Abstract
The PGC-1s (peroxisome-proliferator-activated receptor γ co-activators) are a family of transcriptional regulators that induce the expression of various metabolic genes. PGC-1 proteins stimulate genes involved in mitochondrial biogenesis, fatty acid oxidation and hepatic gluconeogenesis. Previous studies have demonstrated that the PGC-1α and β isoforms interact with nuclear receptors through the conserved LXXLL (leucine-X-X-leucine-leucine) motifs. In the present study, we have investigated the mechanisms by which these PGC-1 isoforms stimulate gene expression. We have determined that the N-terminus of PGC-1 is responsible for transcriptional activation. Two conserved peptide motifs were identified in the N-terminus of PGC-1α and β isoforms. These domains were named AD1 and AD2 (activation domain 1 and 2). Deletion of both of these motifs decreased the induction of various PGC-1-regulated genes including the PEPCK (phosphoenolpyruvate carboxykinase) and CPT-I (carnitine palmitoyltransferase-I) genes. It was determined that amino acids containing a negative charge in AD1 and the leucine residues in AD2 were important for the transcriptional induction of the PEPCK and CPT-I genes. Disruption of the AD motifs did not diminish the ability of the PGC-1α protein to associate with the PEPCK or CPT-I genes. In addition, deletion of the AD domains did not eliminate the ability of PGC-1α to interact with the thyroid hormone receptor. The data indicate that the AD1 and AD2 motifs mediate the induction of many PGC-1- responsive genes, but they do not contribute to the recruitment of PGC-1 to target genes.
Keywords: activation domain, carnitine palmitoyltransferase, peroxisome-proliferator-activated receptor γ co-activator (PGC-1), phosphoenolpyruvate carboxykinase (PEPCK), pyruvate dehydrogenase kinase
Keywords: AD, activation domain; CBP, CREB (cAMP-response-element-binding protein)-binding protein; ChIP, chromatin immunoprecipitation; CMV, cytomegalovirus; CPT, carnitine palmitoyltransferase; FOXO1, forkhead transcription factor; GST, glutathione S-transferase; HNF4, hepatic nuclear factor 4; LXXLL, leucine-X-X-leucine-leucine; PDK4, pyruvate dehydrogenase kinase 4; PEPCK, phosphoenolpyruvate carboxykinase; PGC-1, peroxisome-proliferator-activated receptor γ; co-activator 1; PRC, PGC-related co-activator; PRMT-I, protein arginine methyltransferase I; SRC-1, steroid receptor co-activator-1; T3, thyroid hormone; TR, thyroid hormone receptor; TRAP, thyroid-hormone-receptor-associated protein; TRE, thyroid hormone response element
INTRODUCTION
Transcription of genes is a highly regulated event. In addition to the basal transcription machinery, the control of gene expression involves participation of numerous other proteins including transcription factors, nuclear receptors and co-activators. Co-activators modulate transcription not through directly binding DNA but instead through interactions with nuclear receptors and other transcription factors to increase their ability to stimulate transcription [1]. The common mechanisms by which co-activators facilitate transcription include their inherent enzymatic activities such as histone acetylation and methylation as well as their ability to remodel the chromatin structure [1,2]. Some key co-activators include CBP/p300 [CREB (cAMP-response-element-binding protein)-binding protein], SRC-1 (steroid receptor co-activator-1) and TRAP (thyroid-hormone-receptor-associated protein)/mediator among many others [3].
The PGC-1 (peroxisome-proliferator-activated receptor γ co-activator-1) family of co-activators are crucial regulators of cellular metabolism and energy homoeostasis [4,5]. The three isoforms of PGC-1 include PGC-1α, PGC-1β and PRC (PGC-related co-activator) [6]. These co-activators are unique in the fact that their expression is highly regulated by external stimuli. PGC-1α expression is induced by cold, fasting, exercise and hormones, while thyroid hormone, fasting and high-fat diets elevate PGC-1β [7–10]. PRC is a growth-induced co-activator which is up-regulated during the serum-induced G0 to G1 transition from the quiescence to proliferative growth in fibroblasts [11]. Both PGC-1α and PGC-1β share some common actions, but at the same time there are functions that are exclusively mediated by each isoform. For example, both isoforms induce mitochondrial oxidative metabolism, fatty acid oxidation and lipoprotein secretion [8,12–15]. However, only PGC-1α stimulates hepatic gluconeogenesis and haem biosynthesis [16,17]. Similarly only PGC-1β promotes fatty acid synthesis as well as inhibiting macrophage-mediated inflammation [9,15,18]. The similarities and differences in biological functions probably reflect the various subsets of nuclear receptors interacting with these proteins in a particular gene or pathway. It has been observed that the PGC-1 co-activators lack histone acetylation activity. They are believed to function by docking to specific nuclear receptors and stimulating transcription directly or through the recruitment of other coactivators such as CBP/p300, p160/SRC-1 and TRAP/mediator [19,20].
PGC-1 proteins have a modular structure with different regions of the co-activator required for different functions. Their N-terminus has the characteristic LXXLL (leucine-X-X-leucine-leucine; where X is any amino acid) domain that mediates key interactions of these proteins with the nuclear receptors [7]. The transcriptional activation function of PGC-1α has been ascribed to its N-terminus [23]. Studies in our laboratory and by others have shown that this transcriptional activation function is independent of the LXXLL motif [20,21]. Other domains that have been characterized in PGC-1α are the repression domain (amino acids 200–400), a C-terminal RS (serine/arginine rich) domain and a RRM (RNA recognition motif) which interacts with the RNA-processing machinery [22]. It has been reported that PGC-1α co-activator activity is greatly augmented by arginine methylation by PRMT-I (protein arginine methyltransferase I) on the C-terminus of the protein. However, it was found that an intact activation domain is required for functional synergy by PGC-1α and PRMT-I [24].
PEPCK (phosphoenolpyruvate carboxykinase) is a key regulatory enzyme in the pathway of gluconeogenesis [25]. PGC-1α induces the PEPCK gene via interactions with HNF4 (hepatic nuclear factor 4), the GR (glucocorticoid receptor) and FOXO1 (forkhead transcription factor) [17,26]. In addition, we have found that PGC-1α stimulates the expression of PDK4 (pyruvate dehydrogenase kinase 4) which regulates the activity of the pyruvate dehydrogenase complex [27,28]. PGC-1α directly induces CPT-Iα (carnitine palmitoyltransferase Iα) gene expression and participates in the T3 (thyroid hormone) induction of CPT-Iα [29]. CPT-Iα is a rate-controlling step in the mitochondrial oxidation of long-chain fatty acids [30]. PGC-1α enhances the T3 induction of CPT-Iα both through interactions with the TR (T3 receptor) and elements in the intron [21,31]. In the present study we have defined a common mechanism by which PGC-1α and β isoforms elicit the stimulation of transcription. We identified two conserved regions in the N-terminus of PGC-1α and PGC-1β that are crucial for transcriptional activation. We refer to these peptide motifs as AD1 (activation domain 1) and AD2 (activation domain 2). We have found that deletion of these domains in PGC-1 proteins results in a dramatic loss of function of these proteins. We have further characterized these domains and the effect of their deletions on the ability of PGC-1 to stimulate known target genes.
