Abstract
Island Southeast Asia (ISEA) was first colonized by modern humans at least 45,000 years ago, but the extent to which the modern inhabitants trace their ancestry to the first settlers is a matter of debate. It is widely held, in both archaeology and linguistics, that they are largely descended from a second wave of dispersal, proto-Austronesian–speaking agriculturalists who originated in China and spread to Taiwan ∼5,500 years ago. From there, they are thought to have dispersed into ISEA ∼4,000 years ago, assimilating the indigenous populations. Here, we demonstrate that mitochondrial DNA diversity in the region is extremely high and includes a large number of indigenous clades. Only a fraction of these date back to the time of first settlement, and the majority appear to mark dispersals in the late-Pleistocene or early-Holocene epoch most likely triggered by postglacial flooding. There are much closer genetic links to Taiwan than to the mainland, but most of these probably predated the mid-Holocene “Out of Taiwan” event as traditionally envisioned. Only ∼20% at most of modern mitochondrial DNAs in ISEA could be linked to such an event, suggesting that, if an agriculturalist migration did take place, it was demographically minor, at least with regard to the involvement of women.
Island Southeast Asia (ISEA), the area encompassed by modern Indonesia, East Malaysia, and the Philippines, was colonized by modern humans at least 45,000 years ago1 and possibly >50,000 years ago.2 At that time, the region was split between the Pleistocene continent of Sunda, which stretched from Sumatra to Bali and Palawan, and Wallacea, which included the islands east of Wallace’s line (fig. 1). The Sunda shelf was flooded when sea levels rose in the early-Holocene epoch, spurring the development of maritime exchange between populations on the remnant Sunda islands (especially Borneo and Palawan) and populations in Wallacea.4–6 However, despite this evidence for a dynamic population history in early ISEA, paleoanthropologists tend to classify all early human remains in the region as “Australo-Melanesian” (i.e., related to the indigenous people of Australia and New Guinea) and argue for a mid-Holocene immigration of the ancestors of most of the present-day inhabitants.4
Figure 1. .
Map of Taiwan and Southeast Asia, showing both modern coastlines (darker shading) and the 120-m depth contour below sea level (lighter shading), indicating the extent of Sundaland at the Last Glacial Maximum. Sampling locations or populations are indicated by bold circles and are labeled with short codes if they were from a specific location or population. Taiwan: ATA = Atayal; BUN = Bunun; AMI = Ami; PAI = Paiwan. ISEA: MED = Medan; PAD = Padang; PEK = Pekanbaru; PLB = Palenbang; BGK = Bangka; KK = Kota Kinabalu; BAN = Banjarmasin; MND = Manado; PAL = Palu; TOR = Toraja; UJP = Ujung Padang; MTR = Mataram; WAI = Waingapu. Map outline kindly provided by H. Voris and C. Simpson, Field Museum of Natural History, Chicago.3
The primary justification for a two-tier population history in ISEA is historical linguistics. Almost all indigenous populations in the region speak languages belonging to the Malayo-Polynesian branch of Austronesian, also found in the Pacific and Madagascar, whereas the other nine primary branches of Austronesian (the Formosan languages) are spoken only by aboriginal Taiwanese.7 This is interpreted as implying that the Austronesian languages must have developed in Taiwan and spread, by sea, from there to across their current distribution, with the various branches of Malayo-Polynesian separating along the voyage,8 a model known as “Out of Taiwan.”
In the 1970s, archaeologists began to draw a direct relation between the dispersal of early Austronesian languages and the onset of the Neolithic period in ISEA (and in the Pacific outside of New Guinea). Documentation of the prehistory of ISEA was (and, to an extent, remains) meager,4,9 but several sites have yielded preceramic sequences followed by the introduction of pottery at a mid-Holocene date. The appearance of pottery was used as a proxy for the arrival of the Taiwanese “Austronesians,” whose agricultural subsistence allowed them to supplant or assimilate the indigenous Australo-Melanesian foragers.10 Subsequent archaeological work in China, however, demonstrated the establishment of large settlements dependent on rice agriculture during the early Holocene. Archaeological comparisons suggested that the earliest Neolithic cultures in Taiwan resemble earlier cultures found in South China and also have parallels to Neolithic sites found in the Philippines. According to the model most popular among ISEA archaeologists today, rice agriculturalists in South China dispersed into Taiwan ∼5,500 years ago, where they developed the Austronesian languages before expanding again into the Philippines and the rest of ISEA ∼4,000 years ago.4,11
Vigorous debate continues among archaeologists, however, as to how the evidence should be interpreted. Although the model allows for the assimilation of indigenous populations, the extent of any assimilation cannot be assessed from the archaeological record.12 Moreover, the archaeobotanic evidence in ISEA seems not to fit the simple rice-fueled Out of Taiwan model, placing the motive force for the dispersal in question.13–15 South of northern Luzon, the only possible evidence for Neolithic (pre–Iron Age) rice in ISEA is restricted to western Borneo,16 where it predates the supposed Austronesian arrival. Generally speaking, the Neolithic of ISEA is characterized by tremendous local diversity, rather than a uniform dispersal package,17 and it has even been suggested that Taiwan received some aspects of its Neolithic technology from the mainland and some from ISEA.18
The model, therefore, relies heavily on the linguistic argument—but this is not quite as powerful as often assumed. The Austronesian languages may indeed be most diverse in Taiwan, but assigning a root to the language tree is still not straightforward. Since the reconstructed tree is starlike, with nine Formosan branches and one Malayo-Polynesian branch,7 the root could be anywhere within Taiwan or ISEA. Indeed, some linguists cluster the Philippine languages with those of Taiwan,19 and recent attempts to apply phylogenetic methodologies from biology to the linguistic data have only suggested a root somewhere within the general area of Taiwan, the Philippines, and Borneo.20 A root in ISEA would mesh better with alternative views of the archaeological evidence that place ISEA at the center of population dispersals and Taiwan at the periphery—with only sporadic mainland influence—and with the languages most likely emerging during the Holocene within “Austronesia”18 itself. This would imply an early offshoot to Taiwan and subsequent leveling of language diversity across the Malayo-Polynesian area as a result of the formation of extensive socioeconomic networks.5,6,18
The debates reviewed above can be tested with genetic data. The Out of Taiwan model would predict that at least some ISEA lineages should trace back to ancestral populations in Taiwan and, ultimately, South China, with the age of those ISEA-specific lineage clusters being no more than ∼4,000 years ago. Earlier dispersals, perhaps stimulated by climate change and sea-level rise, as suggested in several alternative models,5,6,18 would, in contrast, predict dispersals centered on ISEA and dating to the late Pleistocene or early Holocene. If several distinct dispersal processes shaped the genetic variation in the region, their signatures should each be evident in the genetic record.
A number of genetic studies have already attempted to address the question of a putative Austronesian expansion, but most mtDNA studies to date have focused on Pacific islanders, with little work on the potential source populations. Because of this and because most studies only examined the fast-evolving first hypervariable segment of the control region (HVS-I), firm conclusions have been elusive. Indeed, the existing mtDNA data have been used both to support21–26 and to contradict27–29 the Out of Taiwan model. Better sampling in Southeast Asia has been achieved in a number of Y-chromosome studies,30–34 and various partitions of the data into Neolithic and pre-Neolithic have been suggested. However, different studies with similar data sets have failed to agree on how the data should be partitioned. This is a consequence of poor phylogenetic resolution—insufficient markers to allow reconstruction of the main branches of the genealogical history. This, in turn, results in poor phylogeographic resolution, since branches of the tree that may be distributed differently in space cannot be distinguished. The use of more markers will be necessary before the Y-chromosome contribution to the debate can be clarified.
In the meantime, we have addressed the poor quality of previous mtDNA data, both by sampling almost 1,000 individuals from locations throughout ISEA and by analyzing the samples at a higher resolution than done previously, by including coding-region as well as control-region variants gleaned from complete sequence data. This dramatically improved data set substantiates recent suggestions that major rethinking is needed with regard to the prehistory of the region.
Material and Methods
Subjects
Sampling locations are shown in figure 1. A total of 929 anonymous, unlinked DNA samples from across ISEA and Taiwan were analyzed and comprised 180 Sumatrans (42 from Medan, 24 from Padang, 52 from Pekanbaru, 28 from Palembang, and 34 from Bangka—previously published by Hill et al.35), 46 Javanese (36 Tengger, 1 from Yogyakarta, 1 from Banjumas, 1 from Garut, 1 from Jakarta, 1 from Probolinggo, 1 from Semarang, 3 from Solo, and 1 from Wonogiri), 157 individuals from Borneo (68 from Kota Kinabalu and 89 from Banjarmasin), 2 individuals from Bajawa in Flores, 82 Balinese (including 67 from Denpasar, 3 from Gianyar, 1 from Nusa Dua, 1 from Semarapura, 4 from Singaraja, 2 from Tabanan, and 2 from Ubud), 44 individuals from Mataram in Lombok, 237 individuals from Sulawesi (46 from Ujung Padang, 38 from Palu, 89 from Manado, and 64 Toraja), 50 individuals from Waingapu in Sumba, 43 individuals from Ambon, 45 individuals from Alor, 61 Filipinos, and 78 Taiwanese aboriginals (21 Ami, 18 Atayal, 18 Bunun, and 21 Paiwan—data updated from Sykes et al.24 and Melton et al.36). All were provided by the Medical Research Council Molecular Haematology Unit, University of Oxford, except for the non-Tengger Javanese samples, three of the Denpasar samples, and the samples from other locations in Bali. The study was approved by the University of Huddersfield Ethics Committee.
Comparative data were taken from the literature, mostly comprising HVS-I sequence data, often with only the 9-bp deletion in the COII/tRNALys intragenic region included in addition (with a few invaluable exceptions37–41). The data used included samples from Thailand, Malaysia, Taiwanese aboriginals, the Philippines, Sabah, East Indonesia, Papua New Guinea, Pacific Islanders, the Nicobars, Taiwanese Han, Hong Kong Han, China, Japan, Mongolia, Korea, and Central Asia and the authors' unpublished data from Singapore, West Papua, and Burmese Moken.22,24,25,36–59
Sequencing and RFLP Typing
HVS-I (minimum length sequenced was nucleotide positions (np) 16080–16370; maximum length sequenced np 15996–16569; average length sequenced np 16020–16500) was sequenced in all samples, and HVS-II (minimum length sequenced np 130–400; maximum length sequenced np 40–429; average length sequenced np 50–420) was also sequenced in selected samples, as described elsewhere.40,60 The samples from Medan and Pekanbaru were sequenced by the University of Dundee sequencing service, by use of an ABI 3700 sequencer; all other samples were sequenced at the University of Huddersfield, by use of a Beckman-Coulter CEQ8000 sequencer, except for the 30 done at Cambridge, which were sequenced using an ABI 3100 sequencer. Sequences were aligned to the revised Cambridge Reference Sequence (CRS)61 and were read by two people; any unusual mutations (e.g., transversions or transitions at sites with a low relative mutation rate) were rechecked. Approximately 10% of the samples were resequenced, to act as quality checks or controls. The sequences were also checked phylogenetically for sites between np 16051 and np 16365,62 and anomalous samples were resequenced. RFLP screening was used to resolve haplogroup status in a hierarchical fashion, as follows: haplogroups M (+10397 AluI; +10394 DdeI), N (−10397 AluI; −10394 DdeI), M7 (+9824 HinfI), D (−5176 AluI), E (−7598 HhaI), G (+4830 HhaI), P (+15606 AluI), U (+12308 HinfI), and I (+10032 AluI). Haplogroup B affiliation was checked by screening for the 9-bp deletion in the COII/tRNALys region,45 haplogroup F affiliation by sequencing position 10310 within the fragment 10270–10991, and macrohaplogroup N affiliation by sequencing position 8701 within the fragment 8196–9163. All haplogroup E samples with the control-region transition at position 16051 were checked by sequencing across the position 8730.
