Abstract
Complex emergent systems of many interacting components, including complex biological systems, have the potential to perform quantifiable functions. Accordingly, we define “functional information,” I(Ex), as a measure of system complexity. For a given system and function, x (e.g., a folded RNA sequence that binds to GTP), and degree of function, Ex (e.g., the RNA–GTP binding energy), I(Ex) = −log2[F(Ex)], where F(Ex) is the fraction of all possible configurations of the system that possess a degree of function ≥ Ex. Functional information, which we illustrate with letter sequences, artificial life, and biopolymers, thus represents the probability that an arbitrary configuration of a system will achieve a specific function to a specified degree. In each case we observe evidence for several distinct solutions with different maximum degrees of function, features that lead to steps in plots of information versus degree of function.
Keywords: origin of life, artificial life, evolution, aptamers, emergent systems
Complex emergent systems, in which interactions among numerous components or “agents” produce patterns or behaviors not obtainable by individual components, are ubiquitous at every scale of the physical universe, for example in neural networks (1), turbulent fluids (2), insect colonies (3), and spiral galaxies (4). Complex systems also appear in a range of artificial symbolic contexts, including genetic algorithms (5), cellular automata (6), artificial life (7), and models of market economies (8).
Life, with its novel collective behaviors at the scale of molecules, genes, cells, and organisms, is the quintessential emergent complex system. Furthermore, the ancient transition from a geochemical world to a living planet may be modeled as a sequence of emergent events, each of which increased the chemical complexity of the prebiotic world (9–11).
Given this ubiquity and diversity, it is desirable to understand the characteristics of emergent complex systems, as well as the factors that might promote complexity in evolving systems. However, complexity has proven difficult to define or measure with precision (12–14). A central objective of this study, therefore, is to examine “functional information” (15) as a quantitative measure of complexity that may be applicable to the analysis and prediction of attributes of a wide range of phenomena in physical and symbolic systems, including evolving biological systems.
An extensive literature explores historical developments and recent advances in the study of complexity and information (14, 16–18) as well as their application to understanding biological systems (3, 13, 19–24). Despite this rich literature, previous discussions of complexity have not generally focused on the relationship between information content and function (25). We propose to measure the complexity of a system in terms of functional information, the information required to encode a specific function.
Systems and Their Functions
In this paper we consider the functional information of both symbolic systems (letter sequences and Avida artificial life genomes) and biopolymers (RNA aptamers). These systems share several characteristics: first, they consist of numerous individual components or “agents”; second, the agents can combine in a combinatorially large number of different configurations; and third, some configurations display functions that are not characteristic of the individual agents. Analyses of these systems address fundamental questions about the relationship between information content and function. For example, How much information does it take to encode a function? Are there multiple distinct solutions? How are solutions distributed in configuration space? How much more information does it take to encode a given improvement in function? What environmental factors might influence these relationships?
The function of some emergent systems is obvious: a sequence of letters communicates a specific idea, a computer algorithm performs a specific computation, and an enzyme catalyzes at least one specific reaction. Less obvious are the functions of systems of many interacting inanimate particles, such as molecules, sand grains, or stars, but these systems may also be described quantitatively in terms of function, for example, in terms of their ability to dissipate energy or to maximize entropy production through patterning (e.g., refs. 26–29). Living systems, by contrast, typically display multiple essential functions (21, 30, 31). This consideration of complexity in terms of the function of a system, as opposed to some intrinsic measure of its patterning or structural intricacy, distinguishes our treatment from many previous efforts.
Quantifying Complexity.
Development of a quantitative measure of complexity has proven difficult for at least three reasons, each of which relates to the diversity of systems that may be labeled “complex.”
Systems may be complex in terms of information content, physical structure, and/or behavior. Consider three stages in the life cycle of a multicellular organism: a fertilized egg, a live adult, and a postmortem adult. All three states are complex, but they are complex in different respects. All three states possess the sequence information (a genome) necessary to grow a living organism. Living and dead adult organisms also display complex anatomical structures, but only living organisms possess behavioral complexity. Any universal definition of complexity must thus have the potential to quantify complexity independently in terms of information, structure, or behavior.
It has been difficult to define complexity in terms of a metric that applies to all complex systems. No obvious common thread exists in comparing the complexity of symbolic systems, such as language, with those of physical agents, such as cells. Parameters useful in characterizing symbolic systems (e.g., algorithm- or information-based complexity metrics) generally differ from those used to analyze systems of interacting particles (e.g., Newtonian dynamics or maximum entropy models). Gell-Mann (12) concludes, “A variety of different measures would be required to capture all our intuitive ideas about what is meant by complexity.”
Complex emergent systems are diverse in terms of their dimensionality. Sequences of letters, computer code, or bipolymers can be treated as one-dimensional strings of symbolic information (or as points in a high-dimensional sequence space). On the other hand, many physical emergent systems, including those composed of many interacting sand grains, cells, organisms, or stars, exhibit time-dependent behaviors in two or three spatial dimensions. It is desirable for a complexity formalism to apply to this range of dimensionalities.
Despite this diversity, a common thread is present: All complex systems alter their environments in one or more ways, which we refer to as functions (32). In the words of von Baeyer (18), “Information gathering by itself, without observable effects on the gatherer's behavior, is a pointless pursuit.” Function is thus the essence of complex systems. Accordingly, we focus on function in our operational definition of complexity. Therefore, although many previous investigators have explored aspects of biological systems in terms of information (e.g., ref. 33), we adopt a different approach and explore information in terms of the function of a system (including biological systems).
Szostak and coworkers (15, 34) introduced “functional information” as a measure of complexity. They proposed that the complexity of an information-rich system, such as RNA aptamers (RNA structures that bind a target molecule), can be quantified in the context of specific functions of the system, in contrast to prior formalisms based on genomic, sequence, or algorithmic information (e.g., refs. 13 and 35). Here we examine applications of this formalism to letter sequences, the artificial life platform Avida (36), and RNA apatmers.
Functional Information as a Measure of System Complexity.