MATERIALS AND METHODS
Transient transfections and luciferase assays
HepG2 cells were transfected using the calcium phosphate transfection technique as described previously [31]. Briefly the cells were cultured in DMEM (Dulbecco's modified Eagle's medium) containing 10% foetal bovine serum and 1% penicillin/streptomycin. The cells were split into 60 mm dishes 4 h prior to transfection. Indicated amounts of plasmid DNA were transfected into the cells. The following day the cells were washed and the medium was changed. After transfection (40 h) the cells were harvested and the luciferase assay was performed using the Promega Dual Luciferase assay kit. The luciferase values were normalized for Renilla activity and protein content. Each experiment was repeated 3–4 times and average values were computed. McA-RH7777 cells were transfected using Lipofectamine™ 2000 following the manufacturer's protocol.
Site-directed mutagenesis
The QuikChange site-directed mutagenesis kit (Stratagene) was used to carry out both the deletion mutagenesis as well as the amino acid substitutions in the PGC-1 proteins. The primers used for deletion mutagenesis were: PGC-1α AD1 sense, 5′-CCAGCCTCTTTGCCCAGTGAATGACTTGGATACA-3′; PGC-1α AD1 antisense, 5′-TGTATCCAAGTCATTCACTGGGCAAAGAGGCTGG-3′; PGC-1α AD2 sense, 5′-GAGAAGATAGATGAAGAGAGTCTCCCCGTGGATG-3′; PGC-1α AD2 antisense, 5′CATCCACGGGGAGACTCTCTTCATCTATCTTCTC-3′; PGC-1β AD1 sense, 5′-GGAACAGCTGTGTGCTGCCAGTGACTTTGACT-3′; PGC-1β AD1 antisense, 5′-AGTCAAAGTCACTGGCAGCACACAGCTGTTCC-3′; PGC-1β AD2 sense, 5′-CCAGATTGACAGTGAGGACATCCCCGAAGACG-3′; PGC-1β antisense, 5′-CGTCTTCGGGGATGTCCTCACTGTCAATCTGG-3′. These deletions were carried out in both the Gal4-PGC-1 or CMV-PGC-1 expression vectors (where CMV is cytomegalovirus).
GST (glutathione S-transferase) pulldown assay
GST-fusion proteins were made by cloning the first 170 amino acids of the N-terminus of PGC-1α into the pGEX 4T.1 vector. Also, we cloned the first 170 amino acids of PGC-1α with AD1 and AD2 deleted into this expression vector. These plasmids along with a GST control were transformed into BL21 bacterial cells. GST-fusion protein expression in bacterial cultures was induced by the addition of 1 mM isopropyl-β-D-1-thiogalactopyranoside. GST proteins were purified with GST-binding resin (Novagen) and eluted with reduced glutathione. Purified proteins were subjected to SDS/PAGE and detected using Coomassie Blue staining. The interaction of PGC-1α proteins with TRβ was tested using [35S]methionine-labelled TRβ prepared by in vitro reticulocyte translation reaction (Promega TNT kit Catalog No. L1170). To perform the pulldown assay, equal amounts of GST proteins (10 μg) were allowed to interact with 10 μl of 35[S]methionine TRβ in a binding buffer [20 mM Hepes (pH 7.5), 75 mM KCl, 0.1 mM EDTA, 2.5 mM MgCl2, 0.05% Nonidet P40, 2 mM DTT and 10% glycerol] overnight at 4 °C. T3 was added at a concentration of 1 μM in this binding reaction. The next morning 40 μl of GST sepharose beads were added to the binding reaction and the bound proteins were collected for 2 h. The resin was washed four times in the binding buffer and the bound proteins were eluted using SDS/PAGE sample buffer. The eluted proteins were resolved on SDS/PAGE (12% gels). The gel was fixed and was treated with fluorescence enhancer (Amersham Biosciences). The gel was exposed to film and the 35[S]TR was detected autoradiographically.
ChIP (chromatin immunoprecipitation) assays
ChIP assays were conducted in McA-RH7777 cells. We infected these cells with adenovirus expressing either a wild-type PGC-1α or a mutant PGC-1α with AD1 and AD2 deletions. After infection (40 h) the cells were cross-linked using 1% formaldehyde. The cross-linking reaction was stopped using 0.125 M glycine. The cells were washed twice with ice-cold PBS and harvested in PBS. The cells were collected by centrifuging at 805 g for 5 min. The pellet obtained was lysed in cell lysis buffer [50 mM Tris/HCl (pH 8.0), 85 mM KCl and 0.5% Nonidet P40) for 0.5 h in ice with occasional vortexing. The samples were spun down at 3200 g for 5 min and the pellet obtained was resuspended in the SDS lysis buffer [1% SDS, 10 mM EDTA and 50 mM Tris/HCl (pH 8.1)]. Lysis was allowed to occur on ice for 10 min. The samples were then sonicated to shear the DNA. The sonication was carried out in a Branson 450 sonicator (12 cycles of 30 s each at 57% amplitude). The DNA was sheared to 500–1000 bp and shearing efficiency was confirmed on a 1% agarose gel. The sonicated and sheared DNA was clarified by centrifuging at 16100 g for 10 min. We diluted 200 μl of the DNA to 1800 μl with ChIP dilution buffer [0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris/HCl (pH 8.1) and 167 mM NaCl] containing protease inhibitor cocktail (Sigma). The samples were precleared using 60 μl of Protein A agarose/salmon sperm DNA (Upstate Biotechnology) for 30 min at 4 °C. The beads were removed by spinning down and the immunoprecipitating antibody (PGC-1α, sc-13067 Santa Cruz) was added. No antibody pulldowns and normal rabbit IgG were used as controls. The pulldown was performed overnight at 4 °C. The next morning 40 μl of Protein A agarose/salmon sperm DNA slurry was added to collect the immunoprecipitated DNA for 2 h. The immunoprecipitated DNA was washed extensively in a series of wash buffers. Finally DNA was eluted using elution buffer (1% SDS and 0.1 M NaHCO3). The cross-linking on the eluted DNA protein complexes was reversed using 30 μl of 5 M NaCl at 65 °C for 4 h. The DNA was precipitated using ethanol at −20 °C. The DNA pellet was resuspended in distilled water and treated with proteinase K to digest the proteins at 45 °C for 1 h. The DNA was purified using a Qiagen PCR purification kit. The final elution was performed in distilled water. The purified eluted DNA was subjected to 35 cycles of PCR using 3–5 μl of DNA. The PCR products were analysed on 2% Nusieve 3:1 agarose (Cambrex, 50094) and visualized with a MultiImage Light Cabinet with Quantity One software.
