone marrow and testis). BLAST searches against the Orthologous Gene Database (Lee et al. 2002) revealed an Esg-1 ortholog only in human and pig, indicating that, like the Oct3/4 gene, Esg-1 seems to occur only in mammals. In addition, the ESTs representing the human Esg-1 ortholog were all from germ-line tumors. Four ESTs for Esg-1 were also found among 1,973 ESTs from E13.5 male primordial germ cells (Abe et al. 1998), indicating a relatively high expression in the germ line (data not shown). cDNA microarray analysis showed that compared with adult stem cells such as hematopoietic stem cells, neural stem cells, and mesenchymal stem cells, ES cells are the only stem cells that express Esg-1 at a high level (data not shown). Taken together, these data establish a remarkable similarity of gene expression patterns between Esg-1 and Oct3/4.
Location of Esg-1 in a Regulatory Pathway
To obtain further insight into a possible involvement of Esg-1 in the well-established Oct3/4 and LIF–Stat3 pluripotency pathways (for review, see Niwa 2001), we performed real-time RT-PCR analysis using RNA from two ES cell lines manipulated genetically in vitro.
First, we measured the expression levels of Esg-1 in ES cells carrying a 4HT-inducible Stat3 expression construct, as Stat3 has been shown to be the key downstream effector of the LIF receptor complex. The presence of constitutively active Stat3 expression has been shown to be sufficient to maintain ES cells in an undifferentiated state even in the absence of LIF. The absence of constitutively active Stat3 expression in LIF-depleted ES cells results in differentiation of ES cells into mainly primitive endoderm-like cells by 5 d (Matsuda et al. 1999). As shown in Figure 4E, the expression levels of both Esg-1 and Oct3/4 were maintained in the Stat3+ ES cells. In the Stat3− ES cells, however, Esg-1 was dramatically down-regulated, whereas Oct3/4 only exhibited a modest decrease in expression. This clearly associates the expression of Esg-1 with the undifferentiated state of ES cells, and indicates that the LIF effector, Stat3, may regulate Esg-1 expression directly or indirectly.
We then followed the temporal expression profile of Esg-1 after suppression of Oct3/4 in ES cells, using a tetracycline-inducible system (Niwa et al. 2000). When the expression level of Oct3/4 was completely shut down at 24 h, expression of Esg-1 was already down-regulated (Fig. 4E). Prompt initiation of Esg-1 down-regulation by the forced reduction of Oct3/4 indicates that Esg-1 is one of the early response genes to Oct3/4 suppression. Complete down-regulation of Esg-1 was slow, however, reaching a 10-fold reduction only after 94 h (Fig. 4E). This is consistent with the delayed down-regulation of Esg-1 in TE cells in vivo; the expression level of Esg-1 showed no significant difference between ICM and TE by semi-quantitative RT-PCR analysis (Fig. 4D), with FGF4, Oct3/4, and EndoA showing the specific expression patterns expected from previous reports (Niswander and Martin 1992; Palmieri et al. 1994). This discrepancy of Esg-1 expression between TE and TS cells indicates that the two types of cells are not equivalent, although TS cells are a direct in vitro derivative of TE.
Taken together, these data indicate that Esg-1 lies downstream of both Oct3/4 and LIF–Stat3 pathways. Although the actual binding and transcriptional activation of Esg-1 by Oct3/4 and Stat3 remains to be tested, analysis of mouse (Ensembl) and human (Ensembl, NCBI) genomic DNA sequences revealed potential binding sites for Oct3/4, Stat3, and Sox2 (a coactivator of Oct3/4) within 10 kb upstream and downstream of the Esg-1 gene (data not shown).
Esg-1 contains a KH-domain, which is conserved among RNA-binding proteins such as the Fragile X mental retardation gene (FMR1), Bicaudal C in Drosophila, and mex-3 and gld-1 in Caenorhabditis elegans (for review, see Adinolfi et al. 1999). The gene may therefore function post-transcriptionally as an RNA-binding protein. This would give Esg-1 a unique position in the Oct3/4 and Stat3 pathways, in which all other known participants are transcription factors. It is notable that the MEX-3 is a component of P-granules, germ-line determinants in C. elegans (Draper et al. 1996). Esg-1 is the first mammalian RNA-binding protein that shows a potential function in pluripotent stem cells and the germ-line.