Many emergent systems of interacting agents can be described in terms of their potential to accomplish one or more quantifiable tasks. Consider a system that can exist in a combinatorially large number of different configurations (i.e., a 100-nt RNA strand comprised of four different nucleotides, A, U, G, and C, with 4100 different possible sequences). Assume that a small fraction of these configurations accomplishes a specified function x to a high degree (corresponding to a high information content). Typically, a significantly greater number of configurations will prove somewhat less efficient in accomplishing function x (corresponding to lower information content), whereas the majority of configurations will display little or no function (34, 35).
Accordingly, “degree of function x” (Ex) is a measure of a configuration's ability to perform the function x. For example, in an enzymatic system Ex might be defined as the increase in a specific reaction rate that is achieved by the enzyme. In the case of a sequence of letters, Ex might represent the probability that the sequence conveys a desired message to a particular recipient. And in a system with water flowing over sand ripples, Ex might be defined as the rate of energy dissipation by turbulence, compared with flow over a smooth, unpatterned surface. The units or scale of Ex may be somewhat arbitrary and will depend on the nature of function x. Thus, for example, catalytic efficiency might be recorded in terms of rate enhancement or in terms of decreased activation energy (proportional to the log of the rate enhancement).
In the formalism of Szostak (ref. 15; see also ref. 19, p. 252), functional information [I(Ex)] is calculated with reference to a specific degree of function x, designated Ex. Typically, a small fraction, F(Ex), of all possible configurations of a system achieves at least the specified degree of function, ≥Ex. Accordingly, we define functional information in terms of F(Ex):
Thus, in a system with N possible configurations (e.g., a sequence of n RNA nucleotides, which has N = 4n discrete possible sequences):
where M(Ex) is the number of different configurations that achieves or exceeds the specified degree of function x, ≥Ex.
In every system, the fraction of configurations, F(Ex), capable of achieving a specified degree of function will generally decrease with increasing Ex (15). The largest possible functional information of a system is exhibited in the case of a single configuration that displays the highest possible degree of function, Emax:
where I is measured in bits. This maximum functional information is thus equivalent to the maximum number of bits necessary and sufficient to specify any particular configuration of the system.
Alternatively, the minimum functional information of a system is zero for configurations with the lowest degree of function, Emin, because all possible states have Ex ≥ Emin:
In this formulation, functional information increases with degree of function, from zero for no function (or minimum function) to a maximum value corresponding to the number of bits necessary and sufficient to specify completely any configuration of that system.
Functional information is defined only in the context of a specific function x. For example, the functional information of a ribozyme may be greater than zero with respect to its ability to catalyze one specific reaction but will be zero with respect to many other reactions. Functional information therefore depends on both the system and on the specific function under consideration. Furthermore, if no configuration of a system is able to accomplish a specific function x [i.e., M(Ex) = 0], then the functional information corresponding to that function is undefined, no matter how structurally intricate or information-rich the arrangement of its agents.
It is important to emphasize that functional information, unlike previous complexity measures, is based on a statistical property of an entire system of numerous agent configurations (e.g., sequences of letters, RNA oligonucleotides, or a collection of sand grains) with respect to a specific function. To quantify the functional information of any given configuration, we need to know both the degree of function of that specific configuration and the distribution of function for all possible configurations in the system. This distribution must be derived from the statistical properties of the system as a whole [as opposed, for example, to the statistical properties of populations evolving in a fitness landscape (37)]. Any analysis of the functional information of a specific functional sequence or object, therefore, requires a deep understanding of the system's agents and their various interactions.
Three examples (letter sequences, the artificial life platform Avida, and RNA aptamers) serve to illustrate the concept of functional information.
The Functional Information of Letter Sequences.
Systems of many interacting components can occur in a combinatorially large number of different configurations. Functional information depends on the fraction of all possible configurations that achieve at least a specified degree of function. Sequences of letters provide a conceptually familiar example.
Consider various sequences of n letters that convey the message: “A fire has just started in a house at the corner of Main Street and Maple Street.” Many different sequences of letters are capable of conveying that information. To determine the functional information of any particular sequence we must specify three parameters:
n, the number of letters in the sequence.
Ex, the degree of function x of that sequence. In the case of the fire example cited above, Ex might represent the probability that a local fire department will understand and respond to the message (a value that might, in principle, be measured through statistical studies of the responses of many fire departments). Therefore, Ex is a measure (in this case from 0 to 1) of the effectiveness of the message in invoking a response.
M(Ex), the total number of different letter sequences that will achieve the desired function, in this case, the threshold degree of response, ≥Ex.
The functional information, I(Ex), for a system that achieves a degree of function, ≥Ex, for sequences of exactly n letters is therefore
Note that 26n is the total number of possible arrangements of 26 letters in a sequence of n letters, and in this treatment we assign equal probability to all possible sequences. The important more general case of configurations of unequal probabilities is a straightforward extension of the treatment of Shannon (38, 39), as discussed by Carothers et al. (34). Greater clarity of expression can be added through additional characters such as “space,” “capital,” and “period”; however, in this example we use only 26 letters. As in all combinatorially large emergent systems, most sequences convey no information (i.e., have no discernable function). Functional information is determined by identifying the fraction of all sequences that achieve a specified outcome.
Consider, for example, sequences of 10 letters that have a high probability (Ex ≅ 1) of evoking a positive response from the fire department. Such sequences might include “FIREONMAIN,” “MAINSTFIRE,” or “MAPLENMAIN.” Additionally, some messages containing phonetic misspellings (FYRE or MANE), mistakes in grammar or usage (FIREOFMAIN), or typing errors (MAZLE or NAPLE) may also yield a significant but lower probability of response (0 ≪ Ex < 1). Given these variants, on the order of 1,000 combinations of 10 letters might initiate a rapid response to the approximate location of the fire. Thus,
Numerous additional 10-letter sequences convey some relevant information but would result in a lower probability of response (0 < Ex < 1): “FIREHELPME,” “DANGERFIRE,” or “BURNINGNOW.” A lower degree of function, Ex, will generally correspond to a larger number of effective letter sequences, M(Ex).
