Abstract
The pentatricopeptide repeat (PPR) proteins form one of the largest families in higher plants and are believed to be involved in the posttranscriptional processes of gene expression in plant organelles. It has been shown by using a genetic approach focusing on NAD(P)H dehydrogenase (NDH) activity that a PPR protein CRR4 is essential for a specific RNA editing event in chloroplasts. Here, we discovered Arabidopsis crr21 mutants that are specifically impaired in the RNA editing of the site 2 of ndhD (ndhD-2), which encodes a subunit of the NDH complex. The CRR21 gene encodes a member of the PPR protein family. The RNA editing of ndhD-2 converts the Ser-128 of NdhD to leucine. In crr21, the activity of the NDH complex is specifically impaired, suggesting that the Ser128Leu change has important consequences for the function of the NDH complex. Both CRR21 and CRR4 belong to the E+ subgroup in the PLS subfamily that is characterized by the presence of a conserved C-terminal region (the E/E+ domain). This E/E+ domain is highly conserved and exchangeable between CRR21 and CRR4, although it is not essential for the RNA binding. Our results suggest that the E/E+ domain has a common function in RNA editing rather than of recognizing specific RNA sequences.
Keywords: Arabidopsis, chloroplast
The plastid genome encodes ≈120 genes that are involved in photosynthesis and housekeeping functions in higher plants. Marked changes in plastid gene expression take place during chloroplast development from undifferentiated proplastids (1). The process is accompanied by the conversion of the transcriptional machinery from nucleus-encoded phage-type RNA polymerase to plastid-encoded bacterial-type RNA polymerase (2). Once plastids are developed, however, plastid gene expression is preferentially regulated at the posttranscriptional level (3) via RNA splicing, RNA processing, RNA editing, RNA degradation, and translation (4). Because the chloroplast genome lacks apparent regulatory genes, chloroplast gene expression is modulated by nucleus-encoded factors. A number of nucleus-encoded factors involved in the posttranscriptional processes have been identified by genetic and biochemical strategies (4).
A growing mass of information supports the idea that the pentatricopeptide repeat (PPR) protein family is involved in posttranscriptional regulation of gene expression in both plastids and mitochondria. The family members are defined by the tandem array of a PPR motif, which is a highly degenerate unit consisting of 35 aa (5). The PPR protein family is extraordinarily large, especially in higher plants, containing 466 members in Arabidopsis and 480 in rice (5). Most PPR proteins are predicted to target plastids or mitochondria (5). Maize CRP1 is required for the translation of petA and psaC and for the processing of petD (6, 7), whereas Arabidopsis CRR2 is required for intergenic RNA processing between rps7 and ndhB (8). Arabidopsis HCF152 is essential for both splicing of petB and intergenic RNA cleavage between psbH and petB (9, 10). Arabidopsis PGR3 is involved in the stabilization of petL RNA operons and may be also involved in the translation of petL and one of the ndh genes (11). Members of the PPR protein family are also involved in the suppression of cytoplasmic male sterility (CMS) possibly via RNA processing or editing of CMS-associated transcripts (12–14).
In Arabidopsis, roughly half of the PPR proteins are in the PLS subfamily, also referred to the plant combinatorial and modular protein (PCMP) subfamily, which is specific to land plants (5, 15). The PLS subfamily exhibits a variable tandem repeat of a standard pattern of three PPR variant motifs (5). The association or not of this repeat with three non-PPR motifs at their C terminus subdivides the PLS subfamily into a further four subgroups; PLS, E, E+, and DYW (5, 15). The expansion of the PLS subfamily in plants may be due to accommodation to the specific characteristics of plant organelles, for example, RNA editing which is unique to land plants (5, 15).
In higher plants, RNA editing is a posttranscriptional process of altering a specific C nucleotide to U in an RNA molecule in mitochondria and plastids (16–19). By contrast, U to C conversion has also been shown to occur frequently in both ferns and hornworts (20, 21). In higher plants, ≈30 editing sites have been detected in the genomes of plastids and >400 in mitochondria (22–25). For site-specific RNA editing, a cis-element is essential and consists of <30 nt surrounding the editing site and, in some cases, a 5′ sequence distal from the editing site in plastids (26–29). A nucleus-encoded factor responsible for the specific RNA editing event was discovered in the genetic study of photosynthetic electron transport. The Arabidopsis crr4 mutants are defective in RNA editing, which creates the translational initial codon of the plastid ndhD gene (the ndhD-1 site) (30). The ndhD gene encodes a subunit of the chloroplast NAD(P)H dehydrogenase (NDH) complex that is involved in cyclic electron flow around photosystem I (31, 32). The CRR4 gene encodes a member of the PPR protein family and belongs to the E+ subgroup (30). We recently showed that the recombinant CRR4 expressed in Escherichia coli directly binds to the 25 nucleotides of the upstream and the 10 nucleotides of the downstream sequences surrounding the editing site of ndhD-1, confirming our model in which a PPR protein is a trans-factor essential to recognition of the RNA editing site (33).
In this study, we report the discovery of a second PPR protein, CRR21, which is essential for a specific RNA editing event in plastids. These findings suggest that the C-terminal domain in the E and E+ subgroups might have a common function among trans-factors of RNA editing in chloroplasts.
Results
Arabidopsis crr21 Mutants Were Defective in NDH Activity.