Western blot analysis
McA-RH777 cells were infected with adenovirus expressing wild-type or mutant PGC-1 for 40 h. After 40 h the cells were harvested and lysed in lysis buffer [40 mM Tris/HCl (pH 8.0), 500 mM NaCl, 0.5% Nonidet P40, 6 mM EDTA, 6 mM EGTA, pH 8.0, 1 mM dithiothreitol and diluted protease inhibitor mixture; P8340, Sigma]. The lysates were quantified and equal amounts of protein were run on a 7.5% acrylamide gel. The proteins were transferred on to 0.45 μm nitrocellulose membrane. The membrane was probed with anti-FLAG antibody (1:1000, Sigma). After probing with an anti-rabbit antibody the immunoreactive proteins were detected using Super Signal chemiluminescence substrate (Pierce).
Real-time PCR
McA-RH7777 cells were transfected with CMV-flagPGC-1wt and CMV-flagPGC-1α ΔAD1/ΔAD2 expression vectors using Lipofectamine™ 2000 (Invitrogen). After 48 h the cells were harvested and RNA was isolated using RNA STAT 60. The RNA obtained was subjected to DNase I treatment. cDNA was made using the Invitrogen SuperScript II cDNA synthesis kit. The cDNA was diluted 1:50 and subjected to real-time PCR using SYBR® Green (Roche) as the fluorescent dye. We used the ΔΔCt method to measure the fold change in gene expression of target genes. 18 S rRNA was used to normalize the gene expression.
Preparation of recombinant viruses
Recombinant adenovirus encoding the wild-type PGC-1α and the AD-deleted PGC-1α were prepared using the AdEasy XL system (Stratagene) as per the manufacturer's instructions. Briefly, the respective cDNAs were amplified by PCR using CMV-tag2a PGC-1α wt and CMV-tag2a PGC-1α ΔAD1/ΔAD2 as templates and NotI/XhoI as restriction sites. Both the fragments were cloned into the shuttle vector pShuttle-IRES-hr-GFP-1. The pShuttle constructs were then linearized with PmeI and transformed into the electrocompetent BJ5183-AD-1 cells pre-transformed with the pAdEasy-1 adenoviral vector. The kanamycin-resistant colonies were tested by PacI restriction enzyme digestion and verified for the correct digestion pattern. Recombinant adenoviral constructs were then purified by transformation into the XL-10 gold ultracompetent cells and transfected into the AD-293 cells. The viral infection was monitored using green fluorescent protein expression. The cells were harvested and another round of amplification was performed in the cells. The viral lysates were purified using the Adeno-X virus purification kit (Clontech, Catalogue No. 631532).
RESULTS
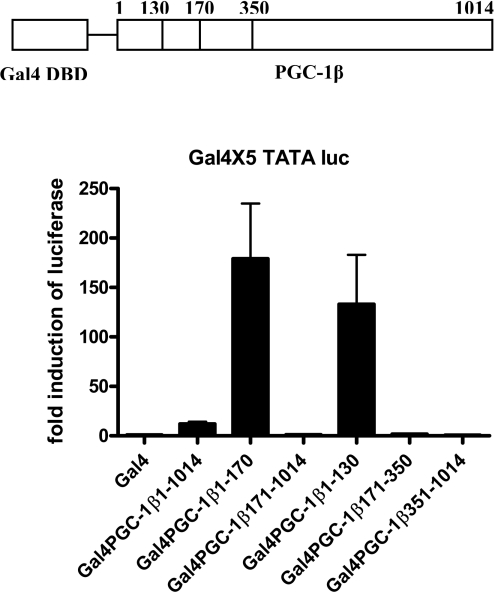
Previous studies from our laboratory found that the N-terminus of PGC-1α contains the transactivation domain [21]. Our first experiments were designed to determine whether the N-terminus of PGC-1β was also responsible for the transcriptional induction. A series of Gal4 expression vectors was created that expressed different portions of PGC-1β ligated to the Gal4 DNA-binding domain, and these vectors were co-transfected with a TATA-luciferase reporter containing five Gal4 sites. The Gal4-PGC-1β-expressing full-length PGC-1β induced the reporter 10-fold (Figure 1). The vectors containing only the N-terminus including amino acids 1–170 or 1–130 very strongly stimulated the reporter suggesting that there are inhibitory domains in the protein. An inhibitory domain has been defined in PGC-1α between amino acids 200 and 400 [5,20]. A similar domain in PGC-1β may lead to the lower transactivation ability of the full-length protein. Deletion of the LXXLL motif did not decrease the ability of the 1–130 truncated protein to induce transcription.
Figure 1. The activation function of PGC-1β is in the N-terminus.
At the top is shown a model of the PGC-1β protein with the Gal4 DNA-binding domain (DBD) at the N-terminus. HepG2 cells were transfected with 2 μg of Gal4 × 5 TATA luciferase (luc), 0.5 μg of TK-Renilla and 50 ng of Gal4 PGC-1β vectors using calcium phosphate precipitation. After transfection (40 h), the cells were harvested and luciferase assays were conducted. The luciferase activity normalized for Renilla activity and protein content. All transfections were performed in duplicate and repeated four times. Data represents the average of four repeats and are expressed as average fold induction of luciferase±S.E.M.
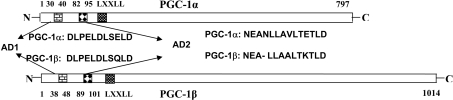
The data in Figure 1 suggested that there might be conserved regions in the first 130 amino acids of the PGC-1 isoforms mediating the transcriptional activity. Comparison of the first 130 amino acids of PGC-1α and PGC-1β revealed that the amino acids 30–40 and 82–95 in PGC-1α are almost completely conserved in PGC-1β (Figure 2). We called these regions AD1 and AD2 respectively and investigated their role in PGC-1 action.
Figure 2. Identification of conserved peptide residues in the N-terminus of PGC-1 isoforms.
PGC-1α and PGC-1β proteins are represented in the Figure as rectangular bars. Conserved AD1 and AD2 are shown along with their amino acid sequence and position in the PGC-1 isoforms. Also shown is the LXXLL motif in the N-terminus.