Toward a Molecular Definition of “Stemness”
The commitment of totipotent embryonic cells to an extraembryonic lineage is unique to mammals and is critical to create the placenta that supports the development of the mammalian embryo. This analysis describes for the first time the global differences between expression profiles of the two stem cell lineages derived from the early embryo, and also identifies genes expressed specifically in TS and ES cells. The signature genes for ES cells are not detected as signature genes of any particular stage of in vivo development. In contrast, the signature genes for TS cells clearly show that this lineage-committed stem cell already expresses lineage-specific genes, in this case for placenta. This indicates that the pluripotent stem cell state may have special attributes not found in lineage-specific stem cells. Further comparison of other pluripotent and restricted stem cells should reveal whether this is a general rule and help define both “stemness” and pluripotency.
METHODS
RNA
Using the QIAGEN RNeasy midi kit (QIAGEN Inc., Santa Clarita, CA), total RNA was extracted from the R1 ES cell line (Nagy et al. 1993), derived originally from F1 hybrid mouse between 129X1 and 129S1, cultured on gelatin with LIF; from diploid-sorted TS cell lines (Tanaka et al. 1998) derived from ICR E3.5 blastocysts (TS3.5) and the extraembryonic ectoderm of E6.5 embryos (TS6.5), cultured in the presence of feeder-conditioned medium with FGF4 (Tanaka et al. 1998); and from mouse embryonic fibroblasts (MEF) derived from C57BL/6 (Hogan et al. 1994). The passage numbers of cells at the time of RNA extraction were as follows: 12 for ES, 40 for TS3.5, 28 for TS6.5, and two for MEF. Total RNA was similarly extracted from dissected embryonic and extraembryonic portions of E8.5, E9.5, E12.5, and E16.5 whole embryos from naturally-mated C57BL/6 females.
Hybridization
Microarray hybridization and Northern hybridization were performed essentially as described (Tanaka et al. 2000). Briefly, for microarray hybridization, 150 μg of each total RNA from ES, TS3.5, TS6.5, and MEF cells, and 10 μg of each total RNA from E8.5 embryonic and extraembryonic parts were primed with oligo(dT)12–18 primers (Amersham Biosciences, Piscataway, NJ). First strand cDNAs were synthesized with 33P-dCTP (Amersham Biosciences) and used as a probe. Prehybridization was carried out at 65°C for 4 h with MicroHyb solution (Invitrogen, Carlsbad, CA) supplemented with yeast tRNA and PolyA RNA, followed by hybridization at 65°C for 20 h with the same hybridization solution plus mouse Cot1 DNA, 8% Dextran Sulfate, and heat-denatured probe. Membranes were washed twice with 2×SSC, 0.1% SDS at room temperature for 30 min, then twice with 2×SSC, 0.1% SDS at 65°C for 30 min, and then twice with 0.1×SSC, 1%SDS at 65°C for 30 min. Dried membranes were exposed to a PhosphorScreen for 10 d at room temperature. Each probe hybridization was performed three times.
For Northern hybridization, 3 μg of each total RNA from ES, TS3.5, and TS6.5 cells, and embryonic fibroblasts, and 10 μg each from dissected embryos and placentas at E9.5, E12.5, and E16.5 were loaded on a formamide-gel and then blotted onto nylon membranes (Oncor, Gaithersburg, MD). For the detection of Esg-1, a multiple cell-line blot containing 1 μg each polyA+ RNA was purchased from Clontech (Clontech, Palo Alto, CA). Expresshyb solution (Clontech) was used for both prehybridization and hybridization. A radiolabeled SalI–NotI fragment containing full-length Esg-1 cDNA from plasmid DNA was used as a probe. After hybridization at 65°C for 20 h, both membranes were washed twice with 2×SSC at room temperature for 30 min, twice with 2×SSC at 65°C for 30 min, and then with 0.1×SSC at 65°C for 30 min, followed by exposure to a PhosphorScreen.