The chloroplast NDH complex catalyzes electron donation to plastoquinone (PQ) from the stromal electron pool. The NDH activity can be monitored as a transient increase in chlorophyll fluorescence after turning off actinic light (AL) (31, 34, 35). The increase in fluorescence is due to reduction of PQ by the stromal electron pool which accumulates during AL illumination. We focused on this change in fluorescence to identify Arabidopsis crr mutants with altered NDH activity (8). More than 10,000 transposon-tagged lines of Arabidopsis were constructed by using the Activator(Ac)/Dissociation(Ds) system, and the flanking sequences of Ds elements were determined (36, 37). In more exhaustive screening for crr mutants, we analyzed a series of mutant lines which have insertions of Ds transposons in the genes encoding putative plastid-targeting proteins directly by using pulse amplitude modulated (PAM) fluorometry. Fig. 1 shows a typical trace of the chlorophyll fluorescence level in the wild type. In crr21 mutants, the transient increase in fluorescence level after the AL illumination was suppressed (Fig. 1), indicating that NDH activity was impaired.
Fig. 1.
Monitoring of NDH activity by using chlorophyll fluorescence analysis. The curve shows a typical trace of chlorophyll fluorescence in the wild type (WT). Leaves were exposed to AL (50 μmol of photons m−2 s−1) for 5 min. AL was turned off and the subsequent change in chlorophyll fluorescence level was monitored. Insets are magnified traces from the boxed area. The fluorescence levels were normalized by Fm levels. ML, measuring light; SP, saturating pulse of white light; crr21+CRR21, crr21 complemented by introduction of the wild-type genomic CRR21.
CRR21 Encodes a Member of the PPR Protein Family.
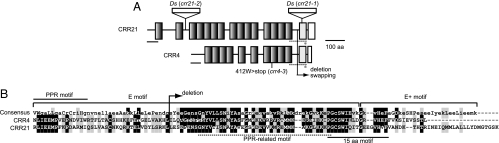
In two independent mutants, crr21–1 and crr21–2, the At5g55740 gene was disrupted by independent insertion of Ds (Fig. 2A). Both mutants had a recessive nature and co-segregated with the Ds insertions (data not shown). To confirm that the crr21 phenotype is due to defects in At5g55740, wild-type genomic DNA covering the sequence encoding At5g55740 was introduced into both crr21–1 and crr21–2. This complementation fully restored the transient increase in chlorophyll fluorescence after AL illumination (Fig. 1). We concluded that the crr21 phenotype was due to the disruption of At5g55740 and that crr21–1 and crr21–2 are allelic. The CRR21 (At5g55740) gene is not interrupted by any introns and encodes a putative protein consisting of 830 aa. The program TargetP 1.1 predicted that the first 59 aa were the target signal to plastids [Fig. 2A and supporting information (SI) Fig. 7]. BLAST and Pfam database searches showed that CRR21 is a member of the PPR protein family and contains 16 characteristic PPR motifs, including one PPR-related motif in the E motif (Fig. 2A and SI Fig. 7). According to the recent bioinformatic classification of PPR proteins (5, 15), CRR21 belongs to the E+ subgroup of the PLS subfamily that contains the E and E+ motifs following a tandem array of PPR motifs (Fig. 2A and SI Fig. 7). In addition, CRR21 contained an unknown motif consisting of 15 aa (PGCSxI/VExxGxV/IHxF), which is highly conserved in some PPR proteins, including CRR4 (Fig. 2 and SI Fig. 7).
Fig. 2.
Predicted motif structure of CRR21. (A) Predicted motif structure of CRR21 as compared with that of CRR4. PPR motifs are depicted as shaded black boxes. The E motif is shown as a dotted line. The PPR-related motif in the E motif and the E+ motif are depicted as gray and white boxes, respectively. The putative transit peptide to plastids is underlined. The 15-amino acid motif is shown by an asterisk. Sites of Ds insertion and the position of the crr4–3 mutation (nonsense) are indicated. Positions of the deletion and domain swapping are indicated. (B) Partial sequence alignment of CRR21 and CRR4. Alignment was performed by using the ClustalW program. The consensus sequence of the E and E+ motifs according to bioinformatics analysis by Lurin et al. (5) is shown at the top of the sequences. The best-conserved residues are in capital letters. Amino acids that are fully conserved or substitutive are respectively shaded black and gray. The PPR-related motif in the E motif and the 15-amino acid motif are respectively underlined by a dotted line and a solid line. The last PPR motifs of CRR4 and CRR21 are indicated. The deletion of the E/E+ domain of CRR4 in the crr4–3+CRR4 (−E/E+) construct is specified.
RNA Editing of the ndhD-2 Site Was Specifically Impaired in crr21 Mutants.