Phylogenetic, Phylogeographic, and Population Analyses
Reduced median networks63 were constructed for each haplogroup by use of the package Network 4.1 (Fluxus-engineering.com). The diversity and the time to the most recent common ancestor of a putative monophyletic lineage cluster was estimated using the statistic ρ, calibrated using a mutation rate of 1 transition every 20,180 years in the region from np 16090 to np 16365.64,65 Inferences concerning the dispersal of particular lineages were made by applying a founder analysis, by use of the f1 criterion to help control for the effects of back-migration.60 Thus, interpretations of the time depth of lineages within a particular region (in this case, Taiwan and/or ISEA) were made on the basis of the distribution of the clade in its potential source region—in this case, the Chinese and/or Southeast Asian mainland. If a certain clade is unique to ISEA, the time depth indicates the minimum age of the lineages within that region. If it derives from a single founder type (matching a type or node in the source phylogeny), with a diversity similar to or reduced with respect to the potential source, the interpretation would be a dispersal from the source around the time of the coalescence of the clade within ISEA. If there were multiple founders, then the time of dispersal would be inferred from the time estimate back to subclades with a single founder type. Haplogroup nomenclature followed Kong et al.66 as much as possible.
Intragroup haplotype diversity was estimated by 1-Σix2i, where xi is the relative frequency in the sample of the ith haplotype,67 for haplotypes defined between np 16090 and np 16365. Principal component (PC) analysis was used to visualize haplogroup frequency profiles.68 HVS-I data alone cannot always be resolved clearly into mtDNA haplogroups and, therefore, were not included in the PC analysis, although, in many cases, sufficient motif information was present to include them in phylogenetic analyses of particular haplogroups or subclades. Analysis of molecular variance (AMOVA), computed with the package Arlequin,69 was used to detect and quantify differences between populations. Frequency distributions were displayed using the Kriging algorithm of Surfer 8, combining the population samples into regions, to ensure sample sizes were adequate.
Results
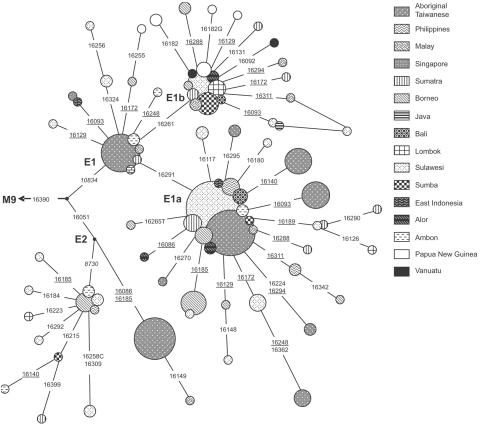
The samples fall into 56 named haplogroups or paragroups (the latter paraphyletic groupings including unclassified lineages within a clade and are marked with an asterisk [*]), all within the three principal non-African haplogroups: M, N, and R (table 1) (GenBank). The phylogenetic relationships between the main known East Eurasian haplogroups and those found in ISEA are shown in figure 2A and 2B. Table 2 shows the haplogroup distribution within the ISEA populations, and table 3 shows age estimates of the main founder clusters within ISEA (indicating likely dispersal and/or arrival times), alongside the overall age of the clade to which they belong. Several new haplogroups and subhaplogroups are defined for the first time in this study, including R23, M21d, M45–M47, F1a4, F1a5, D5d, and E1b (fig. 2).
Table 1. .
Distribution of HVS-I Sequences and RFLP Diagnostic Markers in ISEA and Taiwan
| Coding-Region Marker Statusb | Haplotype Frequency in Population Sample | |||||||||||||||||||||||||||||||||
| Cluster and HVS-I Haplotypea | 9bp del | 10397a | 10394c | 5176a | 9824g | 10310 | 7598f | 4831f | 10032a | 12308g | 15606a | Philippines | Bunun | Atayal | Amis | Paiwan | Medan | Pekanbaru | Bangka | Padang | Palembang | Java | Banjarmasin | Kota Kinabalu | Manado | Palu | Ujung Padang | Toraja | Bali | Mataram | Flores | Waingapu | Alor | Ambon |
| B: | ||||||||||||||||||||||||||||||||||
| 051 189 194C 195 | + | − | − | . | − | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | 2 | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 051 189 362 | + | − | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . |
| 093 189 222 298 299 399 | + | − | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 140 189 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . |
| 189 | + | − | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | ||||||||
| 189 194C 195 | . | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 189 195 286 | + | − | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| B4: | ||||||||||||||||||||||||||||||||||
| 140 189 217 362 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . |
| 189 217 | + | − | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | 1 | . | . | . | . | . |
| 189 217 365 | . | . | . | . | . | . | . | . | . | . | . | . | 2 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |
| B4a: | ||||||||||||||||||||||||||||||||||
| 092 189 217 261 293 | . | . | . | . | . | . | . | . | . | . | . | 3 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 108 189 217 261 | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . |
| 168 189 217 261 311 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . |
| 178 189 217 261 | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 2 | . | . | . | . | . | . | . | . | . | . | . |
| 188 189 217 223 261 355 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . |
| 189 217 223 261 | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | 1 |
| 189 217 223 261 335 | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . |
| 189 217 261 | + | − | − | . | . | . | . | . | . | . | . | 2 | . | . | 1 | 2 | . | 7 | . | 2 | . | . | 5 | 3 | 1 | . | 4 | 3 | 1 | 2 | . | . | . | 2 |
| 189 217 261 278 | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 |
| 189 217 261 286 | . | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 189 217 261 288 | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | |
| 189 217 261 293 | . | . | . | . | . | . | . | . | . | . | . | 2 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 189 217 261 311 | + | − | + | . | . | . | . | . | . | . | . | . | . | . | 4 | . | . | . | . | . | . | . | . | . | 1 | . | . | 1 | . | . | . | . | . | . |
| 189 261 | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . |
| B4a1a1: | ||||||||||||||||||||||||||||||||||
| 093 189 217 247 261 | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . |
| 189 217 247 261 | + | − | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 2 | . | 1 | . | 3 | 5 | . | 1 | . | . | . | 5 | |
| 189 217 247 261 362 | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 |
| B4a2a: | ||||||||||||||||||||||||||||||||||
| 154 189 217 261 324 | . | − | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 189 217 261 272 288 324 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |
| 189 217 261 272 324 | . | − | − | . | . | . | . | . | . | . | . | . | . | . | . | 3 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 189 217 261 324 | . | − | . | . | . | . | . | . | . | . | . | . | . | 2 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| B4b1: | ||||||||||||||||||||||||||||||||||
| 086 136 189 217 | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | 1 | . | . | . | . | . | . | . | . | . | |
| 136 189 213 217 | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 136 189 217 | + | − | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | 1 | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | 2 |
| 136 189 217 261 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . |
| 136 189 217 300 | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . |
| 136 189 217 365 | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| B4c1b: | ||||||||||||||||||||||||||||||||||
| 140 189 217 235 274 | + | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 2 | . | . | . | 1 | . |
| 140 189 217 274 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . |
| B4c1b3: | ||||||||||||||||||||||||||||||||||
| 092 140 189 217 274 283T 311 335 | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . |
| 092 140 189 217 335 | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . |
| 140 188 189 217 261 274 311 335 | + | − | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 140 188 189 217 274 311 335 | + | − | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 4 | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 140 189 217 274 335 | + | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | 1 | 2 | 1 | 1 | . | 3 | 3 | 2 | . | . | . | 1 | . |
| 140 189 217 335 | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 140 189 274 335 | + | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . |
| 189 217 274 335 | . | − | . | . | . | . | . | . | . | . | . | . | 1 | 1 | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| B4c2: | ||||||||||||||||||||||||||||||||||
| 086 147 189 217 | . | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . |
| 092 147 179 189 217 235 | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . |
| 147 184A 189 217 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | ||||
| 147 184A 189 217 235 239 | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 147 184A 189 217 235 260 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . |
| 147 189 217 | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 2 | . | 1 | . | . | . | 2 | . | . | . | . | . |
| 147 189 217 235 | + | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | 1 | . | 1 | . | 4 | . | . | . | 1 | . | 1 | . | . | . | . | . |
| 147 189 217 235 294 360 | + | − | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . |
| 147 189 217 235 294G | . | − | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . |
| B5a: | ||||||||||||||||||||||||||||||||||
| 140 145 189 224 266A | . | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 2 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 140 189 242A 256 261 266A | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . |
| 140 189 248 266A 319 | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 140 189 249 266A | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . |
| 140 189 261 266A | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | 2 | . | . | . | . | . | . | . |
| 140 189 266A | + | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 2 | . | . | . | 1 | 1 | 3 | 1 | 2 | . | 1 | . | 4 | 3 | . | . | . | 2 |
| 140 189 266A 274 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . |
| 140 189 266A 291 | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . |
| 140 189 266A 298 | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . |
| 140 189 266A 362 | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . |
| 140 189 266G | + | . | . | . | . | . | . | . | . | . | . | . | 3 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 140 189 266G 362 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 189 266A | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 |
| 189 266G | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 189 266G 362 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| B5b: | ||||||||||||||||||||||||||||||||||
| 111 129 140 189 234 243 | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 111 129 140 189 234 243 399 | . | . | . | . | . | . | . | . | . | . | . | 3 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 111 140 189 234 243 | . | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 111 140 189 234 243 399 | . | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 140 189 234 243 | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . |
| 140 189 243 | + | − | . | . | . | . | . | . | . | . | . | 2 | . | . | . | . | 2 | . | . | . | . | . | 2 | . | . | 1 | 1 | 1 | . | 1 | . | . | . | . |
| 140 189 243 355 | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 2 | . | . | . | . | . | . | . | . | . |
| C: | ||||||||||||||||||||||||||||||||||
| 051 223 298 327 | . | + | + | + | . | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . |
| 175 223 298 311 327 | . | + | + | + | . | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . |
| 223 298 327 | . | + | + | + | − | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | 1 | . | . | . | . | . | . | . |
| 223 311 327 | . | . | . | + | − | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . |
| D: | ||||||||||||||||||||||||||||||||||
| 093 192 223 271 316 362 | . | + | + | − | − | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . |
| 172 362 | . | + | . | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . |
| 188 223 274 311 362 | . | + | + | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . |
| 192 223 274 362 | . | + | + | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | 1 | . | . | . | . | . | . | . | . |
| 223 274 362 | . | + | . | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . |
| 223 286 362 | . | + | . | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . |
| 223 362 390 | . | + | + | − | . | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 |
| 295 362 | . | + | + | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 |
| D4a: | ||||||||||||||||||||||||||||||||||
| 129 223 274 311 317 362 | . | + | + | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . |
| D5: | ||||||||||||||||||||||||||||||||||