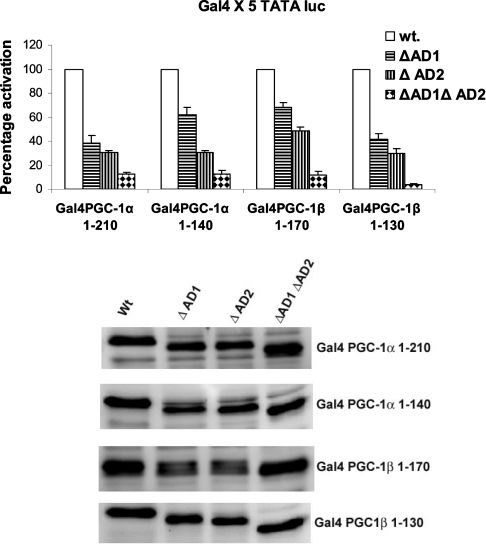
The first experiments tested whether deletion of one or both of these domains altered the ability of Gal4-PGC-1α or Gal4-PGC-1β to induce gene expression. We used two N-terminal amino acid regions of PGC-1α including 1–140 which does not have the LXXLL motif and amino acids 1–210 of PGC-1α which do have the LXXLL motif. Transfection of the Gal4-PGC-1α 1-210 or 1–140 region wild-type proteins induced the Gal4 × 5 TATA-luciferase reporter 300-fold. For PGC-1α 1–210, deletion of AD1 or AD2 decreased the induction of the Gal4 TATA-luciferase vector by 62% and 70% respectively, while deletion of both motifs decreased the induction by 88% (Figure 3). Highly similar results were obtained with the deletion of AD1 (ΔAD1) and AD2 (ΔAD2) in the PGC-1β constructs. The results indicate that each of these domains contributes roughly 50% to the ability of the PGC-1 isoforms to induce transcription. The Gal4 PGC-1 proteins were expressed at similar levels when tested using Western blot analysis as shown in the lower panel of Figure 3.
Figure 3. AD1 and AD2 domains mediate transcriptional activation of PGC-1 isoforms.
Deletion mutagenesis was performed to generate N-terminus Gal4 PGC-1α and Gal4 PGC-1β expression vectors. The ΔAD1 and ΔAD2 indicate deletion AD1 and AD2. These vectors were transfected into HepG2 cells along with Gal4 × 5 TATA luciferase (luc) reporter as described in the legend to Figure 1. Luciferase assays were carried out after 40 h and values were normalized by protein content and Renilla activity. The luciferase activity observed following transfection with the mutant vectors was plotted as a percentage of the wild-type PGC-1 vectors. The data are presented as the average percentage activation of luciferase±S.E.M. All transfections were performed in duplicate and repeated four times. Western blot analysis to test for equal expression of Gal4 PGC-1 proteins is shown in the lower panel.
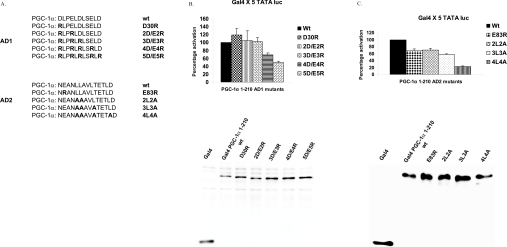
Having identified two critical domains in PGC-1α, the next experiments were designed to determine which amino acids were critical for the transcriptional effects of these peptides. Within AD1, there are five negatively charged aspartic acid (D) or glutamic acid (E) residues (Figure 4A). The negatively charged aspartic acid residues (D) in AD1 were switched to arginine residues (R). As shown in Figure 4(B), the switch of these five amino acids resulted in a 50% reduction in the transcriptional activation suggesting that the negative charge in AD1 contributes to the transcriptional induction through this domain. Within AD2, the glutamic acid residue (E) at position 83 is conserved between PGC-1α and PGC-1β while the glutamic acid residue at position 92 is not found in PGC-1β. The glutamic acid residue (E) at position 83 was switched to an arginine residue (R) resulting in a 32% decrease in the induction of the reporter gene (Figure 4C). Within AD2, there are four leucine residues (L) that are perfectly conserved between the two isoforms. These leucine residues (L) were altered to alanine residues (A) and the ability of PGC-1α to stimulate transcription was assessed. The alteration of all four leucine residues resulted in a 77% decrease in the transactivation by PGC-1α. These data suggest that the negatively charged amino acids in AD1 and AD2 as well as the leucine residues in AD2 form the key components of these motifs as transcriptional regulators.
Figure 4. Identification of amino acids in PGC-1α involved in transcriptional activation.
(A) The amino acid changes in the AD1 and AD2 domains of PGC-1α are shown. The altered amino acids are indicated in bold. (B) HepG2 cells were transfected with Gal4×5 TATA-luciferase and expression vectors for wild-type or AD1 mutant Gal4-PGC-1α 1–210 proteins exactly as described in the legend to Figure 1. Luciferase assays were conducted and transcriptional activation by PGC-1 mutants was calculated as a percentage of the induction by the wild-type PGC-1α±S.E.M. The lower panel shows a Western blot from cells transfected with each of the altered PGC-1α proteins. The Western blot was incubated with an antibody to Gal4 as described in the Materials and methods section. (C) HepG2 cells were transfected with Gal4×5 TATA-luciferase and expression vectors for wild-type or AD2 mutant Gal4-PGC-1α 1–210 proteins. Luciferase assays were conducted and transcriptional activation by PGC-1 mutants was calculated as a percentage of wild-type PGC-1. All transfections were performed in duplicate and repeated four times. The lower panel shows a Western blot from cells transfected with each of the altered PGC-1α proteins as was performed in (B).