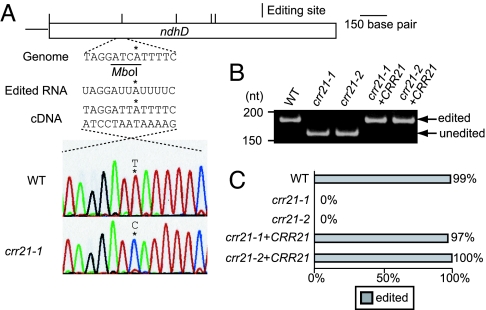
Several PPR proteins are involved in the RNA maturation processes such as RNA processing, stabilization, translation, and editing in organelles in higher plants (7–9, 11). This fact suggests that the RNA maturation process of the chloroplast ndh gene(s) would be impaired in crr21. To assess this possibility, RNA editing of ndh genes was analyzed by directly sequencing RT-PCR products. We confirmed that 16 editing sites present in the ndh genes in another Arabidopsis ecotype, Columbia (38, 39), were also conserved in the Nössen ecotype that is the background for crr21 (SI Table 1). Fig. 3A shows the result of the direct sequencing of RT-PCR products around the RNA editing site 2 of ndhD (ndhD-2). In the wild type, cDNA carrying T at ndhD-2 predominated and corresponds to the edited RNA encoding serine rather than leucine. In contrast, this site was not edited in transcripts from crr21 mutants (Fig. 3A and SI Table 1). Fig. 3B shows a semiquantitative analysis of RNA editing extent using a restriction enzyme that digests only cDNAs derived from unedited RNA molecules. RT-PCR products subjected to the analysis covered the region from −154 to +29 (taking the ndhD-2 site as +1). In the wild type, almost all cDNA was resistant to the restriction enzyme, indicating that the ndhD-2 site is fully edited. In contrast, almost all cDNA was digested in both alleles of crr21, indicating that the editing of ndhD-2 was totally inhibited in crr21 mutants (Fig. 3B). To estimate more accurate efficiency of RNA editing, cDNA including the editing site was amplified by PCR and cloned in E. coli. One hundred independent clones were analyzed by digestion with MboI to detect cDNA originating from the edited RNA molecules. Consistent with the estimation of semiquantitative analysis, 99% of molecules were edited in the wild type (Fig. 3C). In contrast, no molecules were edited in crr21 mutants (Fig. 3C), supporting the idea that crr21 mutants are a null allele. Twenty-eight editing sites have been identified in the plastid genome of Arabidopsis to date (39). To investigate whether other sites are edited in crr21 mutants, we analyzed RNA editing in all of the editing sites. Except for the ndhD-2 site, we did not find any defects in RNA editing in crr21 mutants (SI Table 1).
Fig. 3.
Analysis of RNA editing of ndhD-2. (A) Direct sequencing of RT-PCR products containing the ndhD-2 editing site. Positions of RNA editing are indicated. The restriction enzyme MboI specifically cleaves cDNA derived from unedited molecules. The ndhD-2 site is shown by an asterisk. (B) Semiquantitative analysis of the extent of RNA editing. RT-PCR products were digested with MboI. Fragments originating from edited and unedited RNA molecules are indicated. (C) The editing efficiency of ndhD-2 was analyzed as described in Materials and Methods. Ratio of clones originated from edited RNA molecules is indicated by gray bars. crr21+CRR21, crr21 complemented by introduction of the wild-type genomic CRR21.
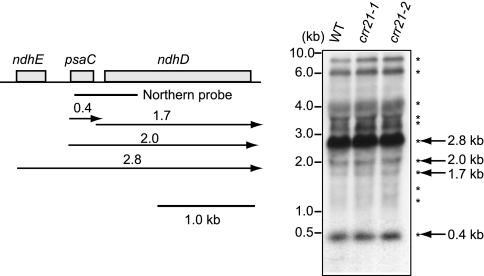
In Arabidopsis, ndhD is polycistronically transcribed with the upstream gene psaC, and then processed into monocistronic RNA (40, 41). The crr21 defect may indirectly affect RNA editing via aberrant RNA processing. To test this possibility, the intergenic RNA cleavage between psaC and ndhD in crr21 mutants was analyzed in an RNA gel blot. The hybridization pattern was identical between the wild type and crr21 mutants (Fig. 4), indicating that the defect in RNA editing is not caused by a primary defect in RNA processing between psaC and ndhD. We also analyzed other plastid-encoded ndh transcripts. All of the hybridization patterns were identical between the wild type and crr21 mutants (SI Fig. 8). These results suggest that the crr21 defect is specific to the RNA editing in the ndhD-2 site.
Fig. 4.
Transcript pattern of psaC-ndhD operon. Total RNA (5 μg) isolated from leaves of 3-week-old wild type (Nössen), crr21–1, and crr21–2 was analyzed by RNA gel blot hybridization. The positions of RNA size markers are indicated on the left. The positions and sizes of RNA detected by RNA gel blot hybridization indicated by asterisks and arrows with letters on the right. Transcript map was deduced from this and the previous report (41).
Activity of the NDH Complex Was Specifically Impaired in crr21 Mutants.
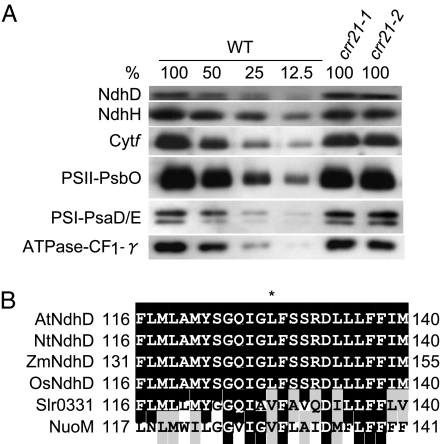
The RNA editing of ndhD-2 converts Ser-128 of NdhD (uCa) to leucine (uUa). This amino acid alteration may destabilize the NdhD subunit. It is also possible that the defect in RNA editing may interfere with translation. To assess whether the mutant form of NdhD is stable in vivo, a protein blot was performed by using an antibody against NdhD. In both alleles of crr21, the NdhD level was not affected (100–50% of the wild-type level) (Fig. 5A). The NDH complex is unstable without the NdhD subunit (30) and the antibody against NdhH can be used to monitor the accumulation of NDH complex. In both alleles of crr21, the NdhH level was also comparable to the wild-type level (100–50%) (Fig. 5A). Because the editing of ndhD-2 was below the detection limit (Fig. 3C), the majority (probably almost all) of NdhD accumulating in crr21 was translated from unedited RNA. The Leu-128 remains as serine in NdhD of crr21, and the residue is essential for activity, but not for stability of the NDH complex. Consistent with our idea, the Leu-128 of NdhD is generated by RNA editing in most dicot plants and is also encoded by the genome of monocot plants (Fig. 5B) (22). This leucine is conserved as a valine that is structurally similar in NdhD homologs of other organisms such as cyanobacteria and E. coli (Fig. 5B).