| 126 189 223 362 | − | − | . | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . |
| 172 189 223 259 362 | − | − | − | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . |
| 189 223 311 362 | − | + | + | − | − | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | 1 | . | . | . | . | . |
| 189 223 362 | − | − | − | − | − | . | . | . | . | . | . | . | . | . | . | 2 | . | . | . | . | . | . | . | . | . | . | . | 3 | . | . | . | . | . | . |
| D5d: | ||||||||||||||||||||||||||||||||||
| 093 148 189 223 362 | − | − | − | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . |
| 148 189 223 309 362 | − | . | . | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . |
| 148 189 223 362 | − | − | − | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 4 | . | 1 | . | . | . | . | . | . | 1 |
| D5d1: | ||||||||||||||||||||||||||||||||||
| 092 129 148 189 223 362 | . | − | − | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . |
| 092 148 184 189 223 362 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 092 148 185 189 223 362 | . | − | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 092 148 189 223 311 362 | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . |
| 092 148 189 223 362 | − | − | − | − | . | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | 3 | . | . | 4 | . | . | . | . | . | . |
| E1: | ||||||||||||||||||||||||||||||||||
| 086 185 223 362 390 | − | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |
| 215 223 362 390 | . | + | + | + | − | . | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . |
| 223 248 362 390 | . | + | . | + | − | . | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 |
| 223 256 324 362 390 | . | + | . | . | . | . | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . |
| 223 324 362 390 | . | + | + | + | − | . | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 2 | . | . | . | . | . | . |
| 223 362 390 | . | + | + | . | . | . | . | . | . | . | . | . | . | 1 | 2 | . | . | . | 1 | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . |
| E1a: | ||||||||||||||||||||||||||||||||||
| 086 223 291 362 465 | . | + | + | + | . | . | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . |
| 117 223 291 362 390 | . | + | + | . | − | . | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 2 | . | . | . | . | . | . |
| 126 189 223 291 362 390 | . | + | . | + | − | . | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . |
| 129 148 223 291 362 390 | . | + | + | . | − | . | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . |
| 129 223 291 362 390 | . | + | . | + | − | . | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . |
| 172 223 248 291 390 | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 172 223 291 362 390 | . | + | . | + | . | . | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 3 | . | . | . | . | . | . | . | . | . |
| 180 223 291 362 390 | . | + | + | . | − | . | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . |
| 185 223 291 362 390 | . | + | + | + | − | . | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | 7 | . | 1 | . | . | . | . | . | . | . | . |
| 189 223 290 291 362 390 | . | + | . | + | − | . | − | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 189 223 291 362 390 | . | + | . | + | . | . | − | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . |
| 223 224 291 294 362 390 | . | + | + | . | . | . | . | . | . | . | . | . | . | . | . | 3 | . | . | . | . | . | . | . | . | . | . | . | . | . | |||||
| 223 265T 291 362 390 | . | + | . | + | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | |
| 223 288 291 362 390 | . | + | . | . | . | . | − | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | |||||
| 223 291 311 342 362 390 | . | + | . | . | − | . | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . |
| 223 291 311 362 390 | . | + | + | . | . | . | − | . | . | . | . | 2 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 223 291 362 390 | . | + | + | + | − | . | − | . | . | . | . | 3 | 1 | . | . | . | . | . | 2 | 1 | . | . | 1 | 2 | 15 | 3 | 5 | 10 | 3 | . | . | 1 | 2 | 2 |
| E1b: | ||||||||||||||||||||||||||||||||||
| 093 223 261 311 362 390 | . | + | . | + | − | . | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . |
| 093 223 261 362 390 | . | + | . | + | − | . | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | 1 | . | . | . | . | . | . | . |
| 131 223 261 362 390 | . | + | . | + | − | . | − | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 172 223 261 362 390 | . | + | . | . | . | . | − | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 223 261 288 362 390 | . | + | . | + | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 2 | . | . | . | . | . | . | . | . | . | . | . |
| 223 261 362 390 | . | + | + | + | − | . | − | . | . | . | . | . | . | . | . | . | . | . | . | 1 | 1 | . | 1 | . | 3 | 1 | 1 | 2 | 1 | 3 | . | 6 | 2 | . |
| E2: | ||||||||||||||||||||||||||||||||||
| 037 051 086 185 223 362 390 | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 051 086 185 223 362 390 | . | + | − | . | . | . | . | . | . | . | . | . | . | . | 2 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 051 184 223 362 | . | . | . | . | . | . | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . |
| 051 185 223 362 | . | + | + | + | − | . | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . |
| 051 215 223 362 | . | + | . | + | − | . | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . |
| 051 215 223 362 390 399 | . | + | + | + | − | . | − | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 051 223 258C 309 362 390 | . | + | + | + | − | . | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | 1 | . | . | . | . | . | . | . |
| 051 223 292 362 390 | . | . | . | + | . | . | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . |
| 051 223 362 390 | . | + | + | + | − | . | − | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | 3 | 2 | 2 | . | . | . | . | . | . | . | . | 2 |
| F: | ||||||||||||||||||||||||||||||||||
| 129 304 362 359 390 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . |
| 168 172 189 311 362 | − | − | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . |
| 304 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . |
| 304 362 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | 1 | . | . | . | . | . |
| F1a: | ||||||||||||||||||||||||||||||||||
| 037 129 172 304 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 3 | . | . | . | . | . |
| 109C 129 172 304 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 129 132 172 304 | . | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 129 172 304 | . | − | − | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | 1 | 1 | 2 | . | . | . | 1 | 1 | . | 1 | . | . | 1 | 1 | 1 | 1 | . |
| 129 172 304 309 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . |
| 129 172 304 352 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . |
| 129 172 304 362 | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . |
| 129 172 304 362 381 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . |
| 129 172 304 362 400 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . |
| 172 189 304 | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 172 304 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . |
| F1a?: | ||||||||||||||||||||||||||||||||||
| 129 172 223 291 305 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . |
| F1a1: | ||||||||||||||||||||||||||||||||||
| 118 129 162 172 304 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . |
| 129 145 162 172 304 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . |
| 129 153 162 172 304 311 399 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . |
| 129 162 172 189 304 311 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 2 | . | . | . | . | . |
| 129 162 172 304 | . | − | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | 1 | . | . | . | . | . | . | . | . | 1 | . | . |
| 129 162 172 304 311 399 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . |
| F1a1a: | ||||||||||||||||||||||||||||||||||
| 108 111 129 162 172 189 304 | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . |
| 108 129 162 170 172 304 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 2 | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 108 129 162 172 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 108 129 162 172 184 304 398 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 108 129 162 172 234 299 304 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . |
| 108 129 162 172 293 304 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . |
| 108 129 162 172 304 | . | − | − | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | 2 | 1 | . | 1 | 2 | . | . | 1 | 1 | . | 1 | 1 | 3 | . | 1 | 1 | 1 |
| 108 129 162 172 304 391 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | ||||
| F1a3: | ||||||||||||||||||||||||||||||||||
| 129 172 214 304 311 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . |
| 129 172 243 304 311 | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 129 172 271 304 311 | . | − | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . |
| 129 172 304 311 | . | − | − | . | − | . | . | . | . | . | . | 4 | . | . | . | . | . | . | . | 1 | 1 | 2 | 1 | 1 | 3 | . | . | . | . | . | . | 2 | . | 3 |
| 129 172 304 311 362 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . |
| F1a4: | ||||||||||||||||||||||||||||||||||
| 092 129 172 294 304 362 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . |
| 093 129 172 294 304 362 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . |
| 104 129 172 294 304 362 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 2 |
| 129 172 173 294 304 362 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . |
| 129 172 192 294 304 362 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 2 | . | . | . | . | . | . | . | . | . | . |
| 129 172 294 304 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . |
| 129 172 294 304 362 | . | − | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 2 | . | . | . | . | 1 | . | 3 | 1 | . | 7 | . | . | 1 | 3 | 2 | . |
| F1a5: | ||||||||||||||||||||||||||||||||||
| 129 134 172 301 304 400 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 129 172 301 304 362 400 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . |
| 129 172 301 304 400 | . | − | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | 1 | . | 6 | . | . | . | . | . | . | . | . | . | . | . | . |
| F1b: | ||||||||||||||||||||||||||||||||||
| 189 284 304 | . | − | − | + | . | A | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . |
| F3: | ||||||||||||||||||||||||||||||||||
| 066 290 298 357 362 | . | − | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . |
| F3b: | ||||||||||||||||||||||||||||||||||
| 093 220c 265 298 311 362 | . | − | + | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 093 220C 265 298 362 | . | − | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 2 | . | . | . | . | . | . | . | . | . | . | . |
| 093 220C 298 362 | . | − | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 4 | . | . | . | . | . | . | . | . | . | . | . |
| 220C 240 265 298 335 362 | . | . | . | . | . | . | . | . | . | . | . | 2 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 220C 261 265 298 362 | . | − | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . |
| 220C 265 274 298 311 362 | . | − | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . |
| 220C 265 298 311 362 | . | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | |
| 220C 265 298 335 362 | . | . | . | . | . | . | . | . | . | . | . | 9 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 220C 265 298 362 | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . |
| 220C 298 311 362 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 2 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| F4b: | ||||||||||||||||||||||||||||||||||
| 069 218 304 311 | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 170 218 304 311 | . | − | − | . | . | A | . | . | . | . | . | . | . | . | . | . | 1 | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 218 304 311 | . | − | − | . | . | A | . | . | . | . | . | . | 5 | 5 | 3 | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| I: | ||||||||||||||||||||||||||||||||||
| 129 223 311 391 | . | − | + | . | . | . | . | . | + | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| M: | ||||||||||||||||||||||||||||||||||
| 0 | . | + | + | + | − | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . |
| 048 162del 214 223 | . | + | + | + | . | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . |
| 093 129 223 234 290 311 384 | . | + | + | + | − | . | + | − | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 093 133 176 223 | . | + | . | + | − | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . |
| 093 193 | . | + | + | + | − | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . |
| 093 223 231 319 | . | + | . | + | − | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 093 223 311 362 | . | + | . | + | − | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 4 | 1 | . | . | . | . | . | . | . | . | . | . | . |