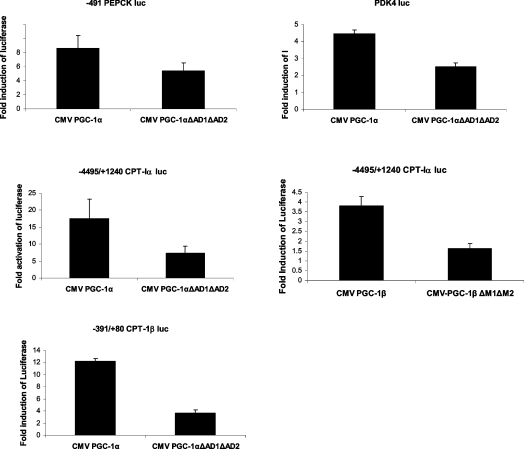
Our next experiments investigated whether these domains participated in the induction of various naturally occurring promoters. The AD1 and AD2 deletions were introduced into CMV-PGC-1α which expresses the full-length cDNA. We tested both PGC-1α as well as PGC-1β on a panel of target genes. For these experiments, we used promoters that we or others had previously identified as PGC-1-responsive genes including PEPCK, CPT-Iα, CPT-Iβ and PDK4 [17,21,27,29]. Since PGC-1β does not induce gluconeogenic genes such as PEPCK [8], we tested CPT-Iα as a PGC-1β target gene. The AD1 and AD2 deletions were introduced into the CMV-PGC-1α expression vector which expresses the full-length PGC-1α. Induction of the PEPCK promoter was reduced by 38% by the removal of the two short peptides, while stimulation of the CPT-Iα and CPT-Iβ genes was decreased by 57% and 70% respectively (Figure 5). The ability of PGC-1α to stimulate the CPT-I isoforms was more profoundly reduced than their induction of PEPCK or PDK4. These results suggest that for the more complex promoters additional regions of PGC-1α are involved in the transcriptional induction or recruitment of other co-activators. Deletion of the AD1 and AD2 domains in PGC-1β reduced the stimulation of CPT-Iα by 57% demonstrating the significance of these domains for the activity of PGC-1β in the context of a target gene.
Figure 5. ADs mediate the induction of PGC-1 target genes.
(A) HepG2 cells were transfected with luciferase reporter genes for PEPCK, CPT-Iα, CPT-Iβ and PDK4. The transfections contained 2 μg of luciferase reporter, 50 ng of CMV-PGC-1α or CMV-PGC-1β and 1.0 μg of TK-Renilla. Luciferase assays were carried out after 40 h and values were normalized by protein content and Renilla activity. The data are expressed as the fold induction by the wild-type or mutant PGC-1α protein±S.E.M. All transfections were performed in duplicate and repeated four times.
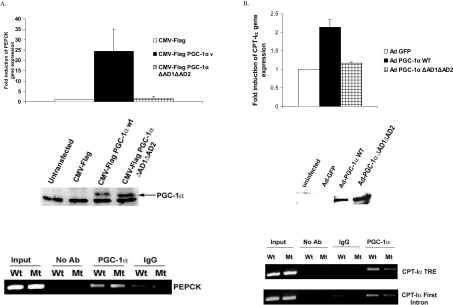
As was seen in the luciferase reporter experiments, the PEPCK-luciferase and CPT-Iα luciferase genes were modestly induced by PGC-1α ΔAD1/ΔAD2. Next we performed experiments to test whether the deletion of AD1 and AD2 domains reduced the induction of endogenous target gene expression using real time PCR. Numerous studies have demonstrated that among hepatic genes, PEPCK is one of the most highly regulated by PGC-1α [17,32,33]. McA-RH7777 cells were transfected with PGC-1α (wild-type, wt) or PGC-1α ΔAD1/ΔAD2 (mutant, mt). PGC-1α induced the PEPCK gene but the ΔAD1/ΔAD2 vector did not (Figure 6A). The mutant and wild-type PGC-1α proteins were equally expressed following transfection. To determine whether PGC-1α ΔAD1/ΔAD2 was associated with the PEPCK gene, we conducted ChIP assays on McA-RH7777 cells infected with the wild-type and mutant PGC-1α-expressing viruses. The pulled-down DNA was subjected to PCR analysis using primers amplifying the region of PEPCK promoter. The ChIP results in the lower panel of Figure 6(A) indicate that both the wild-type and mutant PGC-1α proteins are associated with the PEPCK promoter. We tested the effect of AD1 and AD2 deletion on the ability of PGC-1α to induce the CPT-Iα target gene in H4IIE cells using infection by wild-type or AD1/AD2-deleted PGC-1α adenoviruses. The mutant PGC-1α failed to stimulate CPT-Iα even though the mutant protein was expressed to a similar extent as wild-type, as shown in the Western blot in the middle panel of Figure 6(B). These results indicate that the endogenous PEPCK and CPT-Iα genes are regulated in a similar manner to the transfected luciferase reporters shown in Figure 5. Next we performed a ChIP assay in McA-RH777 cells following infection by wild-type and AD1/AD2-deleted adenoviruses as described for the PEPCK gene. AD1- and AD2-deleted PGC-1α was seen associated with the CPT-Iα TRE (thyroid hormone-response element) and the first intron just as well as the wild-type protein (lower panel, Figure 6B). The data suggest that these motifs are not important for recruiting PGC-1α to the CPT-Iα and PEPCK gene but are essential for transcriptional stimulation.
Figure 6. AD deletions inhibit the induction of endogenous target genes by PGC-1α but not recruitment to the genes.
(A) McA-RH7777 rat hepatoma cells were transfected with CMV-flag PGC-1α wild-type or AD1/AD2-deleted (ΔAD1/ΔAD2) PGC-1α. After 48 h cells were harvested and RNA was isolated. PEPCK mRNA abundance was determined using real-time PCR as described in the Materials and methods section. 18 S rRNA was used as a housekeeping gene to normalize the gene expression values. Western blotting was performed for McA-RH7777 cells transfected with flag PGC-1α wild-type and FLAG PGC-1α ΔAD1/ΔAD2 to show equal expression of wild-type (WT) and mutant proteins (middle panel). McA-RH7777 cells were infected with adenoviruses expressing either the wild-type PGC-1α protein (Wt) or PGC-1α ΔAD1/ΔAD2 (Mt). Cells were cross-linked with formaldehyde for ChIP assay analysis. Immunoprecipitations were conducted with no antibody added (No Ab), rabbit IgG (IgG) or PGC-1α antibody (PGC-1α) as indicated at the top of the panel. The PCR reactions were conducted on the TRE or the first intron of the CPT-Iα gene. PCR products on the gel are shown (lower panel). (B) H4IIE cells were infected with wild-type and mutant viruses to test for CPT-Iα endogenous gene expression and Western blot analysis (upper and middle panels). Similarly, cells were infected with adenoviruses expressing either the wild-type PGC-1α protein (Wt) or PGC-1α ΔAD1/ΔAD2 (Mt) and ChIP assay was carried for the PEPCK promoter as described above. PCR products are shown in the gel panels (lower panel).