Fig. 5.
Protein blot analysis of the NDH complex and the major photosynthetic complexes. (A) Immunodetection of NDH subunits, NdhD and NdhH; a subunit of the cytb6f complex, Cytf; subunits of the photosystem I, PsaE/D; a subunit of the photosystem II, PsbO; and γ-subunit of the chloroplast F0F1-ATPase, CF1-γ. Proteins were extracted from the thylakoid membrane fractions. Lanes were loaded with protein samples corresponding to 0.5 μg chlorophyll for Cytf, PsaD/E, PsbO, and CF1-γ, 5 μg chlorophyll for NdhH, and 10 μg chlorophyll for NdhD (100%) and the series of dilutions indicated. (B) Partial sequence alignment of NdhD containing the editing site. Arabidopsis NdhD protein was aligned with their homologous proteins. Alignment was performed by using the ClustalW program. Sequences (GenBank accession numbers) shown here are as follows: Arabidopsis thaliana (AtNdhD, BAA84437), Nicotiana tabacum (NtNdhD, CAA77432), Zea mays (ZmNdhD, CAA31558), Oryza sativa (OsNdhD, NP_039444), Synechocystis sp. PCC 6803 (Slr0331, NP_441967), and Escherichia coli K12 (NuoM, NP_416780). Amino acids that are fully conserved or substitutive are shaded black and gray, respectively. The asterisk indicates the position of an edited codon. The Ser-128 of AtNdhD and NtNdhD is converted to leucine by RNA editing of ndhD-2. Numbers indicate amino acid positions in protein.
The protein blot experiments were also performed to assess the accumulation of the major photosynthetic complexes by probing their representative subunits. The levels of the subunits of photosystem I, photosystem II, cytochrome (Cyt) b6ƒ complex, and chloroplast F0F1-ATPase in crr21 mutants were the same of those in the wild type (Fig. 5A). Consistent with these results, crr21 mutants did not show any phenotypic changes in photosynthetic electron transport except for NDH activity (data not shown). Thus, crr21 mutants are specifically impaired in the RNA editing of ndhD-2.
The E/E+ Domain Has a Conserved Function Between CRR4 and CRR21.
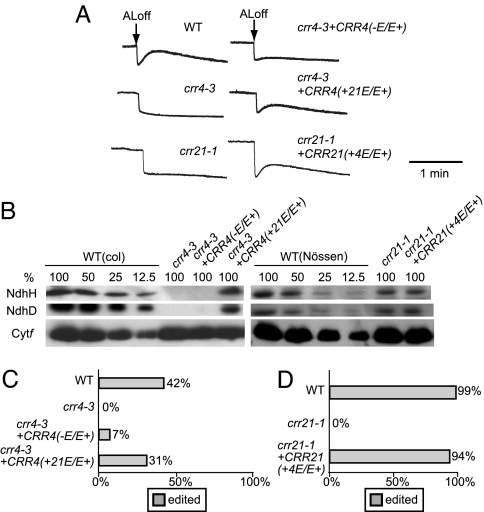
We previously reported that CRR4, a PPR protein, is essential for the RNA editing of ndhD-1 (30). CRR21 does not show a significant sequence similarity to CRR4 with respect to PPR motifs; this is consistent with the idea that a tandem array of PPR motifs is involved in the recognition of a specific RNA sequence. However, the array of PPR motifs is followed by the E and E+ motifs, which are well conserved between CRR4 and CRR21 as well as some members of the PPR protein family. Fig. 2B shows the alignment of the C-terminal region of CRR4 and CRR21. Although CRR4 lacks the C-terminal 16 aa of E+ motif, the remaining sequence of the E and E+ motifs is well conserved. Notably, the boundary of the E and E+ motifs that includes a PPR-related motif and the 15-amino acid motif is highly conserved (Fig. 2B). This region showed 44% sequence identity (61% similarity) between CRR4 and CRR21. The highly conserved C-terminal region (E/E+ domain) might have a common function between CRR4 and CRR21, rather than the specific function of recognizing distinct RNA sequences. To assess this possibility, CRR4 truncated in the E/E+ domain was expressed in crr4–3, in which RNA editing of ndhD-1 is completely impaired (30). NDH activity was drastically impaired in the transgenic lines, although a trace amount of the transient increase in chlorophyll fluorescence was detected after AL illumination (Fig. 6A). In these plants, the amount of NdhD and NdhH was undetectable level (at least less than 12.5% of the wild type) (Fig. 6B). Efficiency of ndhD-1 RNA editing was also significantly reduced in the plants (7% of molecules were edited) (Fig. 6C). It is noted that the ndhD-1 site is partially edited, even in the wild type (42%). We considered that the E/E+ domain is required for activity of the editing. It is possible that the E/E+ domain is required for RNA editing via binding to the target RNA. To test this possibility, an RNA binding activity for the target RNA sequence (RB3 probe) that includes the 25 upstream nucleotides and the 10 nucleotides in the downstream sequence surrounding the ndhD-1 site was determined by using the recombinant CRR4 lacking the E/E+ domain. As shown in SI Fig. 9, the retarded band was detected with increasing amount of CRR4 lacking the E/E+ domain and the Kd value of 1.8 nM. This binding affinity was almost the same as that of 1.6 nM for the entire CRR4. These results indicate that the E/E+ domain is not important for high-affinity binding to RNA.