| 093 223 362 | . | + | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 118 129 192 223 256 272 | . | + | + | + | − | . | + | − | . | . | − | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . |
| 124 166del 214 223 | . | + | + | + | . | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . |
| 126 147 153 223 | . | + | . | . | − | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |
| 129 140 271 | . | + | + | + | − | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . |
| 129 142 166 223 255 274 294 327A | . | + | + | + | − | . | + | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . |
| 129 145 223 291 304 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . |
| 129 148 172 223 256 305 309 | . | + | + | + | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . |
| 129 172 223 234 290 312 | . | + | . | + | − | . | + | − | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 129 189 218 223 | − | + | + | + | − | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . |
| 129 223 | . | + | . | + | − | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 129 223 234 290 311 | . | + | . | + | − | . | + | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . |
| 129 223 362 390 | . | + | + | + | . | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . |
| 145 176 224 233 311 | . | . | . | . | − | . | + | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 148 189 223 246T | . | + | + | + | − | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . |
| 148 189 223 246T 311 362 | − | + | + | + | − | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . |
| 148 189 362 | . | + | + | + | − | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |
| 153 223 362 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 172 189 223 234 249 290 | − | + | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 172 223 245A | . | + | + | + | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . |
| 172 223 362 390 | . | + | . | + | − | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . |
| 179 223 294 | . | + | . | + | − | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 184A 223 | . | + | + | + | − | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 |
| 189 192 223 291 362 | . | + | . | . | − | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 189 195A 241 265C 311 | . | + | + | + | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . |
| 189 213 223 271 311 | . | + | + | + | − | . | + | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 189 223 | − | + | + | + | − | . | + | . | . | . | . | . | . | . | 1 | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 189 223 227 291 362 | . | + | . | + | − | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . |
| 192 362 | . | + | . | + | − | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |
| 219 223 290 291 | . | + | + | + | . | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . |
| 223 | . | + | . | + | − | . | . | . | . | . | . | . | . | . | 1 | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 223 234 362 | . | + | . | + | − | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . |
| 223 243 311 362 | . | + | + | + | − | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . |
| 223 246T 311 362 | . | + | . | + | − | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 223 254 362 | . | + | + | + | − | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 2 | . | . | . | . | . |
| 223 266 284 290 | . | + | . | + | − | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . |
| 223 269 271 | . | . | . | . | . | . | . | . | . | . | . | 2 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 223 269 271 311 | . | + | + | . | − | . | + | . | . | . | . | 3 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 223 293 311 362 | . | + | . | + | − | . | + | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 223 299 311 | . | + | + | + | − | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . |
| 223 299 311 362 | . | + | . | + | − | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . |
| 223 304 325 344 362 381 | . | + | + | . | − | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . |
| 223 311 | . | + | . | + | − | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | 1 | . | . | . | . | . | 1 | . | . | . | . |
| 223 311 335 362 | . | + | . | . | − | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . |
| 223 311 362 | . | + | . | + | − | . | + | . | . | . | . | . | . | . | . | . | . | . | 2 | . | . | 3 | 3 | . | . | 2 | . | . | 2 | . | . | 1 | . | . |
| 223 362 390 | . | + | + | + | − | . | + | . | . | . | . | . | . | . | . | 1 | 1 | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | 2 |
| 234 256 294 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | |||||
| M12a: | ||||||||||||||||||||||||||||||||||
| 051 093 145 223 234 249 290 399 | . | + | . | + | − | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . |
| 223 234 261 290 | . | + | + | + | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . |
| M21a: | ||||||||||||||||||||||||||||||||||
| 093 129 223 256 271 | . | + | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . |
| M21b: | ||||||||||||||||||||||||||||||||||
| 129 223 263 381 | . | + | + | + | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 2 | . | . | . | . | . | . | . | . | . | . | . |
| 172 223 239 263 325 381 | . | + | . | + | − | . | + | − | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . |
| M21d: | ||||||||||||||||||||||||||||||||||
| 145 181 192 223 266 291 304 | . | + | . | + | − | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . |
| 145 181 192 223 291 304 | . | + | + | + | − | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . |
| M45: | ||||||||||||||||||||||||||||||||||
| 093 209 223 325 | . | + | + | + | − | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . |
| M45a: | ||||||||||||||||||||||||||||||||||
| 086 129 209 223 237 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . |
| 086 129 209 223 272 | . | + | + | . | . | . | + | − | . | . | . | . | . | . | . | . | . | . | 2 | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 086 129 209 223 272 311 | . | + | + | + | − | . | + | − | . | . | . | . | . | . | . | . | . | . | 2 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 092 129 209 223 325 | . | + | + | + | − | . | . | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . |
| 093 129 209 223 272 | + | + | + | − | . | + | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 2 | . | . | . | . | . | . | . | . | . | . | |
| 129 179 209 223 272 | . | + | . | + | − | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 129 209 223 272 | . | + | + | + | − | . | + | − | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | 1 | 1 | . | . | 2 | . | 1 | . | . | . | . | . |
| 129 209 223 311 325 | . | + | + | + | − | . | + | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . |
| 129 209 223 325 | . | + | . | + | − | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . |
| M45b: | ||||||||||||||||||||||||||||||||||
| 093 189 209 223 | . | + | . | + | − | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . |
| M45b?: | ||||||||||||||||||||||||||||||||||
| 168 189 209 223 233 304 | . | + | . | + | − | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 |
| 189 209 223 300 | − | + | + | + | − | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . |
| M46: | ||||||||||||||||||||||||||||||||||
| 086 148 223 259 278 319 399 | . | + | + | + | − | . | + | − | . | . | . | . | . | . | . | . | . | . | . | . | . | 2 | 1 | . | . | . | . | . | . | . | . | . | . | . |
| 086 172 173 223 278 311 | . | + | + | . | − | . | + | − | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | ||||||
| 086 223 243 262 278 311 319 | . | + | + | + | − | . | + | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . |
| 092 140 172 189 223 278 | − | . | . | . | . | . | + | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . |
| 092 209 223 224 263 278 319 | . | + | . | + | − | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 093 189 222 223 278 | . | + | + | + | . | . | + | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . |
| 093 189 223 265 278 | . | + | + | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 |
| 093 189 223 278 319 | − | + | + | + | . | . | + | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | ||||||
| 093 209 223 224 263 265 278 319 | . | + | + | . | − | . | + | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . |
| 093 209 223 224 263 274 278 319 356 | . | + | . | + | − | . | + | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 093 209 223 224 263 278 319 | . | + | . | + | − | . | . | . | . | . | . | . | . | . | . | . | . | 2 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 093 223 249 259 278 291 362 | . | + | + | + | − | . | + | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . |
| 093 223 278 | . | + | . | + | − | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . |
| 093 223 278 310 | . | + | . | + | − | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . |
| 124 189 278 292 362 | − | + | + | + | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . |
| 126 129 189 278 | − | + | + | + | − | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 3 | . | . | . | . | . | . | . | . | . | . |
| 126 214A 223 271 278 298 | . | + | . | . | − | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |
| 140 172 189 223 278 | − | . | . | . | . | . | . | − | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | |
| 172 173 223 278 311 | . | + | + | + | − | . | + | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 2 | . | . | 1 | . | . | . | . | . | . | . | . |
| 184A 213 223 278 | . | + | + | + | − | . | + | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . |
| 188 189 223 278 288 | − | + | + | + | − | . | + | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . |
| 189 222 223 278 352 | − | + | + | . | . | . | + | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . |
| 189 223 278 | . | + | + | . | . | . | + | − | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | 1 | 1 | . | 1 | . | . | . | . |
| 196 223 274 278 290 | . | + | + | + | − | . | + | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . |
| 223 278 | . | + | . | + | − | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | |
| 223 278 294 | . | + | + | + | − | . | + | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . |
| 223 278 311 | . | + | . | + | − | . | + | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . |
| 234 256 278 294 | . | + | + | + | − | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . |
| 234 278 294 | . | + | + | + | − | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . |
| M47: | ||||||||||||||||||||||||||||||||||
| 129 166C 189 223 287 319 | . | + | + | + | − | . | + | − | . | . | . | . | . | . | . | . | . | 1 | 1 | . | . | . | . | . | . | . | . | . | ||||||
| M7: | ||||||||||||||||||||||||||||||||||
| 129 155 219 223 356 362 | . | + | + | + | + | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . |
| 189 223 362 | . | + | + | + | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . |
| 223 291 362 | . | + | . | + | + | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 223 362 | . | + | + | + | + | . | + | . | . | . | . | . | . | . | . | . | 3 | . | . | 1 | . | . | . | . | . | 3 | . | . | . | . | . | . | . | 1 |
| 354 | . | + | + | + | + | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . |
| M7b: | ||||||||||||||||||||||||||||||||||
| 126 129 223 297 | . | + | + | + | + | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . |
| 129 172 189 223 297 | . | + | + | + | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . |
| 129 189 223 297 | . | + | . | . | + | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | 1 |
| M7b1: | ||||||||||||||||||||||||||||||||||
| 126 129 192 223 297 | . | + | + | + | + | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | 1 | . | . | . | 1 |
| 129 140 189 192 223 265 297 | − | + | . | + | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . |
| 129 189 192 215 223 297 | − | + | + | . | + | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 2 | 1 | . | . | . | . |
| 129 189 192 223 297 | . | + | + | . | + | . | . | . | . | . | . | . | . | . | . | . | 3 | . | . | . | . | . | . | . | . | 1 | . | . | . | 1 | . | . | . | . |
| 189 192 223 294G 297 | . | + | . | + | + | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . |
| 189 192 223 297 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | |||||
| M7b3: | ||||||||||||||||||||||||||||||||||
| 086 126 129 297 324 | . | + | + | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . |
| 086 129 189 192 297 324 | . | + | − | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 086 129 192 297 324 | . | − | . | . | . | . | . | . | . | . | . | . | . | 2 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 086 129 295 297 324 | . | + | . | + | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 4 | . | . |
| 086 129 297 324 | . | + | + | + | + | . | . | . | . | . | . | . | 1 | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | 2 | . | |||||
| M7c1: | ||||||||||||||||||||||||||||||||||
| 223 224 287 295 | . | . | . | + | + | . | + | . | . | . | − | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . |
| M7c1a: | ||||||||||||||||||||||||||||||||||
| 093 223 319 | . | + | . | + | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . |
| 223 249 319 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| M7c1c: | ||||||||||||||||||||||||||||||||||
| 093 223 291 295 337 362 | . | + | . | + | + | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . |
| 093 223 295 337 362 | . | + | . | . | + | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 2 | . | . | . | . | . | . | . | . | . | . |
| 093 223 295 362 | . | + | . | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . |
| 129 223 295 362 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 145 223 295 362 | . | + | . | + | + | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |
| 150 223 274 295 311 362 | . | + | + | + | + | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . |
| 168 223 295 362 | . | + | + | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 2 | . | . | . | . | . | . |
| 179 223 295 362 | . | + | . | . | + | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 185 223 295 362 | . | + | . | . | + | . | . | . | . | . | . | . | . | . | . | . | 1 | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 189 213 223 295 362 | − | + | + | + | + | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . |
| 192 223 295 362 | . | + | . | + | + | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 2 | . | . | . | . | . |
| 223 278 295 362 | . | + | + | + | + | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | 1 | . | . | . | . | . | . | . | . |
| 223 292 295 362 | . | + | . | . | + | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 223 295 311 362 | . | + | + | + | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 2 | . | . |
| 223 295 346C 362 | . | + | + | + | + | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 2 | . | . | . | . | . | . | . | . | . | . |
| 223 295 356 362 | . | . | . | + | + | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 3 | . | . | . | . | . | . | . | . | . |
| 223 295 362 | . | + | + | + | + | . | + | . | . | . | . | 7 | . | . | . | 1 | 1 | 3 | 1 | . | 6 | 5 | 1 | 3 | 8 | 6 | 1 | 4 | 3 | 1 | . | 3 | 1 | 1 |
| 295 362 | . | . | . | + | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . |
| N: | ||||||||||||||||||||||||||||||||||
| 094 129 223 263 274 311 343 357 | . | − | + | + | − | . | + | . | . | . | − | . | . | . | . | . | . | . | . | . | . | 6 | . | . | . | . | . | . | . | . | . | . | . | . |
| 111 168 172 189 223 242 263 311 362 | − | − | − | . | . | . | . | . | . | . | − | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . |
| 111 168 172 189 223 311 319 362 | − | − | − | . | . | . | . | . | . | . | − | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . |
| 111 168 172 189 223 311 362 | − | − | − | + | . | . | . | . | . | . | − | . | . | . | . | . | . | . | . | . | . | . | 2 | . | . | . | . | . | . | . | . | . | . | . |
| 111 172 189 223 311 362 | − | − | . | . | . | . | . | . | . | . | − | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . |
| 189 223 | − | − | − | + | . | . | . | . | . | . | − | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 223 263 274 311 343 357 | . | − | + | + | − | . | + | . | . | . | − | . | . | . | . | . | . | . | 4 | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . |
| 223 294 | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | |||||||||||
| N21: | ||||||||||||||||||||||||||||||||||
| 193 223 | . | − | − | + | . | . | + | . | . | . | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | 1 | . | . | . | . | . |
| 193 223 249 291 319 | . | − | − | . | . | . | . | . | . | . | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . |
| 193 223 291 319 | . | − | . | . | . | . | . | . | . | . | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . |
| 193 223 311 344 | . | − | . | . | . | . | . | . | . | . | − | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . |
| N22: | ||||||||||||||||||||||||||||||||||
| 093 168 172 223 249 | . | − | − | . | . | . | . | . | . | . | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . |
| 093 168 223 249 278 295 | . | − | − | . | . | . | . | . | . | . | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . |
| 168 172 223 249 | . | − | − | . | . | . | . | . | . | . | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 2 | . | . |
| N9a: | ||||||||||||||||||||||||||||||||||
| 129 223 257A 261 | . | − | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 223 257A | . | − | . | . | . | . | . | . | . | . | . | . | . | . | 2 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| N9a6: | ||||||||||||||||||||||||||||||||||
| 189 223 257A 261 292 | − | − | − | + | . | . | . | . | . | . | . | . | . | . | . | . | . | 3 | 3 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |
| 223 257A 261 292 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . |
| N9a6a: | ||||||||||||||||||||||||||||||||||
| 086 187 223 257A 261 292 294 | − | − | − | . | . | . | . | . | . | . | − | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . |
| 223 257A 261 292 294 | . | − | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . |
| 223 257A 261 292 294 357 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . |
| P: | ||||||||||||||||||||||||||||||||||
| 093 172 266 270 | . | − | − | . | . | . | . | . | . | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . |
| 093 176 266 270 357 | . | − | − | . | . | . | . | . | . | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . |
| 176 221 266 325 357 | . | . | . | . | . | . | . | . | . | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | 2 | . | . | . | . | . | . | . | . | . |
| 249 319 390 | . | − | + | + | − | . | + | . | . | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . |
| Q: | ||||||||||||||||||||||||||||||||||
| 066 129 172 173 223 241 | . | + | . | + | − | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . |
| 086 129 148 223 241 265C 311 343 | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . |
| 092 129 144 148 169 223 265C 311 343 | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . |
| 129 144 148 153 162 192 223 241 249 265C 311 343 | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . |
| 129 144 148 154 209 222 241 265C 311 343 | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 |
| 129 144 148 172 223 241 242 265C 311 343 | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | 1 | . |
| 129 144 148 172 223 241 256 265C 311 343 | . | + | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 |
| 129 144 148 172 223 241 265C 311 343 | . | + | + | + | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | 3 | . |
| 129 144 148 172 223 242 265C 311 343 | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . |
| 129 144 148 193 223 241 265C 311 343 362 | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 2 |
| 129 144 148 209 223 241 265C 311 343 | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . |
| 129 144 148 222/3insA 223 241 265C 311 343 | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . |
| 129 144 148 223 241 265C 299 311 343 362 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . |
| 129 144 148 223 241 265C 311 343 | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | 1 | . | 1 |
| 129 144 148 223 265C 311 343 | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . |
| 129 144 148 241 265C 311 343 | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 5 | . |
| 129 223 241 311 | . | + | + | + | − | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . |
| R: | ||||||||||||||||||||||||||||||||||
| 187 241 269 319 342 | − | − | − | + | . | . | . | . | . | . | − | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . |
| 293 311 355 | . | − | − | . | − | . | + | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| R21/R9: | ||||||||||||||||||||||||||||||||||
| 093 168 187 288 304 | . | − | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . |
| R22: | ||||||||||||||||||||||||||||||||||
| 189 249 286 288 | . | − | − | . | . | . | . | . | . | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . |
| 249 288 | . | − | − | . | . | . | . | . | . | − | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 249 288 295 304 | . | − | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 2 | . | . | . | . | . | . | . | . | . | . | . |
| 249 288 301 304 390 | . | − | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | 2 | . | . |
| 249 288 304 | . | − | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . |
| 249 288 304 344 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . |
| 249 288 304 390 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | 4 | 2 | . | 1 | . | . |
| 249 288 317C | . | − | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . |
| 249 288 317C 319 | . | . | . | . | . | . | . | . | . | . | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . |
| 288 304 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | 1 | . |
| R23: | ||||||||||||||||||||||||||||||||||
| 129 256 290 465 | . | − | − | . | . | . | . | . | . | . | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . |
| 213 256 278 290 465 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . |
| 256 290 465 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 3 | . | . | 1 | . | . |
| R9: | ||||||||||||||||||||||||||||||||||
| 066 209 304 311 399 | . | − | − | . | . | G | . | . | . | . | . | . | . | . | . | . | . | 2 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 124 189 209 293C 304 362 | − | . | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . |
| 129 209 223 233 259 274 290 304 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . |
| 209 223 233 274 304 | . | − | − | . | . | G | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 209 223 234 261 290 304 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| R9? | ||||||||||||||||||||||||||||||||||
| 140 189 304 | − | − | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . |
| R9b: | ||||||||||||||||||||||||||||||||||
| 093 192 288 304 309 390 | . | − | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 192 234 288 293 304 309 390 | . | − | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 2 | . | . | . | . | . | . | . | . | . | . | . | . |
| 192 234 288 304 309 390 | . | − | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | 2 | . | . | . | . | . | . | . | . | . |
| 192 288 304 309 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . |
| 192 288 304 309 390 | . | − | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| R9c: | ||||||||||||||||||||||||||||||||||
| 086 157 256 304 335 | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . |
| 157 169 256 304 311 335 | . | . | . | . | . | . | . | . | . | . | . | 2 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 157 256 304 335 | . | − | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 3 | 1 | . | . | . | . | . | . | 1 | 2 | . |
| 157 256 304 362 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . |
| 157 256 335 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 2 | . |
| U7: | ||||||||||||||||||||||||||||||||||
| 069 207 309 318T | . | − | − | . | . | . | . | . | . | + | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| Y2: | ||||||||||||||||||||||||||||||||||
| 069 231 311 | . | − | + | + | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 093 126 231 311 | . | − | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 126 192 231 311 | . | − | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . |
| 126 231 284 311 | . | . | . | . | . | . | . | . | . | . | . | 4 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 126 231 311 | − | − | + | + | . | . | . | . | . | . | . | 3 | . | 1 | . | . | 5 | 3 | 2 | . | . | 1 | 1 | 2 | 1 | 1 | . | 2 | . | . | . | . | . | . |
| Z: | ||||||||||||||||||||||||||||||||||
| 129 185 189 223 260 298 | − | + | + | + | . | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . |
| 185 223 260 298 | . | + | + | + | − | . | + | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . |
Variant positions from the CRS61 are shown minus 16000. These are transitions, unless a letter suffix explicitly indicates the nucleotide present.
A plus sign (+) indicates the presence of the restriction site (or the presence of the 9-bp deletion), a minus sign (−) indicates the absence, and a dot (.) indicates that the status was not determined. RFLPs are indicated by the nucleotide position at the start of recognition sequence and by a code for the restriction enzyme, as follows: a = AluI; c = DdeI; f = HhaI; g = HinfI. The intragenic 9-bp deletion is indicated by “9bp del.” When an HVS-I haplotype or motif occurred more than once in the sample, the coding-region assays were sometimes performed on only a subset of the samples.
Figure 2. .
Schematic tree of East Eurasian mtDNA haplogroups, displaying the various branches of macrohaplogroup M (A) and macrohaplogroup N (B) and indicating diagnostic control-region positions and coding-region positions tested. We here rename R1270 as R22, since R12 was used in a previous study.71 F1a3 is here defined solely by 16311 because the two other control-region sites mentioned as diagnostic by Kong et al.66 do not appear in our data set.
Table 2. .