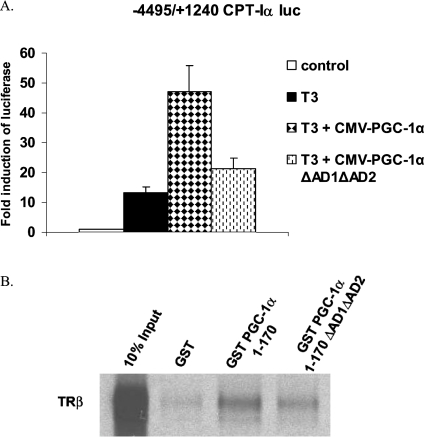
Previously, we demonstrated that PGC-1α enhanced the induction of the CPT-Iα gene by T3 [21]. To determine whether the AD1 and AD2 motifs participated in the T3 induction, we co-transfected CPT-Iα-luciferase with CMV-PGC-1α wild-type or ΔAD1/ΔAD2. T3 increased CPT-Iα-luciferase activity 10-fold. Overexpression of wild-type PGC-1α increased the T3 induction 45-fold but the PGC-1α without the AD1/AD2 motifs did not (Figure 7A). These results indicate that in addition to enhancing the basal expression of several genes, the AD1/AD2 regions participate in the T3 response. We tested whether PGC-1α 1–170 ΔAD1/AD2 still interacted with the TRβ. The first 170 amino acids of PGC-1α contain the LXXLL motif that is critical for the interaction of PGC-1α with the AF-2 domain of the TRβ [34]. Using GST-pulldown assays, it was shown that PGC-1α ΔAD1/ΔAD2 interacted with TRβ (Figure 7B). These data indicated that the AD motifs are not interacting with the TR. Previously, we had found that PGC-1α was associated with both the TRE and the first intron in the CPT-Iα gene [29]. These results along with the ChIP data indicate that AD-deleted PGC-1α is recruited to the promoter of target genes but fails to stimulate transcription.
Figure 7. ADs enhance the T3 induction of CPT-Iα.
(A) HepG2 cells were transfected with CPT-Iα luciferase, an expression vector for the thyroid hormone receptor (RSV-TRβ) and either wild-type or ΔAD1/ΔAD2 CMV-PGC-1α. T3 was added at a concentration of 100 nM after 16 h. After transfection (40 h) cells were harvested and luciferase assays were performed as described in the legend to Figure 4. All transfections were performed in duplicate and repeated four times. (B) GST-pulldown assays were performed using GST-PGC-1α proteins and 35[S]methionine-labelled TRβ. GST-PGC-1α proteins were allowed to interact with 35[S]TRβ overnight at 4 °C. GST was used as a control. GST-sepharose beads were added to collect the proteins, extensively washed and run on a SDS/PAGE (12% gel). The gel was fixed and treated with a fluorescence enhancer. After drying, the signal was detected autoradiographically.
DISCUSSION
PGC-1 induces the expression of numerous genes involved in mitochondrial metabolism [35]. We have identified two conserved peptide sequences in the PGC-1 isoforms that are novel modulators of PGC-1 function. Previous studies have shown that the first 120 amino acids of PGC-1α contain the transactivation domain [12], and we have further dissected this region to localize two peptides which possess significant transactivation potential. In addition, these amino acid motifs participate in the T3 induction of the CPT-Iα gene.
Both AD1 and AD2 are short peptide sequences in the N-terminus of the PGC-1α and β proteins. Homology searches revealed that these peptide motifs are conserved between the two isoforms and across mouse, rat and human species. AD1 is an 11 amino acid peptide that has considerable negative character with five negatively charged residues in the 11 amino acid sequence. AD2 on the other hand has leucine residues arranged in the sequence LLXXL which is an inverted LXXLL motif. AD2 is in fact the L1 peptide which is the first leucine-rich motif in PGC-1α protein that has been described previously [23,36]. The inverted LXXLL motif in PGC-1α and PGC-1β mediates their interaction with the ERRα (oestrogen related receptor α) [23,37]. The LLXXL motif in MyoD is required for the binding of the MAFbx ubiquitin ligase suggesting that this sequence may mediate a variety of protein interactions [38]. Secondary structure prediction suggests that AD2 forms an α-helix secondary structure. Our amino acid substitution studies indicate that the charge on the AD1 motif and the leucine residues in the AD2 motif are the crucial residues. The leucine residues in AD1 are not crucial for the transcription stimulation function as alteration of the leucine residues in AD1 did not decrease the transactivational ability of PGC-1α (results not shown). These data suggest that AD1 and AD2 may mediate key interactions for PGC-1 proteins but that they are mechanistically different.
We speculate that the ADs interact with other transcription factors although the proteins that interact with these domains have not been identified. Other co-activators form one group of candidate proteins. In fact, AD2 resembles the consensus sequence found in nuclear proteins that interact with CBP including SRC-1 [39]. CBP has been shown to interact with the N-terminus of PGC-1 though specific domains have not been elucidated [20]. However, in our experiments we found that although the interaction between PGC-1 and CBP was mediated through the N-terminus neither AD2 nor AD1 domains were involved (results not shown). It has been suggested that SRC-1 contributes to the transcriptional stimulation by PGC-1α and that it interacts with the N-terminus of PGC-1α [20]. In addition, the AD domains may be interacting with the basal transcriptional machinery. Co-activators are bridging proteins between the nuclear receptors and basal transcription factors. It is possible that the AD domains serve this bridging function between the PGC-1 co-activators and basal transcription proteins. Studies are currently underway to identify and characterize the potential interactions of the AD domains.
Stimulation of fatty acid oxidation and gluconeogenesis are two well-characterized actions of PGC-1. We have found that PGC-1α enhances the T3 induction of CPT-Iα [21]. PGC-1α is associated with the TRE and the first intron of the CPT-Iα gene and stimulates its expression [21]. In the present study, we showed that while wild-type PGC-1α is able to synergize with T3 in the induction of CPT-Iα, the ΔAD1/ΔAD2 PGC-1α fails to do so suggesting a role for these domains in the PGC-1α enhancement of T3 responsiveness of CPT-Iα. PGC-1α binding to the TRE and the first intron in vivo is maintained for the mutant PGC-1α indicating that the loss of function of PGC-1α is not due to loss of binding to the promoter. Further evidence that mutant PGC-1α is able to associate with the CPT-Iα promoter is provided in our in vitro pulldown assay in which mutant PGC-1α is able to bind to TRβ.
The induction of gluconeogenesis is a classical biological action of PGC-1α and hepatic PGC-1α levels are up-regulated in mouse models of Type 2 diabetes [32]. Our finding that mutant PGC-1α is unable to stimulate PEPCK gene expression indicates that these domains are required for PGC-1 stimulation of PEPCK. However, even though mutant PGC-1 does not stimulate PEPCK, it is still bound to the promoter in vivo supporting the notion that the AD domains in PGC-1 mediate interaction with the basal transcription machinery. Our studies with the two target promoters of PGC-1 i.e. CPT-Iα and PEPCK, yielded similar results in terms of promoter occupancy. This observation suggests that the loss of function of these proteins upon AD deletion is not due to loss of a gene-specific interaction with nuclear receptors or transcription factors and further supports the hypothesis that the AD domains in PGC-1 may be interacting with the basal transcription machinery. It should be pointed out that even though the AD domains are conserved in PGC-1β, this isoform does not stimulate PEPCK or gluconeogenesis [8]. PGC-1β does not interact with HNF4 and FOXO1, which are the transcriptional proteins that bind to the PEPCK promoter and through which PGC-1α stimulates PEPCK [8]. Our experiments demonstrate that being associated with the promoter alone does not produce transactivation function for these co-activators. Recruitment to the promoter and transactivation function are independent events in the action of PGC-1α.