The formulation of functional information also applies to systems in which sequences of varied lengths are combined. For letter sequences of any length from 1 to n letters,
Varying the maximum length, n, of the letter sequence has a significant effect on the maximum possible degree of function, Ex, as well as the number of states, M(Ex), that achieve that degree of function. Sequences of 1, 2, or 3 letters are unlikely to convey sufficient information to achieve any response. With 4 letters, however, a few suggestive configurations exist (FIRE, MAIN, or MAPL), although all such sequences possess a high degree of ambiguity (i.e., Ex ≪ 1).
On the other hand, with longer letter sequences (n ≫ 10), the number of messages of a given degree of function increases dramatically, with new opportunities for explicit instructions (and hence maximum degree of function, Ex = 1). With a sufficient number of letters, any arbitrary degree of accuracy and precision in a message can be communicated. Note, however, that arbitrarily long sequences are not necessarily more effective at conveying information and thus may not increase the functional information of a system. For example, consider sequences of letters that begin with the following 22 letters:
FIREATMAINSTANDMAPLEST …
Such a sequence should invariably summon the fire department, no matter what or how many additional letters are placed at the end of the sequence. Thus, for this admittedly contrived fire department scenario, the fraction of sequences that achieve the desired outcome attains a maximum value at ≈20 letters. In competitive systems, notably genetic information constrained by length-selective pressure in living systems (e.g., refs. 40–43), longer sequences may prove inefficient and do not necessarily confer an advantage. (Indeed, in the case of reporting a fire, an overly long and detailed message might delay response time.)
Note that in this formulation of functional information the maximum possible value, I(Emax), arises when a message is so specific that only a single letter sequence out of all possible letter sequences achieves a desired outcome. In the case of a sequence of n letters, that maximum functional information occurs when M(Ex) = 1:
Although this conceptual example is qualitative, it introduces key concepts that are required to quantify functional information in any emergent system with numerous configurations. Of special interest is the relationship between information and degree of function. Letter sequences point to the existence of discrete “classes” of functional configurations, based in this case on the appearance of familiar words (“FIRE” and “MAIN”) as well as their mutations (“FYRE” and “MANE”). We explore the role of such multiple classes of solutions in the subsequent sections on Avida and RNA aptamers.
We conclude that rigorous analysis of the functional information of a system with respect to a specified function x requires knowledge of two attributes: (i) all possible configurations of the system (e.g., all possible sequences of a given length in the case of letters or RNA nucleotides) and (ii) the degree of function x for every configuration.
These two requirements are difficult to meet in many systems. In the case of letter sequences, for example, the sequence is obvious, but it is difficult to determine quantitatively the degree of function of many sequences. By contrast, it is relatively straightforward to determine the degree of function (for example, the ligand affinity) of any given RNA sequence, but impossible with present technology to measure all sequences in a large population, e.g., ≈1014 randomly generated 100-mers as used in some aptamer evolution studies (although single-molecule methods may ultimately provide a technical solution to this challenge). However, these concepts may be placed on a firmer footing in the case of computational systems, such as the artificial life platform Avida.
The Functional Information of Avida Populations.
We have adapted the artificial life platform Avida (35–36) to explore the distribution of function in an emergent system. The digital organisms that populate the virtual world of Avida are “computer programs that self-replicate, mutate, and adapt by natural selection” (44) and as such share many (although not all) of the attributes ascribed to biological life. Accordingly, artificial life models have been used as a means of exploring ideas about organic biology that are not readily amenable to experimentation. Here we explore the functional information of randomly generated populations of Avida organisms. Understanding the origin and evolution of complex biological systems motivates this work; however, the first task is to demonstrate an approach for quantifying the relationship between information and functional behavior in a well characterized emergent system, whether or not unambiguous biological insight is immediately revealed.
Avida organisms consist of multiple lines of machine instructions, termed its “genome.” Each organism operates as a formal computer similar to that outlined by Turing (45), and the computational properties of each organism are determined by the sequence of machine instructions stored in its memory. A population of Avida organisms can be thought of as a multitude of identical computers running many different simple programs, where differences between any two members of the population arise solely from the differences in the programs being run.
This research focuses on the ability of a small fraction of all randomly generated Avida organisms to perform computational tasks that arise through the coordinated execution of multiple machine instructions (35). None of these computational tasks can be performed by the execution of a single instruction; indeed, the shortest functional program requires five instructions. The computational ability (function) of Avida organisms thus emerges from the interaction of instructions (the agents), making Avida an ideal model for characterizing complex emergent systems.
In a typical Avida experiment, we generate 107 random instruction sequences (i.e., 107 different individual genomes), each sequence 100–500 instructions in length, from the default set of 26 different machine instructions. Although most sequences display no function, a small subset of sequences code for the ability to compute logic operations (such as “not” or “and”) or arithmetic functions (addition and subtraction).
The set of computational tasks Avida organisms can perform allows for varied solutions, analogous to variations seen in nature. This characteristic is underscored by the fact that in its evolution apparatus Avida does not consider how a task is accomplished but only the resulting function, i.e., whether or not it is executed. The Avida platform does not specify preferred approaches to problem solving, which allows novel solutions to appear through evolution. There may be great variety among these solutions, and they may be very different from those that might have been arrived at by design (44).
Measures of Avida Function.
Just as there is no unique measure of function in natural systems, there is no unique measure of the degree of function in an Avida sequence population. We chose to consider three distinct measures of function: (i) the number of times a sequence is able to compute a specific task, for example, addition or not/and; (ii) the total number of all tasks the sequence is able to compute, because many sequences can perform multiple distinct operations; and (iii) the total number of different tasks the sequence is capable of computing.
Each of these measures of function correlates to strategies that biological organisms employ to increase their fitness. Some organisms rely on the ability to perform one action very well, others rely on the ability to perform multiple actions moderately well, and still others take advantage of flexibility, the ability to do many different tasks (46). However, unlike with living organisms, quantifying the extent of these traits in Avida is straightforward and unambiguous. Most of the discussion that follows, however, focuses on execution of a single task.