For in situ hybridization, digoxigenin (DIG)-labeled sense and antisense riboprobes were synthesized from linearlized plasmid DNA of H3001A06 and hybridized to paraffin sections of placentas at E9.5 and E11.5 as described earlier (Tanaka et al. 2000).
Data Analysis
Signal intensities of single spots were quantified by ImageQuant V. 5.1 (Amersham Biosciences). Other calculations, including statistical analysis, were done using Microsoft Excel. After subtraction of global background, normalization of microarray hybridization experiments was done by adjusting the total sum of signal intensities for all individual clones to 1010. Statistical analysis allowed us to distinguish signals with significantly higher intensities than background (P < 0.05). Expression levels of individual clones within the same condition were averaged, followed by either pair-wise comparisons between two conditions based on Student's t-test, or comparisons across particular sets of conditions based on analysis of variance (ANOVA) tests. Scatter plots were drawn by Spotfire DecisionSite 6.1 (Spotfire, Somerville, MA) to see the global expression profiles for each probe. Within the statistically significant, sequence-verified clones, averaged expression levels of each clone for individual conditions were divided by average expression level among all the conditions to calculate the relative expression levels of clones (expression ratio). These levels were further log-transformed and normalized to perform k-means cluster analysis by J-Express (Dysvik and Jonassen 2001). Cluster number was predicted by the Partek Pro 5.0 program (Partek, St. Charles, MO). Centered data sets of 346 signature genes described above were further grouped by a hierarchical clustering method (Eisen et al. 1998). All of the expression data are available at the National Institute on Aging Web site (http://lgsun.grc.nia.nih.gov/microarray/data.html).
Semi-Quantitative RT-PCR
Embryos were collected at preimplantation stages and mRNA extraction was followed by semi-quantitative RT-PCR analysis as described below. Treatment of fertilized eggs with α-amanitin (Sigma, St. Louis, MO) was performed essentially as described previously (Christians et al. 1995). ICM and TE were isolated by immunosurgery (Solter and Knowles 1975), and FITC-ConA (Sigma) labeling was followed by trypsinization (Collins and Fleming 1995). Briefly, pooled embryos were lysed with extraction buffer containing yeast tRNA as a carrier, followed by mRNA extraction using a MicroPrep mRNA kit (Amersham Biosciences). First-strand cDNA synthesis was carried out by priming with oligo(dT) (Amersham Biosciences), followed by an extension reaction with Superscript II reverse transcriptase (Invitrogen). Half of the mRNA for individual samples was used in each case for an RT-negative reaction. Reaction mixtures for PCR, including Taq polymerase (Invitrogen), were prepared as suggested by the manufacturer. Unless noted, PCR conditions were as follows: first, denaturing at 95°C for 1 min, 40 cycles of denaturing at 94°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 1 min, followed by a final extension reaction at 72°C for 3 min. Primer pairs used in this study have been described: Oct3/4 and Fgf4 (Nichols et al. 1998) and EF1α (Chambers et al. 1998), except for Esg-1 (501-bp product, forward ATAAGCT TGATCTCGTCTTCC, reverse CTTGCTAGGATGTAACAAA GC); EndoA (289-bp product, forward AAAGGCCAGAGG GCATCGTTGG, reverse TCAGTCCTCCTGAGTAGCCGCT); Hsp70.1 (564-bp product, forward AAGAACGCGCTCG AGTCCTATGC, reverse CTGGTACAGTGCACAGTGCTGCT).