Acknowledgments
We thank Dr B. Spiegelman (Department of Cell Biology, Dana Farber Cancer Institute, Boston, MA, U.S.A.) for the PGC-1α and PGC-1β expression vectors. This work was supported by grants from the NIH (National Institutes of Health) DK-059368 (EAP) and American Diabetes Association (EAP). P. S. was supported by a fellowship from the American Heart Association.
References
- 1.Rosenfeld M. G., Lunyak V. V., Glass C. K. Sensors and signals: a coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev. 2006;20:1405–1428. doi: 10.1101/gad.1424806. [DOI] [PubMed] [Google Scholar]
- 2.Lonard D. M., O'Malley B. W. The expanding cosmos of nuclear receptor coactivators. Cell. 2006;125:411–414. doi: 10.1016/j.cell.2006.04.021. [DOI] [PubMed] [Google Scholar]
- 3.Rosenfeld M. G., Glass C. K. Coregulator codes of transcriptional regulation by nuclear receptors. J. Biol. Chem. 2001;276:36865–36868. doi: 10.1074/jbc.R100041200. [DOI] [PubMed] [Google Scholar]
- 4.Puigserver P., Spiegelman B. M. Peroxisome proliferator-activated receptor-γ coactivator 1α (PGC-1α): transcriptional coactivator and metabolic regulator. Endocr. Rev. 2003;24:78–90. doi: 10.1210/er.2002-0012. [DOI] [PubMed] [Google Scholar]
- 5.Lin J., Handschin C., Spiegelman B. M. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab. 2005;1:361–370. doi: 10.1016/j.cmet.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 6.Finck B. N., Kelly D. P. PGC-1 coactivators: inducible regulators of energy metabolism in health and disease. J. Clin. Invest. 2006;116:615–622. doi: 10.1172/JCI27794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Puigserver P., Wu Z., Park C. W., Graves R., Wright M., Spiegelman B. M. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 8.Lin J., Tarr P. T., Yang R., Rhee J., Puigserver P., Newgard C. B., Spiegelman B. M. PGC-1β in the regulation of hepatic glucose and energy metabolism. J. Biol. Chem. 2003;278:30843–30848. doi: 10.1074/jbc.M303643200. [DOI] [PubMed] [Google Scholar]
- 9.Lin J., Yang R., Tarr P. T., Wu P. H., Handschin C., Li S., Yang W., Pei L., Uldry M., Tontonoz P., Newgard C. B., Spiegelman B. M. Hyperlipidemic effects of dietary saturated fats mediated through PGC-1β coactivation of SREBP. Cell. 2005;120:261–273. doi: 10.1016/j.cell.2004.11.043. [DOI] [PubMed] [Google Scholar]
- 10.Weitzel J. M., Hamann S., Jauk M., Lacey M., Filbry A., Radtke C., Iwen K. A., Kutz S., Harneit A., Lizardi P. M., Seitz H. J. Hepatic gene expression patterns in thyroid hormone-treated hypothyroid rats. J. Mol. Endocrinol. 2003;31:291–303. doi: 10.1677/jme.0.0310291. [DOI] [PubMed] [Google Scholar]
- 11.Andersson U., Scarpulla R. C. Pgc-1-related coactivator, a novel, serum-inducible coactivator of nuclear respiratory factor 1-dependent transcription in mammalian cells. Mol. Cell Biol. 2001;21:3738–3749. doi: 10.1128/MCB.21.11.3738-3749.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vega R. B., Huss J. M., Kelly D. P. The coactivator PGC-1 cooperates with peroxisome proliferator-activated receptor α in transcriptional control of nuclear genes encoding mitochondrial fatty acid oxidation enzymes. Mol. Cell Biol. 2000;20:1868–1876. doi: 10.1128/mcb.20.5.1868-1876.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu Z., Puigserver P., Andersson U., Zhang C., Adelmant G., Mootha V., Troy A., Cinti S., Lowell B., Scarpulla R. C., Spiegelman B. M. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98:115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- 14.Rhee J., Ge H., Yang W., Fan M., Handschin C., Cooper M., Lin J., Li C., Spiegelman B. M. Partnership of PGC-1α and HNFα in the regulation of lipoprotein metabolism. J. Biol. Chem. 2006;281:14683–14690. doi: 10.1074/jbc.M512636200. [DOI] [PubMed] [Google Scholar]
- 15.Wolfrum C., Stoffel M. Coactivation of Foxa2 through Pgc-1β promotes liver fatty acid oxidation and triglyceride/VLDL secretion. Cell Metab. 2006;3:99–110. doi: 10.1016/j.cmet.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 16.Handschin C., Lin J., Rhee J., Peyer A. K., Chin S., Wu P. H., Meyer U. A., Spiegelman B. M. Nutritional regulation of hepatic heme biosynthesis and porphyria through PGC-1α. Cell. 2005;122:505–515. doi: 10.1016/j.cell.2005.06.040. [DOI] [PubMed] [Google Scholar]
- 17.Yoon J. C., Puigserver P., Chen G., Donovan J., Wu Z., Rhee J., Adelmant G., Stafford J., Kahn C. R., Granner D. K., Newgard C. B., Spiegelman B. M. Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature. 2001;413:131–138. doi: 10.1038/35093050. [DOI] [PubMed] [Google Scholar]
- 18.Vats D., Mukundan L., Odegaard J. I., Zhang L., Smith K. L., Morel C. R., Greaves D. R., Murray P. J., Chawla A. Oxidative metabolism and PGC-1β attenuate macrophage-mediated inflammation. Cell Metab. 2006;4:13–24. doi: 10.1016/j.cmet.2006.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wallberg A. E., Yamamura S., Malik S., Spiegelman B. M., Roeder R. G. Coordination of p300-mediated chromatin remodeling and TRAP/mediator function through coactivator PGC-1α. Mol. Cell. 2003;12:1137–1149. doi: 10.1016/s1097-2765(03)00391-5. [DOI] [PubMed] [Google Scholar]