Functional sequences constitute a tiny minority of the Avida genome space. Therefore, to explore fully the distribution of function within a sequence space, a large number of randomly generated sequences (i.e., equal probability) must be surveyed (see Methods). Such random explorations of genome space are similar to the strategies used in the directed evolution of RNA structures (e.g., refs. 47–48). Note, however, that this type of random sampling is not possible with living organisms because the portion of genome space explored in an evolution experiment will be constrained by the topology of the underlying fitness landscape and the particular configuration of the environment maxima (25, 49–51).
Avida Results.
Random sampling of genome space has yielded several interesting results related to the frequency and distribution of functional configurations. By using Avida's default set of 26 machine instructions, a randomly generated sequence with length of a magnitude of ≈102 lines was found to be functional (i.e., was able to perform at least one logic or arithmetic operation at least once) with probability P ≈ 10−3. The functional fraction of a population decreases with decreasing sequence length until it reaches zero for populations with sequences of a length of four machine instructions or less.
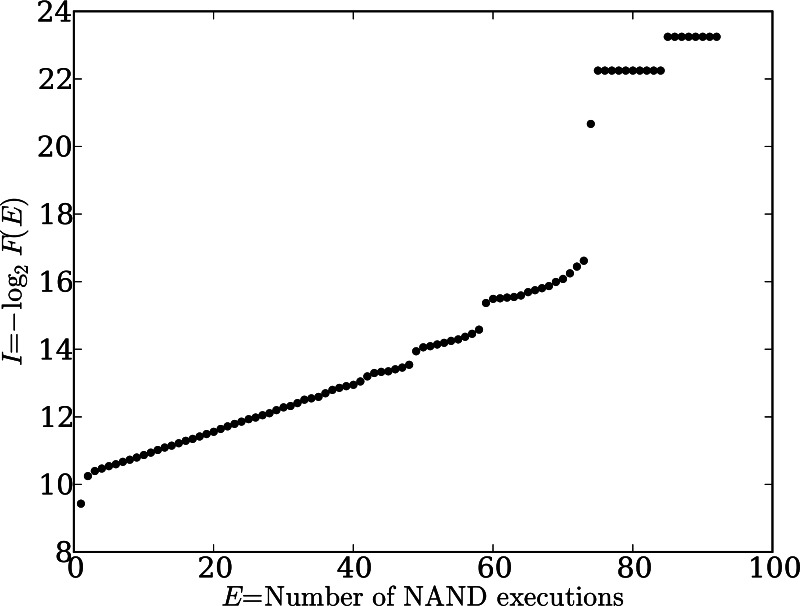
We observe regular, reproducible structure in the distribution of task execution frequency, for example, in the number of not/and or addition operations executed (Ex) versus functional information (Fig. 1). This plot, which illustrates the distribution of function for 107 randomly generated 300-instruction genomes, is continuous over most values of Ex, for example, between 2 and 48. However, at several values of Ex, discontinuities appear. At Ex > 73 these discontinuities point to isolated individual genomes of high functionality; such outliers always appear, but they may occur at different values of Ex for repetitions of this experiment. However, other discontinuities (notably those between 48/49 and 58/59) are robust, always appearing in experiments on 300-instruction genomes. Thus these gap-like features reflect an intrinsic behavior of Avida genomes.
Fig. 1.
Distribution of the not/and (NAND) function in 300-line Avida genomes in a randomly generated sample of 107 genomes. The degree of function, E, is the number of times NAND is executed by the genome, whereas functional information, I (in bits), is −log2 of the fraction of all sequences that achieves at least that degree of function, F(E). Note the discontinuities, which are a recurrent feature in these experiments.
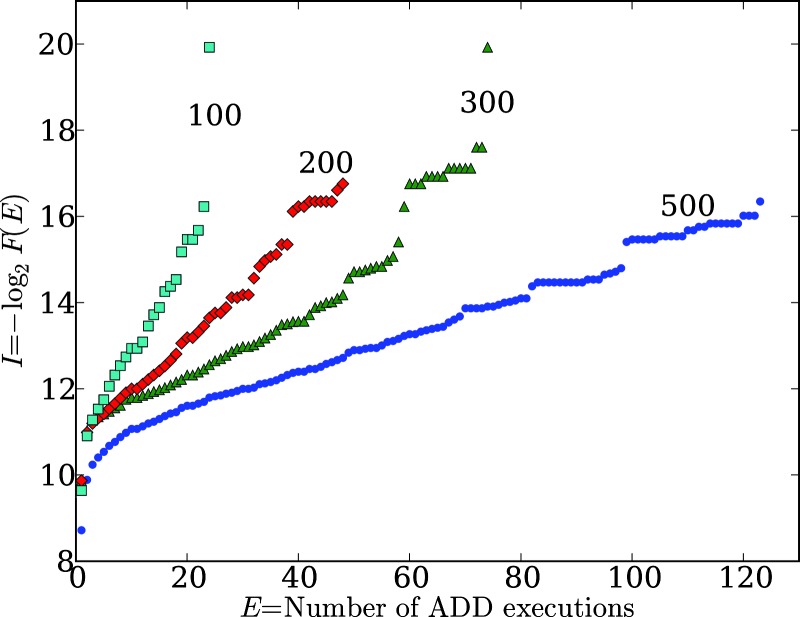
We also find that the number and specific location of these gaps, as well as the maximum values of I(Ex) and Ex, depend on the length of the sequences being studied (Fig. 2). For example, we examined the number of executions of the addition function for 106 randomly generated genomes of 100, 200, 300, and 500 instructions. We find that the maximum number of addition executions, Ex, increases with genome length. We often observe discrete highly functional genomes, representing outlier solutions, as well as reproducible gaps. For randomly generated genomes of 100, 200, 300, and 500 instructions, the first significant gap in addition execution frequency occurs at 19, 39, 59, and 69 executions, respectively.
Fig. 2.
The frequency of the ADD function in 100-, 200-, 300-, and 500-line linear Avida genomes in randomly generated samples of 106 genomes. Degree of function, E, is the number of times the ADD function is executed by the genome, whereas functional information, I (in bits), is −log2 of the fraction of all sequences that achieves at least that degree of function, F(E). Note that maximum E increases with genome length.
Islands of Function.