Fig. 6.
Effects of the deletion and conversion of the E/E+ domain in CRR4. (A) Analysis of the transient increase in chlorophyll fluorescence after turning off AL. Leaves were exposed to AL (50 μmol of photons m−2 s−1) for 5 min. AL was turned off, and the subsequent change in chlorophyll fluorescence level was monitored. The fluorescence levels were normalized by Fm levels. (B) Protein blot analysis of the NDH complex. Immunodetection of NDH subunits, NdhD and NdhH, and a subunit of the cytb6f complex, Cytf. Proteins were extracted from the thylakoid membrane fractions. Lanes were loaded with protein samples corresponding to 0.5 μg of chlorophyll for Cytf, 5 μg of chlorophyll for NdhH, and 10 μg of chlorophyll for NdhD (100%) and the series of dilutions indicated. (C) Analysis of the extent of RNA editing in the ndhD initiation codon. The editing efficiency of the ndhD-1 was analyzed as described in Materials and Methods. Ratio of clones originated from edited RNA molecule is indicated by gray bars. (D) Analysis of the extent of RNA editing in the ndhD-2 site. Ratio of clones originated from edited RNA molecule is indicated by gray bars. crr4–3+CRR4 (−E/E+), crr4–3 transformed with the CRR4 truncated in the E/E+ domain; crr4–3+CRR4 (+21E/E+), crr4–3 transformed with CRR4, in which the E/E+ domain was replaced by that of CRR21; crr21–1+CRR21(+4E/E+), crr21–1 transformed with CRR21, in which the E/E+ domain was replaced by that of CRR4.
Although CRR4 and CRR21 are involved in different editing events by recognizing distinct target RNAs for editing, conservation of the E/E+ domains suggests a common function for these domains. If this interpretation is true, the domain might be exchangeable between two PPR proteins. The E/E+ domain of CRR4 was therefore exchanged with that of CRR21, and the chimeric gene was introduced into the crr4–3 allele. Both the transient increase in chlorophyll fluorescence after AL illumination (Fig. 6A), and the NdhD and NdhH levels (Fig. 6B) were restored by introducing this chimeric gene. We also confirmed that the introduction of the chimeric gene restored the RNA editing of ndhD-1, although the efficiency of RNA editing in transgenic plants (31%) was slightly lower than that in the wild type (42%) (Fig. 6C). We also performed a reciprocal experiment in which the E/E+ domain of CRR21 was exchanged with that of CRR4, and the chimeric gene was introduced into crr21–1. The transient increase in chlorophyll fluorescence after AL illumination was restored by introducing this chimeric gene (Fig. 6A). We confirmed that this change in NDH activity was not accompanied by any increase in the level of NdhD and the NDH complex (Fig. 6B). We also confirmed that the introduction of the chimeric gene restored the RNA editing of ndhD-2 (Fig. 6D). These results suggest that the E/E+ domain in CRR4 and CRR21 have a common function in RNA editing.
Discussion
Here, we showed that a PPR protein, CRR21, is specifically required for RNA editing in the ndhD-2 site in Arabidopsis chloroplasts. This finding follows our initial discovery of a PPR protein, CRR4, which is required for RNA editing in the ndhD-1 site (30). The hypothesis that a PPR protein functions as a site-recognition factor in plastid RNA editing was confirmed biochemically (33). Discovery of the second PPR protein functioning in RNA editing in chloroplasts improved our knowledge of the editing machinery in chloroplasts.
CRR4 was discovered to be a site-recognition factor for RNA editing of the ndhD-1 site. The RNA editing of ndhD-1 has characters that distinguish it from those in other sites (i) This RNA editing creates a translational initiation codon rather than altering the coding amino acid; and (ii) this site is partially edited and its editing extent is developmentally regulated, suggesting that it may play a role in the regulation of ndhD translation (30, 40). Although we have provided solid genetic and biochemical evidences to indicate that this PPR protein is a site-recognition factor for RNA editing, it may still be possible that CRR4 is essential for stabilizing the edited RNA molecule via regulating translation rather than by direct involvement in RNA editing. However, CRR21 is involved in the RNA editing which alters the coding amino acid, and the editing takes place in all RNA molecules. It is unlikely that the RNA editing is secondarily affected via a defect in RNA stabilization in crr21. The intergenic RNA cleavage between psaC and ndhD was also not affected in crr21 mutants (Fig. 4), as was the case in crr4 (30). Translation is also unlikely to be primarily affected by the crr21 defects, because the ndhD-2 site is fully edited in crr4, in which ndhD mRNA does not have a translational initiation codon (30). These facts indicate that RNA editing is independent from the translation. From these results, it is clear that the PPR proteins are directly involved in RNA editing.
Translation preceding RNA editing may produce mutant versions of proteins. There may be a form of regulation that suppresses the translation of unedited RNA molecules. However, the mutant phenotypes strongly suggest that almost all of the NdhD accumulating in crr21 had been translated from unedited transcripts (Fig. 5A), suggesting that the status of editing is not a determinant of translation. Consistent with our observations, both edited and unedited rps12 transcripts are translated in plant mitochondria, but only the edited translation products accumulate in mitochondrial ribosomes (42). These results indicate that translation products from unedited transcripts are selectively degraded. However, it is unlikely that unedited ndhD mRNA is subjected to translation in the wild type, because it would lead to the accumulation of the inactive NDH complex. We believe that RNA editing of ndhD-2 precedes the translation in the wild type, but that editing is not necessarily essential for translation. Thus, we conclude that crr21 is directly defective in RNA editing of ndhD-2, rather than indirectly through other effects that might influence the editing process. There is also no evidence to suggest that CRR4 is involved in translation that would consequently affect the editing process.