Real-Time RT-PCR
Quantitative real-time PCR (Q-PCR) was performed on total RNA from two different ES cell lines. (1) ES cells expressing active Stat3 conditionally in the presence of 4-hydroxytamoxifen (4-HT), a ligand for the modified estrogen receptor whose ligand-binding domain was fused to the entire coding sequence of Stat3 (Matsuda et al. 1999). Cells were cultured for 5 d without LIF in the presence or absence of 4-HT; and (2) ES cells repressing Oct3/4 conditionally in the presence of tetracycline (Tet) (Niwa et al. 2000). Cells were cultured with LIF in the presence of Tet for 0, 24, 48, and 96 h. RNA from each cell line was treated with RNase-free DNase (Ambion, Inc., Austin, TX) to remove genomic DNA contamination, and the concentration of total RNA was determined using an Agilent Labchip Bioanalyzer (Agilent Technologies, Palo Alto, CA). cDNA was synthesized from 2 μg of total RNA with Retroscript reverse transcriptase (Ambion) using random decamer primers. Q-PCR was performed on an ABI Prism 7700 sequence detection system (Applied Biosystems, Foster City, CA) using SYBR green chemistry. Gene expression was normalized relative to the endogenous gene EF1α and expressed relative to untreated controls using the comparative CT method (Applied Biosystems User Bulletin #2, P/N 4303859). The sequences of primers are as follows: (1) EF1α, forward primer ONO44F CGTCGTTCTGGTAAGAAGCTGGAAG, reverse primer ONO43R AACAGCAAAGCGACCAAGTGGAG; (2) Oct3/4, forward primer Oct3/4F GGCGTTCTCTTTGGA AAGGTGTTC, reverse primer Oct 3/4R CTCGAACCACAT CCTTCTCT; and (3) Esg-1 (175-bp product), forward primer GCCGTGCGTGGTGGATAAGC, reverse primer GCCAAACA GATATTTCAGCACCAGC.
Acknowledgments
We thank David Schlessinger for discussion and critical reading of the manuscript; Kevin Becker and William Wood III for microarray production; Robert Herrington for technical assistance; Dawood Dudekula and Yong Qian for gene annotation; and Kuniya Abe for the collaboration on PGC EST project. T.S.T. was a JSPS Research Fellow in Biomedical and Behavioral Research in NIH.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL KoM@grc.nia.nih.gov; FAX (410) 558-8331.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.670002.
REFERENCES
- Abe K, Ko MSH, MacGregor GR. A systematic molecular genetic approach to study mammalian germline development. Int J Dev Biol. 1998;42:1051–1065. [PMC free article] [PubMed] [Google Scholar]
- Adinolfi S, Bagni C, Castiglione Morelli MA, Fraternali F, Musco G, Pastore A. Novel RNA-binding motif: The KH module. Biopolymers. 1999;51:153–164. doi: 10.1002/(SICI)1097-0282(1999)51:2<153::AID-BIP5>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Astigiano S, Barkai U, Abarzua P, Tan SC, Harper MI, Sherman MI. Changes in gene expression following exposure of nulli-SCCl murine embryonal carcinoma cells to inducers of differentiation: Characterization of a down-regulated mRNA. Differentiation. 1991;46:61–67. doi: 10.1111/j.1432-0436.1991.tb00866.x. [DOI] [PubMed] [Google Scholar]
- Beddington RS, Robertson EJ. An assessment of the developmental potential of embryonic stem cells in the midgestation mouse embryo. Development. 1989;105:733–737. doi: 10.1242/dev.105.4.733. [DOI] [PubMed] [Google Scholar]
- Bierbaum P, MacLean-Hunter S, Ehlert F, Moroy T, Muller R. Cloning of embryonal stem cell-specific genes: Characterization of the transcriptionally controlled gene esg-1. Cell Growth Differ. 1994;5:37–46. [PubMed] [Google Scholar]
- Boguski MS, Lowe TMJ, Tolstoshev CM. dbEST-database for “expressed sequence tags.”. Nat Genet. 1993;4:332–333. doi: 10.1038/ng0893-332. [DOI] [PubMed] [Google Scholar]