What is the source of the reproducible discontinuities in Figs. 1 and 2? We suggest that the population of random Avida sequences contains multiple distinct classes of solutions, perhaps with conserved sequences of machine instructions similar to those of words in letter sequences or active RNA motifs (52). Each class has a maximum possible degree of function; therefore, the discontinuities occur at degrees of function below which a major class of sequences is represented and above which it is not represented.
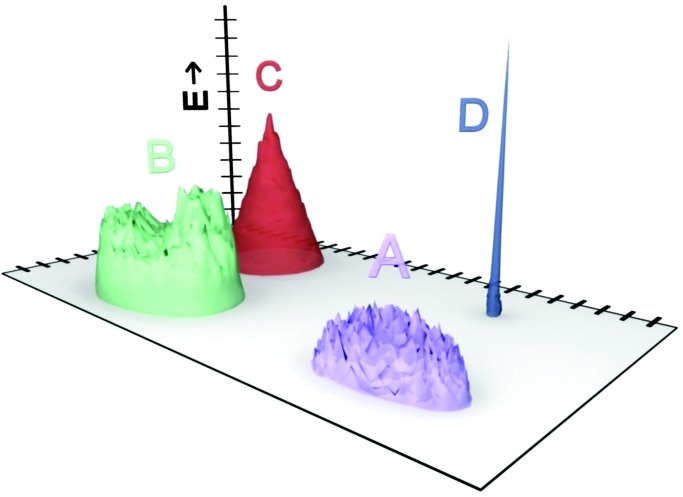
Fig. 3 demonstrates one possible model for this stepped behavior, based on discrete “islands” of solutions. In Fig. 3, the islands, each of which represents a specific distinct set of solutions to the function [i.e., fitness (z axis)], are conceptually represented as being close to each other in sequence space (projected on the x–y plane). Note, however, that these islands are a visual simplification. For example, in the case of RNA sequences, any given “island” of closely related functional solutions may be more realistically represented by a densely interconnected network that spans all of sequence space (25, 53, 54). Similar consideration of function topologies has been applied to neural network connections (55) and viroid solutions to infecting the same plant host (56). Avida may be similar, because the commands relevant to a given solution do not necessarily need to appear sequentially at a specific location in the string but can occur in different registers and can be spread apart by neutral commands.
Fig. 3.
Schematic representation of four discrete functional classes, or “islands,” of solutions that display function. The vertical axis is degree of function, E, whereas the horizontal plane represents a two-dimensional projection in sequence space. The number of sequences with degree of function ≥E corresponds to the area intersected by the horizontal plane at that height along the E axis. Increasing E above the heights of the flat-topped islands A and B will result in discontinuities in the function E versus I, as illustrated in Figs. 1 and 2. Island C is a cone-shaped distribution, and island D represents a discrete solution of the type that might not be discovered in random sampling experiments.
Consider a case where four classes of solutions for the same function, labeled A–D, occur in the population (Fig. 3). Each class may contain a normal distribution of degrees of function, but each has a different topology in sequence space and a different maximum degree of function, Ex. For relatively low values of Ex, all four islands contribute functional sequences. As the value of Ex increases to just above the heights of flat-topped islands A and then B, discontinuities in the plot of Ex versus I(Ex) will occur (i.e., in Fig. 1 the height corresponding to island A would be Ex = 48 and the height of island B would be Ex = 58). This model also matches the observation that the continuous stretches of Ex versus I(Ex) are longest for populations of long sequences: Longer sequences allow for a greater number of distinct solutions whose superposition would serve to drown out individual discontinuities.
This model for generating discontinuities is plausible because multiple distinct solutions may exist in sequence space for a given task. For example, the shortest possible sequence (“gene”) for accomplishing subtraction is five lines long (35). However, an alternative unrelated subtraction gene 10 lines long can be constructed within the Avida language using two's-complement arithmetic (57). This second class of solutions reinforces the concept of “islands” of function in sequence space, where two or more types of solutions exist that achieve the same task but do so in an unrelated fashion.
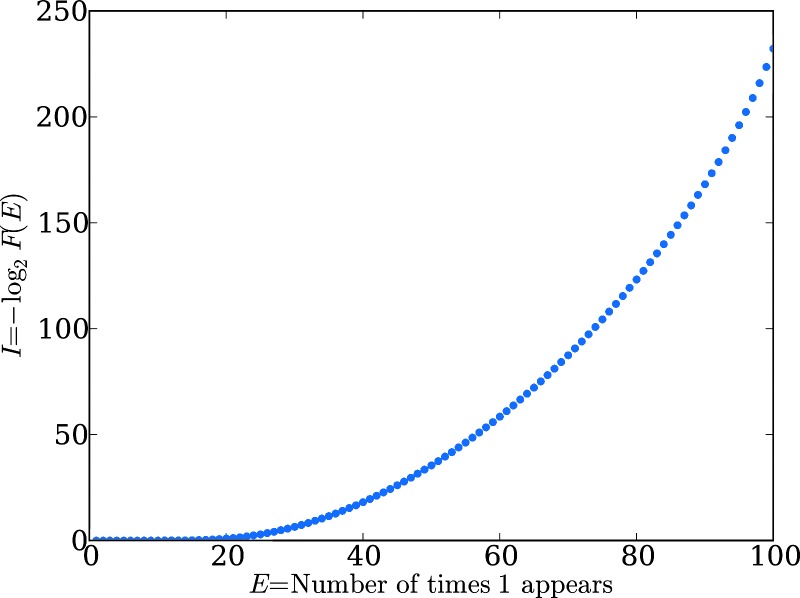
We note, by contrast, that purely random statistical functions do not display steps. For example, if the degree of function is defined as the frequency of the appearance of the number “1” in randomly generated sequences of 100 digits, then functional information follows a well behaved smooth curve (Fig. 4). Maximum functional information arises for the solitary state with 100 consecutive 1s, whereas an obvious uniform distribution follows for lesser degrees of function. This statistically random case is not stepped. By comparison, the structures depicted in Figs. 1 and 2 suggest that the tasks being considered as functions are neither trivial, nor are they achieved by essentially arbitrary or random, albeit rare, configurations of the system. The interactions in the Avida system, and perhaps many other complex systems, lead to distribution of function that prove far richer than in systems possessing statistically trivial function. It remains to be seen, however, whether the observed stepped relationship between I(Ex) and Ex is a general feature of functional information or an idiosyncratic characteristic of Avida genomes.