Both CRR4 and CRR21 belong to the E+ subgroup of the PLS subfamily. Although the tandem array of PPR motifs shows some diversity as to length and the sequence between CRR4 and CRR21, the E/E+ domain is highly conserved (Fig. 2). Biochemical analyses of PPR proteins have suggested that the PPR motif acts as an RNA-binding motif (5, 10, 43). The sequence of the E motif is also related to the PPR motifs. However, the sequence of this C-terminal PPR motif in the E motif diverges to some degree from the N-terminal PPR motifs, suggesting that it has distinct function. In consistent with these observations, truncation of the E/E+ domain of CRR4 did not affect to an RNA binding activity (SI Fig. 9), suggesting that the N-terminal PPR motifs of CRR4 are sufficient for binding to target RNA. Moreover, we demonstrated that truncation of the E/E+ domain of CRR4 drastically decreased an RNA editing activity of ndhD-1 in vivo and the E/E+ domain is exchangeable between CRR4 and CRR21 (Fig. 6), suggesting that the E/E+ domain in CRR4 and CRR21 have a common function in RNA editing.
We cannot eliminate the possibility that the E/E+ domain is essential for stabilizing CRR4 rather than the CRR4 function in vivo. To test the case, we constructed a chimeric gene so that the N-terminal end of CRR4 truncated in the E/E+ domain was fused with the HA-epitope tag and introduced it into crr4–3. Transgenic lines showed the same phenotypes in NDH activity, NdhD level, and RNA editing extent of ndhD-1 with the lines transformed with the same construct without HA-epitope tag (data not shown). However, although the accumulation of mRNA was confirmed by RT-PCR, the antibody against HA-epitope tag could not detect the truncated CRR4-HA protein (data not shown). This result is consistent with our previous report that CRR4 protein level remained low even by over-expression possibly via unknown regulatory mechanism (30). Although we could not show the stability of the truncated CRR4 in vivo, it is likely to be stable, because the truncated CRR4 expressed in E. coli is active for RNA binding as the full version (SI Fig. 9).
Another PPR protein CRR2 (a member of the DYW subgroup) that contains all of the motifs present in CRR4 and CRR21 is specifically involved in the RNA processing but not RNA editing (8). Furthermore, truncation of the E/E+ domain did not cause complete loss of an RNA editing activity in vivo (Fig. 6). These observations suggest that the E/E+ domain is unlikely to be domains that catalyze the reaction of RNA editing. This is contrasting to crr4–3, in which the final three PPR motifs are also lost (Fig. 2A) resulting in a complete lack of the RNA editing activity (30). Bioinformatic evidence indicates a different evolution for the PPR region and the C-terminal PPR motifs in the PLS subfamily, pointing to a distinct function (15). These results suggest that several C-terminal PPR motifs might be involved in a common function in RNA editing as well as the E/E+ domain. We propose that the C-terminal region of CRR4 and CRR21 including the E/E+ domain might interact with an editing enzyme catalyzing C to U, which is still unclear, or another component of the editing machinery.
Materials and Methods
Chlorophyll Fluorescence Analysis.
Chlorophyll fluorescence was measured by using a MINI-PAM portable chlorophyll fluorimeter (Walz, Effeltrich, Germany). The transient increase in chlorophyll fluorescence after turning off AL was monitored as described (31).
Plant Transformation.
For complementation of the crr21 mutation, the wild-type genomic sequence surrounded by 5′-GAGCTCATGTCACCTTCTTCTTCTG-3′ and 5′-GAATTCGCAATGTCATCAGTGTCAG-3′ was cloned in pBIN19 vector. The resultant plasmid was introduced into crr21–1 and crr21–2 via Agrobacterium tumefaciens MP90. For the expression of CRR4 truncated in the E/E+ domain to be deleted, the wild-type genomic sequence surrounded by 5′-GGTACCCGTCAGAAGGGTAGCTAG-3′ and 5′-GAATTCAAGGTGTTTTGCGACAAGC-3′ was cloned in the pBIN19 vector. For the expression of CRR4, in which the E/E+ domain of CRR4 is replaced by that of CRR21, the nucleotide sequence encoding CRR4 truncated in the E/E+ domain was ligated to the sequence encoding the E/E+ domain of CRR21 surrounded by 5′-GAATTCCTTGAATCCGAGCCTG-3′ and 5′-GAATTCTTAAAGTTTGATTAGACCCG-3′. The resultant chimeric gene was finally cloned into the pBIN19 vector. The resultant plasmids were introduced into the crr4–3. For the expression of CRR21, in which the E/E+ domain of CRR21 is replaced by that of CRR4, the wild-type genomic sequence surrounded by 5′-GAGCTCATGTCACCTTTCTTCTTCTG-3′ and 5′-GAATTCCAATTTCCTTGATAAGTAGTC-3′ was amplified by PCR and was ligated to the sequence encoding the E/E+ domain of CRR4 surrounded by 5′-GAATTCATTTTGCAGGCTGGATATAAC-3′ and 5′-GAATTCCTGCACTCATGAATCCTC-3′. The resultant chimeric gene was finally cloned into the pBIN19 vector. The resultant plasmid was introduced into crr21–1.
Immunoblot Analysis.
Chloroplasts were isolated from the leaves of 4-week-old plants as described (8). The protein samples were separated by 12.5% SDS/PAGE and used for immunodetection.
Analysis of RNA Editing.