Haplogroup Frequencies in China and Southeast Asia[Note]
| Haplogroup | NW Chinaa | NE Chinab | SW Chinac | SE Chinad | Thailand | Melayu Malays | Orang Asli | Taiwan | Philippines | Sumatra | Borneo | Java | Bali | Lombok | Sumba | Sulawesi | Alor | Ambon | Total ISEA |
| B* | .2 | .5 | .8 | .5 | .4 | … | 1.2 | … | … | 3.9 | … | 2.2 | … | … | … | .4 | … | … | .9 |
| B4* | 1.2 | 2.1 | 2.8 | 2.0 | 3.7 | … | .8 | .4 | … | … | .6 | … | 1.2 | … | … | .4 | … | … | .3 |
| B4a* | 1.0 | 3.4 | 6.5 | 6.1 | 5.3 | .9 | … | 9.2 | 11.3 | 6.1 | 7.0 | 2.2 | 2.4 | 4.5 | 2.0 | 5.1 | … | 9.3 | 5.5 |
| B4a1a1 | … | … | … | … | … | … | … | … | … | … | 1.3 | … | … | 2.3 | … | 3.8 | 2.2 | 14.0 | 2.0 |
| B4a2 | … | .2 | .3 | .2 | … | … | … | 11.5 | … | … | … | … | … | … | … | … | … | … | … |
| B4b1 | 2.0 | 2.7 | 1.8 | 3.1 | … | .9 | … | 6.6 | 1.6 | .6 | 1.9 | … | … | … | … | .8 | 6.6 | 4.7 | .9 |
| B4c1b | .2 | 2.1 | .3 | 1.6 | … | 2.7 | … | 3.3 | … | 4.4 | 1.9 | 2.2 | 6.0 | … | … | 3.8 | … | … | 3.0 |
| B4c2 | … | … | .8 | 1.0 | 2.3 | 2.8 | … | … | … | 2.2 | 5.7 | … | 7.2 | … | … | .8 | … | … | 2.2 |
| B5a | 2.2 | 3.4 | 6.8 | 4.9 | 10.5 | 9.2 | .8 | 5.1 | … | 3.9 | 4.5 | 2.2 | 4.8 | 6.8 | … | 2.5 | 4.4 | 7.0 | 3.5 |
| B5b | 1.0 | 1.4 | .6 | 2.6 | … | .9 | 5.8 | … | 9.7 | 2.2 | 1.3 | … | … | 2.3 | … | 2.5 | … | … | 2.0 |
| C | 4.7 | 4.3 | 5.7 | 5.2 | 3.6 | .9 | … | … | … | … | 1.3 | 2.2 | … | 2.3 | … | .4 | … | … | .5 |
| D* | 9.9 | 12.6 | 4.9 | 6.3 | 4.0 | … | … | .2 | … | … | .6 | … | … | … | … | 2.5 | … | 4.7 | .9 |
| D4 | 3.2 | 3.4 | 4.6 | 4.7 | .4 | … | … | .9 | … | … | … | … | … | … | … | … | 2.2 | … | .1 |
| D5 | 3.0 | 7.1 | 2.6 | 5.6 | 1.6 | .9 | … | 4.1 | … | … | 4.5 | … | 1.2 | 2.3 | … | 8.4 | … | 2.3 | 3.2 |
| E1* | .2 | … | .1 | .2 | … | … | … | 2.4 | … | .6 | .6 | … | 1.2 | … | … | 2.1 | … | 7.0 | … |
| E1a | … | … | … | … | … | .9 | … | 6.2 | 8.1 | 3.3 | 8.9 | … | 3.6 | 2.3 | 2.0 | 17.7 | 6.6 | 4.7 | 8.0 |
| E1b | … | … | … | … | … | 3.7 | … | … | … | 2.2 | 1.9 | 2.2 | 1.2 | 6.8 | 12.0 | 3.8 | 4.4 | … | 3.0 |
| E2 | … | … | … | … | … | .9 | … | 3.1 | 1.6 | .6 | 3.2 | … | … | … | 2.0 | 2.9 | … | 4.7 | … |
| F* | .7 | 1.4 | .6 | 2.4 | … | … | … | … | … | … | 2.5 | … | 1.5 | 2.3 | … | … | … | … | .6 |
| F1a* | 1.0 | 2.1 | 4.9 | 2.9 | 4.7 | 3.8 | … | .2 | 1.6 | 4.4 | .6 | 2.8 | 6.0 | 9.1 | 2.0 | 1.7 | 2.2 | … | 2.8 |
| F1a1* | 2.5 | 2.5 | 1.9 | 3.3 | 1.2 | 3.7 | … | 2.5 | … | … | 1.3 | 2.2 | 4.8 | … | 2.0 | .4 | … | … | .9 |
| F1a1a | .7 | .2 | 3.7 | 2.7 | 8.9 | 8.3 | 10.9 | … | … | 5.0 | .6 | 4.4 | 2.4 | 9.1 | 4.0 | 1.3 | 2.2 | 2.3 | 2.6 |
| F1a3 | … | .2 | … | … | … | 1.9 | … | .7 | 8.2 | 1.1 | 1.9 | 5.6 | … | 2.2 | 4.0 | 1.7 | … | 7.0 | 2.3 |
| F1a4 | … | .2 | .2 | … | .5 | … | … | .7 | … | 1.1 | 1.9 | … | … | 2.3 | 6.0 | 5.1 | 6.7 | 4.7 | 2.9 |
| F1a5 | … | … | … | … | … | … | … | … | … | 1.7 | … | 13.0 | … | … | … | .4 | … | … | 1.0 |
| F1b | 2.0 | 2.7 | 2.8 | 3.3 | .8 | … | … | … | … | … | … | … | … | … | … | .4 | … | … | .1 |
| F2 | 1.0 | 1.8 | 2.9 | 1.9 | 2.4 | … | … | .5 | … | … | … | … | … | … | … | … | … | … | … |
| F3a | .7 | .9 | 1.1 | 1.4 | 1.6 | .9 | … | … | … | … | … | … | … | … | … | … | … | … | … |
| F3b | … | … | .01 | … | .2 | … | … | 7.1 | 19.0 | … | 5.7 | … | … | … | 2.0 | … | … | … | 2.5 |
| F4 | .2 | … | .2 | … | .8 | … | … | 11.8 | … | 1.7 | … | … | … | … | … | … | … | … | .9 |
| I | … | … | … | … | … | … | … | … | … | .6 | … | … | … | … | … | … | … | … | .1 |
| M* | 11.1 | 4.1 | 8.0 | 7.0 | 4.7 | 14.2 | 1.6 | .7 | 2.4 | 10.1 | 8.3 | 13.0 | 12.0 | 11.4 | 8.0 | 3.0 | 2.2 | 2.3 | 6.9 |
| M7* | .7 | .2 | .9 | 1.2 | .8 | … | … | … | … | 2.8 | .6 | … | 1.2 | … | 2.0 | 1.3 | … | 2.3 | 1.2 |
| M7b* | 1.0 | 1.1 | 3.4 | 3.0 | 2.0 | … | … | .1 | … | .6 | 1.3 | … | 1.2 | … | … | … | … | 2.3 | .5 |
| M7b1 | 1.7 | 3.2 | 4.6 | 2.1 | 2.8 | 3.7 | … | .7 | … | 1.7 | .6 | 2.2 | 4.8 | 6.8 | … | .4 | … | 2.3 | 1.4 |
| M7b3 | .2 | … | … | … | … | … | … | 10.2 | … | … | … | … | … | … | 8.0 | 1.3 | … | … | .8 |
| M7c1* | 1.2 | .2 | .3 | .9 | 1.2 | .9 | … | … | … | … | … | … | … | … | … | .4 | … | … | .1 |
| M7c1a | … | .5 | … | .7 | … | … | .4 | 1.2 | … | … | .6 | … | … | … | … | … | … | … | .2 |
| M7c1c | … | .5 | .2 | … | .4 | 4.6 | 3.1 | 4.2 | 11.3 | 8.9 | 7.0 | 10.9 | 6.0 | 2.3 | 12.0 | 11.0 | 4.4 | 2.3 | 8.3 |
| M12 | … | … | .2 | .7 | 2.4 | 2.8 | … | … | … | … | .6 | … | 1.2 | … | … | … | … | … | .2 |
| M21a | … | … | … | … | 6.9 | 4.6 | 21.0 | … | … | … | .6 | … | … | … | … | … | … | … | .1 |
| M21b | … | … | … | … | .4 | .9 | 3.5 | … | … | .6 | 1.3 | … | … | … | … | .4 | … | … | .4 |
| M21d | … | … | … | … | … | .9 | … | … | … | … | … | … | 2.4 | … | … | … | … | … | .2 |
| M45 | … | … | .5 | … | .5 | 1.9 | … | … | … | 3.9 | 2.5 | 2.2 | 4.8 | 2.3 | 2.0 | 1.3 | … | 2.3 | 2.3 |
| M46 | … | … | … | … | … | 3.8 | … | … | … | 5.0 | 6.4 | 4.3 | 4.8 | 9.1 | … | 3.0 | 4.4 | 2.3 | 4.0 |
| M47 | … | … | … | … | … | … | … | … | … | 1.1 | … | … | … | … | … | … | … | … | .2 |
| N* | 2.0 | 1.8 | .4 | .8 | … | 1.8 | … | … | … | 2.8 | .6 | 15.2 | … | … | … | … | … | … | 1.3 |
| N9a6 | … | … | .3 | … | .8 | 2.8 | 6.2 | … | … | 3.3 | .6 | 2.2 | … | … | … | .8 | … | … | 1.1 |
| N21 | … | … | … | … | .4 | 1.8 | 9.3 | … | … | .6 | … | … | 1.2 | … | … | .4 | 4.4 | … | .5 |
| N22 | … | … | … | … | … | … | 1.6 | … | … | … | … | … | … | … | 8.0 | … | … | … | .4 |
| P | … | … | … | … | … | .9 | … | … | … | … | … | … | … | … | 4.0 | 1.3 | … | … | .5 |
| Q | … | … | … | … | … | 1.8 | … | … | … | … | 1.3 | … | 1.2 | … | 4.0 | 2.1 | 28.9 | 11.6 | 2.9 |
| R* | … | .2 | .7 | .6 | … | 1.8 | … | … | 1.6 | … | .6 | … | … | … | … | … | … | … | .2 |
| R9* | … | .2 | .3 | … | … | .9 | … | … | … | 2.2 | … | … | 1.2 | … | … | … | … | … | .6 |
| R9b | .7 | .2 | 1.7 | .9 | 1.2 | .9 | 9.3 | … | … | 1.7 | .6 | 4.4 | … | 2.3 | … | .8 | … | … | .9 |
| R9c | .2 | .2 | .3 | .4 | .9 | … | … | 1.8 | 3.3 | … | 1.9 | … | … | … | 2.0 | .8 | 11.1 | … | 1.3 |
| R22 | … | … | … | … | .8 | … | … | … | … | .6 | 1.9 | 2.2 | 7.2 | 11.4 | 8.0 | .4 | 2.2 | … | 2.3 |
| R23 | … | … | … | … | … | … | … | … | … | … | … | … | 6.0 | … | 2.0 | … | … | … | .6 |
| U7 | … | … | … | … | … | … | … | … | … | .6 | … | … | … | … | … | … | … | … | .1 |
| Y2 | … | .7 | … | … | … | 1.8 | … | 1.2 | 12.9 | 6.7 | 1.9 | 2.2 | 1.2 | … | … | 1.7 | … | … | 2.9 |
| Z | 3.2 | 4.6 | 1.3 | 1.2 | .4 | … | … | … | … | .6 | .3 | … | … | … | … | … | … | … | .4 |
Note.— An ellipsis (…) indicates a zero frequency.
Northwest China: Gansu, Qinghai, Sichuan, Shanxi, and Xinjiang.
Northeast China: Zhejiang, Anhui, Jiangsu, Qingdao, Shanghai, and Wuhan.
Southwest China: Guangxi and Yunnan.
Southeast China: Macau, Hunan, Fujian, Guangdong, and Jiangxi.
Table 3. .
Ages and Diversities of the Most Common ISEA Haplogroups
| Age Range and Haplogroup |
Overall Age (SE)a [years] |
Age (SE) in ISEAb [years] |
| >25,000 years ago: | ||
| M46 | 62,700 (12,400) | 62,700 (12,400) |
| M45 | 47,700 (21,600) | 47,700 (21,600) |
| M45b | 40,400 (18,900) | 40,400 (18,900) |
| N21 | 43,000 (25,000) | 30,300 (15,400) |
| M45a | 30,300 (15,600) | 30,300 (15,600) |
| R22 | 29,800 (19,200) | 28,300 (18,700) |
| E | 25,400 (11,500) | 26,000 (10,900) |
| F3b | 36,100 (14,600) | 25,600 (12,300) |
| 5,000–25,000 years ago: | ||
| B4c2 | 21,100 (12,200) | 17,900 (13,600) |
| B4a | 26,900 (6,800) | 14,800 (6,900) |
| B4c1b3 | 15,900 (5,300) | 13,500 (6,700) |
| R23 | 10,100 (5,800) | 10,100 (5,800) |
| B5a | 16,200 (3,300) | 9,200 (3,000) |
| B4b1 | 22,200 (5,500) | 8,400 (5,600) |
| B5b | 35,300 (11,400) | 8,200 (3,000) |
| M7c1c | 8,200 (2,500) | 8,100 (2,200) |
| E1a | 9,400 (2,800) | 7,500 (3,000) |
| F1a1a | 9,300 (2,800) | 7,300 (2,700) |
| E2 | 15,600 (12,500) | 7,200 (3,800) |
| E1b | 6,400 (2,300) | 6,000 (3,000) |
| B4a1a1 | 7,900 (2,000) | 5,700 (2,700) |
| <5,000 years ago: | ||
| F1a4 | 5,400 (2,600) | 4,100 (1,800) |
| D5d1 | 12,300 (8,200) | 4,000 (2,900) |
| F1a3 | 4,000 (2,900) | 4,000 (2,900) |
| Y2 | 3,600 (2,000) | 3,400 (1,700) |
| F1a2 | 3,200 (1,400) | 3,400 (1,900) |
Age estimates with use of all data.