Fig. 4.
I(E) versus E for the statistically random system, where E is the number of times the digit 1 appears at least that many times in a sequence of 100 digits. This statistically random case is not stepped, in contrast to the topology of Avida genomes.
Functional Information and RNA Polymers.
The previous two examples, sequences of letters and Avida machine commands, illustrate the utility of the functional information formalism in characterizing the properties of symbolic systems that can occur in combinatorially large numbers of configurations. Functional information also has applicability to complex biological and biochemical systems; indeed, it was originally developed (15, 34) to analyze aptamers (RNA structures that bind target ligands) and ribozymes (RNA structures that catalyze specific reactions). Thus, the degree of function, Ex, of these linear sequences of RNA letters (A, C, G, and U) can be defined quantitatively as the binding energy to a particular molecule or the catalytic increase in a specific reaction rate. We can easily specify every possible RNA sequence of length n, and we can (at least in principle) synthesize RNA strands and measure the degree of function of any given sequence. The behavior of aptamers and ribozymes thus lends itself to the type of quantitative analysis that we applied previously to letter sequences and Avida populations (34).
In general, a single RNA nucleotide will display minimal catalytic or binding function, xmin. It follows that a minimum sequence length (nmin nucleotides) will be required to achieve any significant degree of ribozyme or aptamer function, Ex > Emin. Increasing the number of nucleotides (n > nmin) will generally lead to many more functional sequences, some of which will have a greater degree of function. Furthermore, for any given catalytic or binding function there exists an optimal RNA sequence of length nopt that attains the maximum possible degree of function, Emax. That sequence thus possesses the maximum possible functional information:
For degrees of function less than the maximum (Ex < Emax), an intermediate functional information obtains [I(Ex) < Imax(Emax)].
The in vitro evolution of RNA aptamers (e.g., refs. 47 and 48) provides a dramatic illustration of the evolution and selection of systems with high functional complexity. Aptamer evolution experiments begin with large populations (up to 1016 randomly generated RNA sequences), which are subjected to a selective environment, a test tube coated with a target molecule, for example. A small fraction of the random RNA population will selectively bind to the target molecules. Those RNA strands are recovered, amplified with mutations (through reverse transcription, PCR, and transcription), and the process is repeated several times. Each cycle yields a more restricted RNA population with improved binding specificity (i.e., a higher degree of function, Ex).
Carothers et al. (34), who analyzed the distribution of functional RNA aptamers in a random population, provide data on a specific example. They identify 11 distinct classes of GTP-binding RNAs, which are distinguished from each other both by nucleotide sequences (RNA motifs) (52) and secondary stem–loop structures. The degree of function of these aptamers can be defined by a solution dissociation constant, a measure of the binding strength between GTP and the folded aptamer. Carothers and coworkers find that a 10-fold increase in GTP binding strength requires ≈10 additional bits of information (i.e., a 1,000-fold decrease in abundance in a population of random sequences). Such a finding is in accord with studies of biopolymers (58, 59) that show functionally similar peptides with dissimilar primary structures, as well as reports of many distinct classes of protease enzymes (60, 61).
Furthermore, although the data of Carothers et al. (34) are too few to draw definitive conclusions, there is a suggestion of a stepped relationship between binding strength (Ex) and functional information (I), a relationship analogous to that displayed by populations of Avida organisms (e.g., Fig. 1). These steps, if real, are likely caused by the existence of separate classes of GTP-binding solutions. Functional classes with greater numbers of stems represent a significantly smaller fraction of all RNA sequences, but they have the potential to display greater GTP-binding affinities.
Functional Information in Higher-Dimensional Systems.
Functional information provides a measure of complexity by quantifying the probability that an arbitrary configuration of a system of numerous interacting agents (and hence a combinatorially large number of different configurations) will achieve a specified degree of function. This concept was originally discussed in the context of biopolymer sequences that perform specific binding or catalytic functions (15, 34). In the preceding sections we demonstrated that the extension of functional information analysis to one-dimensional systems of letters or Avida computer code is conceptually straightforward, requiring only specification of the degree of function of each possible sequence.
We suggest that the functional information formalism may also be applicable to complex physical structures in higher-dimensional systems. Of special interest in this regard are biological systems that display complex emergent behavior, for example, through long-range chemical signaling among a collection of cells in social amoebas (62–64), cooperation among consortia of host organisms and symbionts (65), or colonies of social insects (3, 22, 66). We propose that functional information can be applied, at least in principle, to any such emergent system that has the ability to perform a function.
Many emergent systems can be analyzed in terms of their ability to dissipate energy or maximize entropy production (27, 29, 67, 68). For example, consider the functional information of an assemblage of sand grains subjected to a steady flow of wind or water (e.g., refs. 69 and 70). The formation of periodic sand dunes or ripples serves to initiate turbulent flow and thus increase energy dissipation. Functional information of the system can thus be measured as the fraction of all possible sand configurations, F(Ex), that achieve at least the corresponding energy dissipation, Ex. Such a problem might be analyzed with Monte Carlo simulations of numerous gravitationally stable sand configurations. The analytical challenge remains to determine the degree of function of a statistically significant random fraction of all possible configurations of the system so that the relationship between I(Ex) and Ex can be deduced.
Conclusions
A complexity metric is of little utility unless its conceptual framework and predictive power result in a deeper understanding of the behavior of complex systems. Analysis of complex systems in terms of functional information reveals several characteristics that are important in understanding the behavior of systems composed of many interacting agents. Letter sequences, Avida genomes and biopolymers all display degrees of functions that are not attainable with individual agents (a single letter, machine instruction, or RNA nucleotide, respectively). In all three cases, highly functional configurations comprise only a small fraction of all possible sequences. Furthermore, these three examples reveal that several discrete classes of functional configurations exist, a situation that can lead to distinctive step features in plots of information versus function.