- Brulet P, Jacob F. Molecular cloning of a cDNA sequence encoding a trophectoderm-specific marker during mouse blastocyst formation. Proc Natl Acad Sci. 1982;79:2328–2332. doi: 10.1073/pnas.79.7.2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers DM, Peters J, Abbott CM. The lethal mutation of the mouse wasted (wst) is a deletion that abolishes expression of a tissue-specific isoform of translation elongation factor 1α, encoded by the Eef1a2 gene. Proc Natl Acad Sci. 1998;95:4463–4468. doi: 10.1073/pnas.95.8.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Jaradat SA, Banerjee N, Tanaka TS, Ko MSH, Zhang MQ. Evaluation and comparison of clustering algorithms in analyzing ES cell gene expression data. Statistica Sinica. 2002;12:241–262. [Google Scholar]
- Christians E, Campion E, Thompson EM, Renard JP. Expression of the HSP 70.1 gene, a landmark of early zygotic activity in the mouse embryo, is restricted to the first burst of transcription. Development. 1995;121:113–122. doi: 10.1242/dev.121.1.113. [DOI] [PubMed] [Google Scholar]
- Collins J, Fleming T. Specific mRNA detection in single lineage-marked blastomeres from preimplantation embryos. Trends Genet. 1995;11:5–7. doi: 10.1016/s0168-9525(00)88975-1. [DOI] [PubMed] [Google Scholar]
- Draper BW, Mello CC, Bowerman B, Hardin J, Priess JR. MEX-3 is a KH domain protein that regulates blastomere identity in early C. elegans embryos. Cell. 1996;87:205–216. doi: 10.1016/s0092-8674(00)81339-2. [DOI] [PubMed] [Google Scholar]
- Dysvik B, Jonassen I. J-Express: Exploring gene expression data using Java. Bioinformatics. 2001;17:369–370. doi: 10.1093/bioinformatics/17.4.369. [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- Hemberger M, Cross JC, Ropers HH, Lehrach H, Fundele R, Himmelbauer H. UniGene cDNA array-based monitoring of transcriptome changes during mouse placental development. Proc Natl Acad Sci. 2001;98:13126–13131. doi: 10.1073/pnas.231396598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan B, Beddington R, Costantini F, Lacy E. Manipulating the mouse embryo: A laboratory manual. 2nd edition. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- Hosler BA, LaRosa GJ, Grippo JF, Gudas LJ. Expression of REX-1, a gene containing zinc finger motifs, is rapidly reduced by retinoic acid in F9 teratocarcinoma cells. Mol Cell Biol. 1989;9:5623–5629. doi: 10.1128/mcb.9.12.5623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kargul GJ, Dudekula DB, Qian Y, Lim MK, Jaradat SA, Tanaka TS, Carter MG, Ko MSH. Verification and initial annotation of the NIA mouse 15K cDNA clone set. Nat Genet. 2001;28:17–18. doi: 10.1038/ng0501-17. [DOI] [PubMed] [Google Scholar]
- Kim SK, Lund J, Kiraly M, Duke K, Jiang M, Stuart JM, Eizinger A, Wylie BN, Davidson GS. A gene expression map for Caenorhabditis elegans. Science. 2001;293:2087–2092. doi: 10.1126/science.1061603. [DOI] [PubMed] [Google Scholar]
- Kunath T, Strumpf D, Rossant J, Tanaka S. Trophoblast stem cells. In: Marshak DR, Gardner RL, Gottlieb D, editors. Stem cell biology. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. pp. 267–287. [Google Scholar]
- Lee Y, Sultana R, Pertea G, Cho J, Karamycheva S, Tsai J, Parvizi B, Cheung F, Antonescu V, White J, et al. Cross-referencing eukaryotic genomes: TIGR orthologous gene alignments (TOGA) Genome Res. 2002;12:493–502. doi: 10.1101/gr.212002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci. 1981;78:7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda T, Nakamura T, Nakao K, Arai T, Katsuki M, Heike T, Yokota T. STAT3 activation is sufficient to maintain an undifferentiated state of mouse embryonic stem cells. EMBO J. 1999;18:4261–4269. doi: 10.1093/emboj/18.15.4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy A, Rossant J, Nagy R, Abramow-Newerly W, Roder JC. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc Natl Acad Sci. 1993;90:8424–8428. doi: 10.1073/pnas.90.18.8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, Scholer H, Smith A. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–391. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- Niswander L, Martin GR. Fgf-4 expression during gastrulation, myogenesis, limb and tooth development in the mouse. Development. 1992;114:755–768. doi: 10.1242/dev.114.3.755. [DOI] [PubMed] [Google Scholar]