- 20.Puigserver P., Adelmant G., Wu Z., Fan M., Xu J., O'Malley B., Spiegelman B. M. Activation of PPARγ coactivator-1 through transcription factor docking. Science. 1999;286:1368–1371. doi: 10.1126/science.286.5443.1368. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Y., Ma K., Song S., Elam M. B., Cook G. A., Park E. A. Peroxisomal proliferator-activated receptor-γ coactivator-1α (PGC-1α) enhances the thyroid hormone induction of carnitine palmitoyltransferase I (CPT-Iα) J. Biol. Chem. 2004;279:53963–53971. doi: 10.1074/jbc.M406028200. [DOI] [PubMed] [Google Scholar]
- 22.Monsalve M., Wu Z., Adelmant G., Puigserver P., Fan M., Spiegelman B. M. Direct coupling of transcription and mRNA processing through the thermogenic coactivator PGC-1. Mol. Cell. 2000;6:307–316. doi: 10.1016/s1097-2765(00)00031-9. [DOI] [PubMed] [Google Scholar]
- 23.Huss J. M., Kopp R. P., Kelly D. P. Peroxisome proliferator-activated receptor coactivator-1α (PGC-1α) coactivates the cardiac-enriched nuclear receptors estrogen-related receptor-α and -γ. Identification of novel leucine-rich interaction motif within PGC-1α. J. Biol. Chem. 2002;277:40265–40274. doi: 10.1074/jbc.M206324200. [DOI] [PubMed] [Google Scholar]
- 24.Teyssier C., Ma H., Emter R., Kralli A., Stallcup M. R. Activation of nuclear receptor coactivator PGC-1α by arginine methylation. Genes Dev. 2005;19:1466–1473. doi: 10.1101/gad.1295005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hanson R. W., Reshef L. Regulation of phosphoenolpyruvate carboxykinase (GTP) gene expression. Annu. Rev. Biochem. 1997;66:581–611. doi: 10.1146/annurev.biochem.66.1.581. [DOI] [PubMed] [Google Scholar]
- 26.Herzog B., Hall R. K., Wang X. L., Waltner-Law M., Granner D. K. Peroxisome proliferator-activated receptor γ coactivator-1α, as a transcription amplifier, is not essential for basal and hormone-induced phosphoenolpyruvate carboxykinase gene expression. Mol. Endocrinol. 2004;18:807–819. doi: 10.1210/me.2003-0384. [DOI] [PubMed] [Google Scholar]
- 27.Ma K., Zhang Y., Elam M. B., Cook G. A., Park E. A. Cloning of the rat pyruvate dehydrogenase kinase 4 gene promoter. J. Biol. Chem. 2005;280:29525–29532. doi: 10.1074/jbc.M502236200. [DOI] [PubMed] [Google Scholar]
- 28.Harris R. A., Bowker-Kinley M. M., Huang B., Wu P. Regulation of the activity of the pyruvate dehydrogenase complex. Adv. Enz. Regul. 2002;42:249–259. doi: 10.1016/s0065-2571(01)00061-9. [DOI] [PubMed] [Google Scholar]
- 29.Song S., Zhang Y., Ma K., Jackson-Hayes L., Lavrentyev E. N., Cook G. A., Elam M. B., Park E. A. Peroxisomal proliferator activated receptor γ coactivator (PGC-1α) stimulates carnitine palmitoyltransferase I (CPT-Iα) through the first intron. Biochim. Biophys. Acta. 2004;1679:164–173. doi: 10.1016/j.bbaexp.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 30.Park E. A., Cook G. A. Differential regulation in the heart of mitochondrial carnitine palmitoyltransferase-I muscle and liver isoforms. Mol. Cell Biochem. 1998;180:27–32. [PubMed] [Google Scholar]
- 31.Jansen M. S., Cook G. A., Song S., Park E. A. Thyroid hormone regulates carnitine palmitoyltransferase Iα gene expression through elements in the promoter and first intron. J. Biol. Chem. 2000;275:34989–34997. doi: 10.1074/jbc.M001752200. [DOI] [PubMed] [Google Scholar]
- 32.Herzig S., Long F., Jhala U. S., Hedrick S., Quinn R., Bauer A., Rudolph D., Schutz G., Yoon C., Puigserver P., Spiegelman B., Montminy M. CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature. 2001;413:179–183. doi: 10.1038/35093131. [DOI] [PubMed] [Google Scholar]
- 33.Herzog B., Cardenas J., Hall R. K., Villena J. A., Budge P. J., Giguere V., Granner D. K., Kralli A. Estrogen-related Receptor α is a repressor of phosphoenolpyruvate carboxykinase gene transcription. J. Biol. Chem. 2006;281:99–106. doi: 10.1074/jbc.M509276200. [DOI] [PubMed] [Google Scholar]
- 34.Wu Y., Delerive P., Chin W. W., Burris T. P. Requirement of helix 1 and the AF-2 domain of the thyroid hormone receptor for coactivation by PGC-1. J. Biol. Chem. 2002;277:8898–8905. doi: 10.1074/jbc.M110761200. [DOI] [PubMed] [Google Scholar]
- 35.Puigserver P. Tissue-specific regulation of metabolic pathways through the transcriptional coactivator PGC1-α. Int. J. Obes. 2005;29:S5–S9. doi: 10.1038/sj.ijo.0802905. [DOI] [PubMed] [Google Scholar]
- 36.Knutti D., Kressler D., Kralli A. Regulation of the transcriptional coactivator PGC-1 via MAPK-sensitive interaction with a repressor. Proc. Natl. Acad. Sci. U.S.A. 2001;98:9713–9718. doi: 10.1073/pnas.171184698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kressler D., Schreiber S. N., Knutti D., Kralli A. The PGC-1-related protein PERC is a selective coactivator of estrogen receptor α. J. Biol. Chem. 2002;277:13918–13925. doi: 10.1074/jbc.M201134200. [DOI] [PubMed] [Google Scholar]
- 38.Tintignac L. A., Lagirand J., Batonnet S., Sirri V., Leibovitch M. P., Leibovitch S. A. Degradation of MyoD mediated by the SCF (MAFbx) ubiquitin ligase. J. Biol. Chem. 2005;280:2847–2856. doi: 10.1074/jbc.M411346200. [DOI] [PubMed] [Google Scholar]
- 39.Matsuda S., Harries J. C., Viskaduraki M., Troke P. J., Kindle K. B., Ryan C., Heery D. M. A conserved α-helical motif mediates the binding of diverse nuclear proteins to the SRC1 interaction domain of CBP. J. Biol. Chem. 2004;279:14055–14064. doi: 10.1074/jbc.M310188200. [DOI] [PubMed] [Google Scholar]