The functional information formalism may also point to key factors in the origin and emergence of biocomplexity. In particular, functional information quantifies the probability that, for a particular system, a configuration with a specified degree of function will emerge. Furthermore, analysis of the relationship between information and function may reveal how much more information is required to encode a given improvement in function. The formalism also points to strategies, such as increasing the concentration and/or diversity of molecular agents, that might maximize the effectiveness of chemical experiments that attempt to replicate steps in the origin of life.
Methods
Determination of the computational properties of a randomly generated instruction sequence is accomplished within Avida's analyze mode. The trace feature in analyze mode generates detailed information on the state of the virtual computer at each step in the processing of a genome, including a notation of when a recognized function has been executed. An automated script parsed these logs to collect all of the data necessary to determine the functional properties of each sequence and cataloged the genomes found to be functional to permit later study. Detailed documentation of the Avida software, including descriptions of the trace function and analyze mode, can be found online at the Digital Evolution Laboratory at Michigan State University web site (http://devolab.cse.msu.edu/software/avida/doc).
Acknowledgments
We thank John Avise and Francisco Ayala for organizing this SacklerColloquium; H. J. Cleaves, K. Esler, R. Lenski, H. Morowitz, C. Ofria and D. Sverjensky for valuable comments and suggestions. This work was supported in part by the National Aeronautics and Space Administration Astrobiology Institute, the National Science Foundation, and the Carnegie Institution. J.W.S. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “In the Light of Evolution I: Adaptation and Complex Design,” held December 1–2, 2006, at the Arnold and Mabel Beckman Center of the National Academies of Sciences and Engineering in Irvine, CA. The complete program is available on the NAS web site at www.nasonline.org/adaptation_and_complex_design.
The authors declare no conflict of interest.
References
- 1.Deamer DW, Evans J. In: Life As We Know It: Cellular Origin, Life in Extreme Habitats and Astrobiology. Seckbach J, editor. Vol 10. New York: Springer; 2006. [Google Scholar]
- 2.Frisch M. Turbulence: The Legacy of A. N. Kolmogorov. Cambridge, UK: Cambridge Univ Press; 1995. [Google Scholar]
- 3.Camazine S, Deneubourg JL, Franks NR, Sneyd H, Theraulaz G, Bonabeau E. Self-Organization in Biological Systems. Princeton: Princeton Univ Press; 2001. [Google Scholar]
- 4.Carlberg R. In: The Astronomy and Astrophysics Encyclopedia. Maran SP, editor. New York: Van Nostrand Reinhold; 1992. pp. 268–270. [Google Scholar]
- 5.Mitchell M. An Introduction to Genetic Algorithms. Cambridge, MA: MIT Press; 1996. [Google Scholar]
- 6.Wolfram S. A New Kind of Science. Champaign, IL: Wolfram; 2002. [Google Scholar]
- 7.Adami C. Physica D. 1995;80:154–170. [Google Scholar]
- 8.Holland JH. Hidden Order. Reading, MA: Helix Books; 1995. [Google Scholar]
- 9.De Duve C. Vital Dust: Life as a Cosmic Imperative. New York: Basic Books; 1995. [Google Scholar]
- 10.Morowitz HJ. The Emergence of Everything. New York: Oxford Univ Press; 2002. [Google Scholar]
- 11.Hazen RM. Genesis: The Scientific Quest for Life's Origins. Washington, DC: Joseph Henry; 2005. [Google Scholar]
- 12.Gell-Mann M. Complexity. 1995;1:16–19. [Google Scholar]
- 13.Adami C. Complexity. 2003;8:49–56. [Google Scholar]
- 14.Shalizi CR. 2006 ArXiv: nlin.AO/0307015. [Google Scholar]
- 15.Szostak JW. Nature. 2003;423:689. doi: 10.1038/423689a. [DOI] [PubMed] [Google Scholar]
- 16.Kåhre J. The Mathematical Theory of Information. Boston: Kluwer; 2002. [Google Scholar]
- 17.Gell-Mann M, Lloyd S. In: Nonextensive Entropy: Interdisciplinary Applications. Gell-Mann M, Tsallis C, editors. New York: Oxford Univ Press; 2003. pp. 387–389. [Google Scholar]
- 18.Von Baeyer HC. Information: The New Language of Science. London: Weidenfeld & Nicolson; 2003. [Google Scholar]
- 19.Morowitz HJ. Foundations of Bioenergetics. New York: Academic; 1978. [Google Scholar]
- 20.Bell G. The Basics of Selection. New York: Chapman & Hall; 1997. [Google Scholar]
- 21.Allen C, Bekoff M, Lauder G, editors. Nature's Purposes: Analyses of Function and Design in Biology. Cambridge, MA: MIT Press; 1998. [Google Scholar]
- 22.Solé R, Goodwin B. Signs of Life: How Complexity Pervades Biology. New York: Basic Books; 2000. [Google Scholar]
- 23.Avery J. Information Theory and Evolution. Singapore: World Scientific; 2003. [Google Scholar]
- 24.Ricard J. C R Soc Biol. 2003;326:133–140. doi: 10.1016/s1631-0691(03)00064-7. [DOI] [PubMed] [Google Scholar]
- 25.Lehman N, Donne MD, West M, Dewey TG. J Mol Evol. 2000;50:481–490. doi: 10.1007/s002390010051. [DOI] [PubMed] [Google Scholar]
- 26.Bertalanffy L. General System Theory: Foundations, Applications, Development. New York: Braziller; 1968. [Google Scholar]
- 27.Nicolis G, Prigogine I. Self-Organization in Non-Equilibrium Systems: From Dissipative Structures to Order through Fluctuations. New York: Wiley; 1977. [Google Scholar]
- 28.Swenson R, Turvey MT. Ecol Psychol. 1991;3:317–348. [Google Scholar]