Total RNA was isolated from rosette leaves by using an RNAeasy Plant Mini Kit (Qiagen, Valencia, CA) and treated with DNase I (Invitrogen, Carlsbad, CA). DNA-free RNA (2.5 μg) was reverse-transcribed with random hexamers. Sequences including the editing sites were amplified by PCR with primers (SI Table 2). The RT-PCR products were sequenced directly. For analysis of editing efficiency of ndhD-2, the sequence including the editing sites was amplified by PCR using the primers ndhD-2-edit-FW and ndhD-2-edit-RV. The RT-PCR products were ligated into the pGEM-T vector (Promega, Madison, WI) and transformed in E. coli. PCR products from 100 independent clones by using the primers ndhD-2-edit-FW and ndhD-2-edit-RV were digested with MboI and analyzed on 8% polyacrylamide gel. The editing efficiency of ndhD-1 was analyzed as described (30).
RNA Preparation and RNA Gel Blot Analysis.
The fragments used as probes were obtained by PCR amplification using oligonucleotides: 5′-GAGCATGCCCTACAGAC-3′ and 5′-CCATCTATTCCCATTCTCC-3′ for psaC and ndhD region. Labeling of the probe was carried out by using a PCR DIG Probe Synthesis Kit (Roche, Indianapolis, IN). Total RNA was isolated from the leaves of 3-week-old plants by using an RNAeasy Plant Mini Kit (Qiagen) and treated with DNase I (Invitrogen). Total RNA (5 μg) was then loaded onto a 1% agarose gel containing 0.6 M formaldehyde and electrophoresed in 1× Mops buffer (pH 7.0) comprising 20 mM Mops, 5 mM sodium acetate, and 2 mM EDTA. After electrophoresis, the RNA was transferred onto a nylon membrane and hybridized with DIG-labeled DNA probe. The signals from the hybridized bands were detected by using a Gene Image CDP-Star Detection Kit (GE Healthcare, Piscataway, NJ).
Acknowledgments
We thank Misato Tanoue for skilled technical support and Tsuyoshi Endo (Kyoto University, Kyoto, Japan), Amane Makino (Tohoku University, Tohoku, Japan), Mercedes Martin (University of Alcalá, Alcalá, Spain), and Toru Hisabori (Tokyo Institute of Technology, Tokyo, Japan) for their gifts of antibodies. This work was supported by Grant-in-Aid 16085206 for Scientific Research on Priority Areas and Grant 17GS0316 for Creative Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Abbreviations
- AL
actinic light
- NDH
NAD(P)H dehydrogenase
- PPR
pentatricopeptide repeat.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0700865104/DC1.
References
- 1.Mullet JE. Annu Rev Plant Physiol Plant Mol Biol. 1988;39:475–502. doi: 10.1146/annurev.arplant.48.1.355. [DOI] [PubMed] [Google Scholar]
- 2.Hedtke B, Börner T, Weihe A. Science. 1997;277:809–811. doi: 10.1126/science.277.5327.809. [DOI] [PubMed] [Google Scholar]
- 3.Deng XW, Gruissem W. Cell. 1987;49:379–387. doi: 10.1016/0092-8674(87)90290-x. [DOI] [PubMed] [Google Scholar]
- 4.Barkan A, Goldshumidt-Clermont M. Biochimie. 2000;82:559–572. doi: 10.1016/s0300-9084(00)00602-7. [DOI] [PubMed] [Google Scholar]
- 5.Lurin C, Andres C, Aubourg S, Bellaoui M, Bitton F, Bruyere C, Caboche M, Debast C, Gualberto J, Hoffmann B, et al. Plant Cell. 2004;16:2089–2103. doi: 10.1105/tpc.104.022236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barkan A, Walker M, Nolasco M, Johnson D. EMBO J. 1994;13:3170–3181. doi: 10.1002/j.1460-2075.1994.tb06616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fisk DG, Walker MB, Barkan A. EMBO J. 1999;18:2621–2630. doi: 10.1093/emboj/18.9.2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hashimoto M, Endo T, Peltier G, Tasaka M, Shikanai T. Plant J. 2003;36:541–549. doi: 10.1046/j.1365-313x.2003.01900.x. [DOI] [PubMed] [Google Scholar]
- 9.Meierhoff K, Felder S, Nakamura T, Bechtold N, Schuster G. Plant Cell. 2003;15:1480–1495. doi: 10.1105/tpc.010397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakamura T, Meierhoff K, Westhoff P, Schuster G. Eur J Biochem. 2003;270:4070–4081. doi: 10.1046/j.1432-1033.2003.03796.x. [DOI] [PubMed] [Google Scholar]
- 11.Yamazaki H, Tasaka M, Shikanai T. Plant J. 2004;38:152–163. doi: 10.1111/j.1365-313X.2004.02035.x. [DOI] [PubMed] [Google Scholar]
- 12.Bentolila S, Alfonso AA, Hanson MR. Proc Natl Acad Sci USA. 2002;99:10887–10892. doi: 10.1073/pnas.102301599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kazama T, Toriyama K. FEBS Lett. 2003;544:99–102. doi: 10.1016/s0014-5793(03)00480-0. [DOI] [PubMed] [Google Scholar]