- Niwa H. Molecular mechanism to maintain stem cell renewal of ES cells. Cell Struct Funct. 2001;26:137–148. doi: 10.1247/csf.26.137. [DOI] [PubMed] [Google Scholar]
- Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- Okazawa H, Okamoto K, Ishino F, Ishino-Kaneko T, Takeda S, Toyoda Y, Muramatsu M, Hamada H. The oct3 gene, a gene for an embryonic transcription factor, is controlled by a retinoic acid repressible enhancer. EMBO J. 1991;10:2997–3005. doi: 10.1002/j.1460-2075.1991.tb07850.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri SL, Peter W, Hess H, Scholer HR. Oct-4 transcription factor is differentially expressed in the mouse embryo during establishment of the first two extraembryonic cell lineages involved in implantation. Dev Biol. 1994;166:259–267. doi: 10.1006/dbio.1994.1312. [DOI] [PubMed] [Google Scholar]
- Pesce M, Gross MK, Scholer HR. In line with our ancestors: Oct-4 and the mammalian germ. BioEssays. 1998;20:722–732. doi: 10.1002/(SICI)1521-1878(199809)20:9<722::AID-BIES5>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Quackenbush J, Cho J, Lee D, Liang F, Holt I, Karamycheva S, Parvizi B, Pertea G, Sultana R, White J. The TIGR Gene Indices: Analysis of gene transcript sequences in highly sampled eukaryotic species. Nucleic Acids Res. 2001;29:159–164. doi: 10.1093/nar/29.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosner MH, Vigano MA, Ozato K, Timmons PM, Poirier F, Rigby PW, Staudt LM. A POU-domain transcription factor in early stem cells and germ cells of the mammalian embryo. Nature. 1990;345:686–692. doi: 10.1038/345686a0. [DOI] [PubMed] [Google Scholar]
- Rossant J, Cross JC. Placental development: Lessons from mouse mutants. Nat Rev Genet. 2001;2:538–548. doi: 10.1038/35080570. [DOI] [PubMed] [Google Scholar]
- Scholer HR, Hatzopoulos AK, Balling R, Suzuki N, Gruss P. A family of octamer-specific proteins present during mouse embryogenesis: Evidence for germline-specific expression of an Oct factor. EMBO J. 1989;8:2543–2550. doi: 10.1002/j.1460-2075.1989.tb08392.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen MM, Leder P. Leukemia inhibitory factor is expressed by the preimplantation uterus and selectively blocks primitive ectoderm formation in vitro. Proc Natl Acad Sci. 1992;89:8240–8244. doi: 10.1073/pnas.89.17.8240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A. Embryonic stem cells. In: Marshak DR, Gardner RL, Gottlieb D, editors. Stem cell biology. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. pp. 205–230. [Google Scholar]
- Solter D, Knowles BB. Immunosurgery of mouse blastocyst. Proc Natl Acad Sci. 1975;72:5099–5102. doi: 10.1073/pnas.72.12.5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szebenyi G, Fallon JF. Fibroblast growth factors as multifunctional signaling factors. Int Rev Cytol. 1999;185:45–106. doi: 10.1016/s0074-7696(08)60149-7. [DOI] [PubMed] [Google Scholar]
- Tanaka S, Kunath T, Hadjantonakis AK, Nagy A, Rossant J. Promotion of trophoblast stem cell proliferation by FGF4. Science. 1998;282:2072–2075. doi: 10.1126/science.282.5396.2072. [DOI] [PubMed] [Google Scholar]
- Tanaka TS, Jaradat SA, Lim MK, Kargul GJ, Wang X, Grahovac MJ, Pantano S, Sano Y, Piao Y, Nagaraja R, et al. Genome-wide expression profiling of mid-gestation placenta and embryo using a 15,000 mouse developmental cDNA microarray. Proc Natl Acad Sci. 2000;97:9127–9132. doi: 10.1073/pnas.97.16.9127. [DOI] [PMC free article] [PubMed] [Google Scholar]