Age estimates within ISEA determined on the basis of geographical specificity and/or identification of plausible founder types by use of the f1 criterion.60
A Bird’s-Eye View: Population Summary Statistics
All the ISEA populations studied have high levels of diversity, suggesting that they have maintained a comparatively large size over time and have not undergone substantial amounts of drift. The most diverse group is from Banjarmasin in Borneo (haplotype diversity 0.979), and the least diverse is the Tenggerese from Java (0.904). The PC analysis shows a clear east-west pattern (fig. 3). Most of the populations east of Wallace’s line are grouped together, the only exceptions being the samples from Mataram and Palu. In the case of the Mataram sample, this is perhaps not surprising, since it is found so close to Wallace’s line and therefore to the western populations. The Palu sample is unusual because of the much reduced level of haplogroup B types found; it is also one of the few eastern populations to contain haplogroups N9a6 and Y2, which could be due to recent arrivals from the west or north. The Taiwanese are outliers in both PCs, whereas the Filipinos are found at the western extreme of PC2 (14.1%) but just within the eastern part of PC1 (15.9%). The pattern was generally robust to a coarser haplotype classification. When Chinese data are included, Taiwan clusters closely with ISEA in PC1 (20.2%) and is clearly distinct from South China, albeit marginally closer than ISEA (data not shown; see also the work of Trejaut et al.41).
Figure 3. .
PC analysis of mtDNA haplogroup frequencies in Taiwan and ISEA (Taiwanese data include that of Trejaut et al.41).
AMOVA showed that the east-west patterning seen in the PC analysis is small but significant. When the populations were split into two groups relative to Wallace’s line, the difference between the two groups was found to be 0.17% of the total variation (P=.0186). This difference was even more significant when a central group (made up of the populations from Java, Borneo, Bali, and Mataram) was separated from the others; in this case, the difference between the three groups was 0.26% of the total variation (P=.0039). However, no significant difference was found when the populations were separated according to language. The difference between those groups that speak western Malayo-Polynesian languages and those that speak central Malayo-Polynesian languages was found to represent 0.06% of the total variation (P=.2893). However, all but two of the groups studied speak western Malayo-Polynesian languages (the exceptions are those from Alor and Waingapu).
Phylogeography of Lineages within Macrohaplogroup M
The most common entirely indigenous haplogroup in ISEA, at ∼14%, is haplogroup E (figs. 4 and 5A). It is also common in Taiwanese aboriginals but is almost absent in China, although its likely sister clade, M9a, is found on the East Asian mainland. It is also virtually absent in the Pacific, so that—although it is largely restricted to Austronesian speakers—it does not span their entire distribution. There are two major subclades: E1 (comprising E1a and E1b) and E2.41 E1a is almost entirely restricted to Taiwan and ISEA. E1b is predominantly found in ISEA but is absent in Taiwan. Haplogroup E as a whole dates to ∼25,000 years ago, with subclades ranging from ∼6,000 to 16,000 years ago.
Figure 4. .
Reduced median network of HVS-I sequences of haplogroup E in the region 16050–16400. The circles represent mtDNA HVS-I sequence types, shaded according to region and with an area proportional to their frequency in each region. Links are labeled with the nucleotide position of mutations; letters following positions indicate transversions, and the others are transitions. Mutations that have occurred more than once in the tree are underlined. Coding-region positions that were sequenced are indicated in italics. Subclades are labeled, and the M9 ancestor is indicated (arrow).
Figure 5. .
Spatial frequency distributions, created using the Kriging algorithm of the Surfer package, of haplogroups E (A), M7c1c (B), D5 (C), and Y2 (D). Samples from each island were merged to a central location, indicated with a point, to reduce any effect of error in small samples.
Haplogroup M7c1c, dating to 8,000 years ago, is also virtually absent from the mainland and is common throughout Taiwan aboriginals and ISEA (figs. 5B and 6). It makes up ∼8% of the sample and, like haplogroup E, is almost exclusively Austronesian in its distribution (although it does not extend into the Pacific farther than a few instances in Fiji and Micronesia). Other M7 subclades probably have a mainland origin (although M7b3 is largely restricted to Taiwan) and total <4% in ISEA.
Figure 6. .
Reduced median network of HVS-I sequences of haplogroup M7c1c. They are labeled as in figure 4. The M7c1 ancestor is indicated (arrow).
Haplogroup D5 is found at ∼3% overall in ISEA, although it reaches >10% in some parts of Sulawesi (fig. 5C). There is a distinct subclade, D5d1, which dates to ∼4,000 years ago in ISEA and belongs to a larger clade (D5d) with a mainland Chinese origin ∼12,000 years ago.
The only other branch of macrohaplogroup M to be found at relatively high levels in ISEA is haplogroup Q. This haplogroup is predominantly found in New Guinea and Near Oceania and has recently been found at low levels in Remote Oceania.26 It falls into three major subclades,72 all of which occur in ISEA. The highest levels of the haplogroup in ISEA are found in Alor (∼30%) and Ambon (∼12%), with far fewer examples elsewhere, although it has been found as far west as Borneo. More data from New Guinea would be needed to assess the time depth of these lineages in Indonesia.
The remaining M types appear largely unrelated to any lineages found elsewhere in the world. At least three new basal M haplogroups can be tentatively identified: M45 and M46, with local time depths of ∼40,000–70,000 years ago, and the very rare M47. In addition, a novel branch of M21, M21d (previously found concentrated in the aboriginal inhabitants of Malaysia35,73), has been found in several individuals in ISEA, the Malay Peninsula, and South China and also occurs at high frequency among the Austronesian-speaking Moken “sea-gypsies” of Burma. All other unclassified haplogroup M* types found in ISEA appear to be only very distantly related to each other, and their shared root dates to 58,900 (±13,600) years ago. Although the HVS-I dating is particularly imprecise in these cases, it seems likely that all these novel lineages are very ancient within ISEA.
Phylogeography of Lineages within Macrohaplogroup N
One of the most common haplogroups in ISEA is haplogroup B, which falls into two main clades, B4 and B5, although the unity of haplogroup B remains conjectural, since it is defined solely by a recurrent 9-bp deletion and a fast transition at an HVS-I site. The majority of B lineages in ISEA fall within haplogroup B4a, which is most frequent among Taiwanese aboriginals and in the Philippines. B4a includes the “Polynesian motif” (now classified as the root type within B4a1a1, formerly B4a166), which approaches fixation in Remote Oceania.22,27
However, only the root type of B4a and one derived type (which may have been generated by recurrent mutation) are shared between ISEA and Taiwan. In fact, most Taiwanese B4a lineages sit on a separate branch (B4a2a41), which has only been found elsewhere in two individuals from South China and which dates to 19,600 (±13,100) years ago. Lineages within B4a date to ∼15,000 years ago in ISEA. Despite the extensive sampling of ISEA, only 19 individuals were found to belong to haplogroup B4a1a1 and hence share the Polynesian motif. These types were found as far west as Kalimantan and Lombok but are most common in Ambon and Sulawesi. In contrast to the high diversity of B4a1a1 in the eastern Indonesian data of Redd et al.,22 all but two of the types found in this study were the root type, with an age of ∼6,000 years ago in ISEA. Other B4 lineages are rather rare in most of ISEA, although B5a is found at ∼4%.
The other major branch of haplogroup N in the region is R9. Most extant lineages belong to haplogroup F, and the most common branch of F in ISEA is haplogroup F1a, dating to 33,900 (±11,300) years ago, which itself contains two hierarchically nested subclades, F1a1 and F1a1a. The three nested subclades are found together at appreciable frequencies only in mainland Southeast Asia, and F1b and F1c are largely restricted to South China, suggesting a possible origin for F1 and F1a in this region. F1a1* types are rare, but both F1a* and F1a1a are common across Southeast Asia. Several geographically restricted subclades in F1a* are found with ages of ∼3,000–4,000 years, similar to the minor haplogroup Y2 (fig. 5D). Haplogroup F1a1a, dating to ∼9,000 years ago, is mainly found in the west and south of ISEA and is elsewhere most common in Thailand and in aboriginal Senoi groups of the Malay Peninsula.35 The only other branch of haplogroup F that is common in ISEA is F3b, which is largely restricted to the Philippines and Borneo and is of a Pleistocene age similar to its mainland sister clade, F3a.
As with haplogroup M, there are also several rare ancient haplogroups within haplogroup N and its subhaplogroup R, which are most common along the southern rim of the archipelago. N21 and N22 have previously been seen only in the Orang Asli of the Malay Peninsula,35,73 and haplogroup R22, also found in mainland Southeast Asia and the Nicobar Islands70 is also found across southern ISEA. Most of these rare lineages appear to date to the Pleistocene.
Discussion
Evidence of the Original Settlers of ISEA
Almost 14% of individuals found in ISEA have mtDNA haplotypes that belong to macrohaplogroup M but that appear unrelated to other M types found outside ISEA and that date to ∼40,000–70,000 years ago. It seems likely that these haplotypes, and others found only in the Malay Peninsula, can be traced back to the original inhabitants of ISEA, who would have colonized the area at around that time.73 Haplogroups N21 and R22 may provide further evidence of the persistence of mtDNAs from the earliest settlers. Today, N21 is more common in the aboriginal populations of the Malay Peninsula,35 but the phylogeographic pattern suggests that it arrived there from Sumatra. Haplogroup R22 now appears to be most common in the Shompen group of the Nicobar Islands70; however, it is most diverse in ISEA, and the root type is only found in Lombok and Alor, suggesting that it could be an indigenous marker for that area. If haplogroups N21, R22, M45, M46, M47, and M21d and the remaining unclassified M* types do indeed represent indigenous haplogroups, then this suggests that about a fifth of the modern inhabitants can trace their maternal ancestry back to the first anatomically modern settlers of ISEA.
Possible Markers for a Neolithic Dispersal Out of Taiwan
Elsewhere, it has been claimed that haplogroup B4a1a1 (containing the Polynesian motif) represents an Austronesian signature because of its high frequency in Polynesia and the presence of one-step ancestral types in Taiwanese aboriginals.22–24 However, the people of Remote Oceania show unusual genetic patterns due to their recent ancestry and the numerous founder events that have occurred during their history. Because of this, certain mtDNA haplogroups, particularly B4a1a1, are raised to extremely high frequencies in Remote Oceania, but this does not appear to be the case in the rest of the Austronesian-speaking world. In this study, B4a1a1 was found to be relatively rare, making up only ∼2% of the population as a whole and reaching a high of ∼14% in Ambon. It is completely absent in most of ISEA and is not found further west of Wallace’s line than southeast Borneo. The clade in which it nests, B4a1a, is indeed restricted to Austronesian-speaking populations but is pre-Holocene in age,41 so that its participation in a mid-Holocene Out of Taiwan event can be ruled out.
A more plausible candidate as a potential signature for a mid-Holocene Out of Taiwan dispersal is M7c1c. It has been found in all locations studied in this investigation and accounts for ∼8% of the ISEA sample. It has also been found in several Chinese individuals, and the clade from which it derives, M7c1, is most common in China. Furthermore, its starlike phylogeny suggests that it has undergone a population expansion through east and west Indonesia. However, the single founder mtDNA for M7c1c dates to ∼8,000 years ago, which is older than would be expected from the traditional Out of Taiwan model. It also appears to be more diverse in Taiwan and Borneo than would be expected if it had arrived from China <6,000 years ago, and its frequency distribution (fig. 5B) centers on Borneo and Sulawesi, resembling that of haplogroup E (fig. 5A), which is more plausibly ascribed to postglacial dispersals. Therefore, although M7c1c is the best candidate we have found for a marker of the Out of Taiwan dispersal, it remains possible that it has been present in Taiwan and/or Borneo since the early Holocene and that its distribution is, in fact, the result of a mid-Holocene dispersal centered on Borneo.