- 14.Wang Z, Zou Y, Li X, Zhang Q, Chen L, Wu H, Su D, Chen Y, Guo J, Luo D, et al. Plant Cell. 2006;18:676–687. doi: 10.1105/tpc.105.038240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rivals E, Bruyère C, Toffano-Nioche C, Lecharny A. Plant Physiol. 2006;141:825–839. doi: 10.1104/pp.106.077826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maier RM, Zeltz P., Kössel H, Bonnard G, Gualberto JM, Grienenberger JM. Plant Mol Biol. 1996;32:343–365. doi: 10.1007/BF00039390. [DOI] [PubMed] [Google Scholar]
- 17.Brennicke A, Marchfelder A, Binder S. FEMS Microbiol Rev. 1999;23:297–316. doi: 10.1111/j.1574-6976.1999.tb00401.x. [DOI] [PubMed] [Google Scholar]
- 18.Bock R. Biochimie. 2000;82:549–557. doi: 10.1016/s0300-9084(00)00610-6. [DOI] [PubMed] [Google Scholar]
- 19.Shikanai T. Cell Mol Life Sci. 2006;63:698–708. doi: 10.1007/s00018-005-5449-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kugita M, Yamamoto Y, Fujikawa T, Matsumoto T, Yoshinaga K. Nucleic Acids Res. 2003;31:2417–2423. doi: 10.1093/nar/gkg327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wolf RG, Rowe CA, Hasebe M. Gene. 2004;339:89–97. doi: 10.1016/j.gene.2004.06.018. [DOI] [PubMed] [Google Scholar]
- 22.Tsudzuki T, Wakasugi T, Sugiura M. J Mol Evol. 2001;53:327–332. doi: 10.1007/s002390010222. [DOI] [PubMed] [Google Scholar]
- 23.Handa H. Nucleic Acids Res. 2003;31:5907–5916. doi: 10.1093/nar/gkg795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giege P, Brennicke A. Proc Natl Acad Sci USA. 1999;96:15324–15329. doi: 10.1073/pnas.96.26.15324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Notsu Y, Masood S, Nishikawa T, Kubo N, Akiduki G, Nakazono M, Hirai A, Kadowaki K. Mol Genet Genomics. 2002;268:434–445. doi: 10.1007/s00438-002-0767-1. [DOI] [PubMed] [Google Scholar]
- 26.Chaudhuri S, Maliga P. EMBO J. 1996;15:5958–5964. [PMC free article] [PubMed] [Google Scholar]
- 27.Hirose T, Sugiura M. EMBO J. 2001;20:1144–1152. doi: 10.1093/emboj/20.5.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reed ML, Peeters NM, Hanson MR. Nucleic Acids Res. 2001;29:1507–1513. doi: 10.1093/nar/29.7.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sasaki T, Yukawa Y, Wakasugi T, Yamada K, Sugiura M. Plant J. 2006;47:802–810. doi: 10.1111/j.1365-313X.2006.02825.x. [DOI] [PubMed] [Google Scholar]
- 30.Kotera E, Tasaka M, Shikanai T. Nature. 2005;433:326–330. doi: 10.1038/nature03229. [DOI] [PubMed] [Google Scholar]
- 31.Shikanai T, Endo T, Hashimoto T, Yamada Y, Asada K, Yokota A. Proc Natl Acad Sci USA. 1998;95:9705–9709. doi: 10.1073/pnas.95.16.9705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Munekage Y, Hashimoto M, Miyake C, Tomizawa K, Endo T, Tasaka M, Shikanai T. Nature. 2004;429:579–582. doi: 10.1038/nature02598. [DOI] [PubMed] [Google Scholar]
- 33.Okuda K, Nakamura T, Sugita M, Shimizu T, Shikanai T. J Biol Chem. 2006;281:37661–37667. doi: 10.1074/jbc.M608184200. [DOI] [PubMed] [Google Scholar]
- 34.Burrows PA, Sazanov LA, Svab Z, Maliga P, Nixon PJ. EMBO J. 1998;17:868–876. doi: 10.1093/emboj/17.4.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kofer W, Koop H-U, Wanner G, Steinmüller K. Mol Gen Genet. 1998;258:166–173. doi: 10.1007/s004380050719. [DOI] [PubMed] [Google Scholar]
- 36.Kuromori T, Hirayama T, Kiyosue Y, Takabe H, Mizukado S, Sakurai T, Akiyama K, Kamiya A, Ito T, Shinozaki K. Plant J. 2004;37:897–905. doi: 10.1111/j.1365.313x.2004.02009.x. [DOI] [PubMed] [Google Scholar]
- 37.Ito T, Motohashi R, Kuromori T, Noutoshi Y, Seki M, Kamiya M, Mizukado S, Sakurai T, Shinozaki K. Plant Cell Physiol. 2005;46:1149–1153. doi: 10.1093/pcp/pci112. [DOI] [PubMed] [Google Scholar]
- 38.Lutz KA, Maliga P. Curr Genet. 2001;40:214–219. doi: 10.1007/s002940100242. [DOI] [PubMed] [Google Scholar]
- 39.Tillich M, Funk HT, Schmitz-Linneweber C, Poltnigg P, Sabater B, Martin M, Maier RM. Plant J. 2005;43:708–715. doi: 10.1111/j.1365-313X.2005.02484.x. [DOI] [PubMed] [Google Scholar]
- 40.Hirose T, Sugiura M. EMBO J. 1997;17:6804–6811. doi: 10.1093/emboj/16.22.6804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.del Campo EM, Sabater B, Martin M. J Biol Chem. 2002;277:36457–36464. doi: 10.1074/jbc.M204500200. [DOI] [PubMed] [Google Scholar]
- 42.Phreaner CG, Williams MA, Mulligan RM. Plant Cell. 1996;8:107–117. doi: 10.1105/tpc.8.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schumitz-Linneweber C, Williams-Carrier R, Barkan A. Plant Cell. 2005;17:2791–2804. doi: 10.1105/tpc.105.034454. [DOI] [PMC free article] [PubMed] [Google Scholar]