- 29.Emanuel K. Phys Today. 2006:74–75. August. [Google Scholar]
- 30.Ayala FJ. Hist Philos Life Sci. 1999;21:3–33. [PubMed] [Google Scholar]
- 31.McShea DW. Biol Philos. 2000;15:641–668. [Google Scholar]
- 32.Bigelow J, Pargetter RP. In: Nature's Purposes: Analyses of Function and Design in Biology. Allen C, Bekoff M, Lauder G, editors. Cambridge, MA: MIT Press; 1998. [Google Scholar]
- 33.Schneider TD, Stormo GD, Gold L, Ehrenfeuch A. J Mol Biol. 1986;188:415–431. doi: 10.1016/0022-2836(86)90165-8. [DOI] [PubMed] [Google Scholar]
- 34.Carothers JM, Oestreich SO, Davis JH, Szostak JW. J Am Chem Soc. 2004;126:5130–5137. doi: 10.1021/ja031504a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lenski RE, Ofria C, Pennock RT, Adami C. Nature. 2003;423:139–144. doi: 10.1038/nature01568. [DOI] [PubMed] [Google Scholar]
- 36.Adami C. Introduction to Artificial Life. New York: Springer; 1998. [Google Scholar]
- 37.Wright S. Bull Am Math Soc. 1942;48:223–246. [Google Scholar]
- 38.Shannon C. Bell System Technical J. 1948;27:379–423. 623–656. [Google Scholar]
- 39.Klir G. Uncertainty and Information: Foundations of Generalized Information Theory. Hoboken, NJ: Wiley; 2006. [Google Scholar]
- 40.Mills DR, Peterson RL, Spiegelman S. Proc Natl Acad Sci USA. 1967;58:217–224. doi: 10.1073/pnas.58.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Andersson JO, Andersson SG. Curr Opin Genet Dev. 1999;9:664–671. doi: 10.1016/s0959-437x(99)00024-6. [DOI] [PubMed] [Google Scholar]
- 42.Shigenobu S, Watanabe H, Hattori M, Sakaki Y, Ishikawa H. Nature. 2000;407:81–86. doi: 10.1038/35024074. [DOI] [PubMed] [Google Scholar]
- 43.Nakabuchi A, Yamashita A, Toh H, Ishikawa H, Dunbar HE, Moran NA, Hattori M. Science. 2006;314:267. doi: 10.1126/science.1134196. [DOI] [PubMed] [Google Scholar]
- 44.Lenski RE, Ofria C, Collier TC, Adami C. Nature. 1999;400:661–664. doi: 10.1038/23245. [DOI] [PubMed] [Google Scholar]
- 45.Turing A. Proc London Math Soc. 1936;42:230–265. [Google Scholar]
- 46.Wilson EO. The Diversity of Life. Cambridge, MA: MIT Press; 1992. [Google Scholar]
- 47.Ellington AE, Szostak JW. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 48.Wilson DS, Szostak JW. Annu Rev Biochem. 1999;68:611–647. doi: 10.1146/annurev.biochem.68.1.611. [DOI] [PubMed] [Google Scholar]
- 49.Van Nimwegen E, Crutchfield JP, Huymen M. Proc Natl Acad Sci USA. 1999;96:9716–9720. doi: 10.1073/pnas.96.17.9716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Taverna DM, Goldstein RA. Biopolymers. 2000;53:1–8. doi: 10.1002/(SICI)1097-0282(200001)53:1<1::AID-BIP1>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 51.Sasaki A, Nowak MA. J Theor Biol. 2003;224:241–247. doi: 10.1016/s0022-5193(03)00161-9. [DOI] [PubMed] [Google Scholar]
- 52.Knight R, Yarus M. RNA. 2003;9:218–230. doi: 10.1261/rna.2138803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huynen MA, Stadler PF, Fontana W. Proc Natl Acad Sci USA. 1996;93:397–401. doi: 10.1073/pnas.93.1.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reidys C, Forst CV, Schuster P. Bull Math Biol. 2001;63:57–94. doi: 10.1006/bulm.2000.0206. [DOI] [PubMed] [Google Scholar]
- 55.Ebner M, Shackleton M, Shipman R. Complexity. 2002;7:19–33. [Google Scholar]
- 56.Codoñer FM, Daròs J-A, Solé RV, Elena SF. PLoS Pathogens. 2006;2:1187–1193. doi: 10.1371/journal.ppat.0020136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zarowski JC. An Introduction to Numerical Analysis for Electrical and Computer Engineers. New York: Wiley; 2004. [Google Scholar]
- 58.Aronson HG, Royer WE, Hendrickson WA. Protein Sci. 1994;3:1706–1711. doi: 10.1002/pro.5560031009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang QS, Unrau PJ. RNA. 2005;11:404–411. doi: 10.1261/rna.7238705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rawlings ND, Barrett AJ. Biochem J. 1993;290:205–218. doi: 10.1042/bj2900205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rawlings ND, Morton FR, Barrett AJ. Nucleic Acids Res. 2006;34:D270–D272. doi: 10.1093/nar/gkj089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Goldbeter A. Biochemical Oscillations and Cellular Rhythms. New York: Cambridge Univ Press; 1996. [Google Scholar]
- 63.Brännström Å, Dieckmann U. Proc R Soc London Ser B. 2005;272:1609–1616. doi: 10.1098/rspb.2005.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schaap P, Winckler T, Nelson M, Alvarez-Curto E, Elgie B, Hagiwara H, Cavender J, Milano-Curto A, Rozen DE, Dingermann T, et al. Science. 2006;314:661–663. doi: 10.1126/science.1130670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Moran NA. Proc Natl Acad Sci USA. 2007;104(Suppl):8627–8633. doi: 10.1073/pnas.0611659104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Strassman JE, Queller DC. Proc Natl Acad Sci USA. 2007;104(Suppl):8619–8626. doi: 10.1073/pnas.0701285104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lorenz RD. Science. 2003;299:837. doi: 10.1126/science.1081280. [DOI] [PubMed] [Google Scholar]
- 68.Whitfield J. Nature. 2005;436:905–907. doi: 10.1038/436905a. [DOI] [PubMed] [Google Scholar]
- 69.Bagnold RA. The Physics of Sediment Transport by Wind and Water. New York: Am Soc of Civil Eng; 1988. [Google Scholar]
- 70.Hansen JL, van Hecke M, Hanning A, Ellegaard C, Andersen KH, Bohr T, Sams T. Nature. 2001;410:324. doi: 10.1038/35066631. [DOI] [PubMed] [Google Scholar]