The only other lineages that can be plausibly ascribed to a mid-Holocene Out of Taiwan event are within D5, Y2, and F1a*. The root type of D5 is most common in China and Taiwan and is also found in a few individuals from ISEA. Many ISEA samples belong to the subgroup D5d1, which dates to ∼4,000 years ago in ISEA. The root type of this branch is not found in Taiwan, but three derived types are found there, suggesting that the root type may have been lost because of drift, and the absence of D5 from the Philippines may be due to insufficient sampling. F1a* includes two starlike subclades, F1a3 and F1a4, that, in their age and distribution, could be reconciled with a mid-Holocene dispersal from South China. The distribution and age of Y2 fits quite well with a proposed movement of some Neolithic groups south and west into Borneo and Sumatra from the Philippines.4
All told, these potential Out of Taiwan lineages (M7c1c, D5, Y2, F1a3, and F1a4) account for only ∼20% of the current data set. This is superficially similar to the results found for the Y chromosome by Capelli et al.,30 who also found that ∼20% of their ISEA sample could be accounted for by possible Taiwanese haplogroups (haplogroup O3, although Kayser et al.33 suggested that haplogroup O1 also took part in the dispersal, increasing the frequency of potential Out of Taiwan Y chromosomes). However, Y-chromosome studies to date suffer from poor phylogenetic resolution and should be interpreted with caution. The results of the present study suggest that if a mid-Holocene Neolithic migration did occur, it was—on the maternal side at least—demographically minor, contributing, at most, only a fifth of modern ISEA mtDNAs. That is, any Neolithic immigrants integrated into the resident population, rather than replacing it.9,13,74,75 Whether the putative immigrants can plausibly be assumed to have brought with them and imposed Austronesian languages9 or whether they themselves assimilated languages already spoken in ISEA would remain an open question.
Holocene Dispersal from Indochina and Near Oceania
Haplogroup F is a candidate for both postglacial and Neolithic dispersals. F1a1a provides a distinctive pattern; it is diverse in both South China and Indochina, dating to ∼9,000 years ago, but is most common in Indochina and some of the indigenous groups of peninsular Malaysia.35 It is not found in Taiwan, the Philippines, or northern Borneo but is found at a frequency of 3%–5% in Sumatra, Bali, and eastern Indonesia, with an estimated founder/dispersal age of ∼7,000 years ago in ISEA. This pattern suggests an expansion from mainland Southeast Asia during the Holocene.15 Another subclade with a similar, if more restricted, southerly distribution is F1a5.
Haplogroup N9a6 also seems to have its origins in Indochina. N9a, as a whole, is common across Japan, Korea, China, and Taiwan; however, the subclade N9a6 is largely restricted to Indochina, the Malay Peninsula, and ISEA, where it is found at a frequency of ∼1%. It is entirely absent in Taiwan and the Philippines. Because of its scarcity, it is difficult to be confident, but an Indochinese origin for N9a6 seems most plausible given the current evidence.
In this study, the Near Oceanian haplogroups P and Q were found at low levels in ISEA. Haplogroup P is rare and has been found only at low levels in Sulawesi and Sumba. Haplogroup Q is most common in the easternmost locations studied (reaching 29% in Alor, where Papuan as well as Austronesian languages are spoken76), but, at 3% of the sample as a whole, it is found as far west as Borneo, indicating long-range gene flow from Near Oceania into ISEA. This may suggest traces of the voyaging corridor proposed by Terrell and Welsch,77 although it is unclear how far back in time this influence extends.9 It is worth considering the possibility that this may be a genetic trace of a conduit into ISEA for the root and tuber crops, perhaps of New Guinean origin,9 that arguably contributed far more to a change in subsistence in the Neolithic period of the region than did the introduction of rice farming from the north or west.
Evidence of Major Postglacial Expansions
Perhaps the most striking result of this study is the signature of another phase of dispersal and settlement, not previously considered by most prehistorians of the region. This is represented by a third set of lineages, of which the most prominent are subclades of haplogroup E. M9, the larger haplogroup in which E nests, is present on the mainland, but E itself seems almost entirely restricted to Taiwan and ISEA and dates to at least 25,000 years ago, with its major subclades dating to between ∼5,000 and 15,000 years ago. It therefore seems that haplogroup E has an ancestry in the Pleistocene and may have originated around the peak of the last glaciation, probably on the east Sunda coastline (fig. 1), with dispersals both north and east as the sea level rose in the early Holocene. Given its distribution, we would not rule out that perhaps the direct ancestor of the Austronesian languages (proto-Austronesian or its hypothetical ancestor, pre-Austronesian) might have been dispersed in the postglacial expansion alongside haplogroup E lineages, rather than with the mtDNAs that we have identified as possible farming-dispersal markers.
Despite its previously assumed role in the putative Out of Taiwan dispersal, it seems that the presence of haplogroup B4a in ISEA can be traced back to the late Pleistocene. B4a is highly diverse in both China and Thailand but is most common in Taiwan and the Philippines. B4a seems to have originated on the mainland, where it is highly diverse and dates to ∼36,000 years ago. However, most of the haplotypes found in Taiwan fall within the restricted branch B4a2,41 which dates to ∼20,000 years ago within Taiwan, implying a probable separate ancestry over this period. These lineages appear to have emerged when Taiwan was a peninsula of the Chinese coastline at the peak of the last glaciation and to have been trapped on Taiwan when it separated from the mainland ∼12,000 years ago.18 B4a in Indonesia (within B4a1a) dates to ∼15,000 years ago, suggesting that the other offshore lineages were also present before the Holocene. The subhaplogroup B4a1a1, defined by the Polynesian motif, dates to ∼6,000 years ago in ISEA, providing further evidence for more-recent expansions within eastern Indonesia. Late-Pleistocene and/or early-Holocene dispersals may also explain the distributions of haplogroups F3b and M7b3, which are also restricted to ISEA and Taiwan and which also both date to the Pleistocene: F3b dates to ∼36,000 years ago and M7b3 to ∼12,000 years ago.
In conclusion, the rather simple “two-layer” settlement model of Australo-Melanesians ∼50,000 years ago followed by “Mongoloid” Austronesians ∼4,000 years ago—even with allowance for considerable survival of indigenous lineages, as in more recent versions—clearly does not capture the complexity of demographic history in the region. This chimes with recent analyses of skeletal remains and burial patterns in the region, which stress heterogeneity rather than an abrupt transition.78 Instead, we have evidence of dispersals across the region of Austronesia throughout the early to mid Holocene. Some of these may trace a mid-Holocene Neolithic dispersal, although, given the imprecision of the dating, they might equally reflect more-recent or even slightly earlier developments that may or may not have involved agriculture.13 In any case, the strongest signals in our data appear to result from the movement and expansion of indigenous, rather than introgressive, mtDNA lineages, dating to between ∼15,000 and ∼5,000 years ago. These lineages relate more closely to those of the Southeast Asian mainland than to those of modern aboriginal Australians and New Guineans.
The most likely driving forces behind such large-scale postglacial redispersals are the huge sea-level rises that flooded much of Greater Sundaland, reducing it to the present day archipelago (fig. 1).3,5,6,18,79,80 This is the first substantial genetic data set indicating support for such a view, and it suggests that a considerable shift of focus and a broadening of perspective may be necessary with regard to the prehistory of this region.
Acknowledgments
We thank the British Academy, the Bradshaw Foundation, the Royal Society, and the University of Huddersfield, for financial support. We also thank John Clegg and A. S. M. Sofro for providing most of the Indonesian samples, Catharina Forster for the Bali samples, Louis Jorio for the Moken samples, Harold Voris and Clara Simpson for the map outline used in figure 1, Graeme Barker and Chris Hunt for a critical reading of the manuscript, and Alessandro Achilli for advice on the use of Surfer. P.B.B. was supported by a St John’s College, University of Cambridge–Ruprecht-Karls Universität, Heidelberg Exchange Scholarship. P.S. was supported by a Marie Curie Early Stage Training Grant.
Web Resources
Accession numbers and URLs for data presented herein are as follows:
- Fluxus-engineering.com, http://www.fluxus-engineering.com/sharenet.htm (for Network 4 software package)
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for sequences [accession numbers EF068415–EF069260])
References
- 1.Barker G, Barton H, Beavitt P, Bird M, Daly P, Doherty C, Gilbertson D, Hunt C, Krigbaum J, Lewis H, et al (2002) Prehistoric foragers and farmers in southeast Asia: renewed investigations at Niah Cave, Sarawak. Proc Prehist Soc 68:147–164 [Google Scholar]
- 2.Détroit F, Dizon E, Falguères C, Hameau S, Ronquillo W, Sémah F (2004) Upper Pleistocene Homo sapiens from the Tabon cave (Palawan, The Philippines): description and dating of new discoveries. C R Palevol 3:705–712 10.1016/j.crpv.2004.06.004 [DOI] [Google Scholar]
- 3.Voris HK (2000) Maps of Pleistocene sea levels in Southeast Asia: shorelines, river systems and time durations. J Biogeogr 27:1153–1167 10.1046/j.1365-2699.2000.00489.x [DOI] [Google Scholar]
- 4.Bellwood P (1997) Prehistory of the Indo-Malaysian archipelago. University of Hawaii Press, Hawaii [Google Scholar]
- 5.Oppenheimer S (1998) Eden in the East. Wiedenfeld and Nicholson, London [Google Scholar]
- 6.Solheim WG (2006) Archaeology and culture in southeast Asia: unraveling the Nusantao. University of Philippines Press, Quezon City [Google Scholar]
- 7.Blust R (1999) Subgrouping, circularity and extinction: some issues in comparative Austronesian linguistics. Symp Ser Inst Linguis Academica Sinica 1:31–94 [Google Scholar]
- 8.Blust R (1996) Austronesian culture history: the window of language. In: Goodenough WH (ed) Prehistoric settlement of the Pacific. American Philosophical Society, Philadelphia, pp 28–35 [Google Scholar]
- 9.Spriggs M (2003) Chronology of the Neolithic transition in Island Southeast Asia and the western Pacific: a view from 2003. Rev Archaeol 24:57–80 [Google Scholar]
- 10.Schutler R, Marck JC (1975) On the dispersal of Austronesian horticulturalists. Archaeol Phys Anthropol Oceania 10:81–113 [Google Scholar]
- 11.Bellwood P (2005) First farmers, the origins of agricultural societies. Blackwell Publishing, Oxford [Google Scholar]
- 12.Pawley A (2002) Austronesian dispersal: languages, technologies and people. In: Bellwood P, Renfrew C (eds) Examining the farming/language dispersal hypothesis. McDonald Institute for Archaeological Research, Cambridge, pp 251–273 [Google Scholar]
- 13.Paz V (2002) Island Southeast Asia—spread or friction zone? In: Bellwood P, Renfrew C (eds) Examining the farming/language dispersal hypothesis. MacDonald Institute for Archaeological Research, Cambridge, pp 275–285 [Google Scholar]
- 14.Hunt CO, Rushworth G (2005) Cultivation and human impact at 6000 cal yr B.P. in tropical lowland forest at Niah, Sarawak, Malaysian Borneo. Quaternary Res 64:460–468 10.1016/j.yqres.2005.08.010 [DOI] [Google Scholar]
- 15.Anderson A (2005) Crossing the Luzon Strait: archaeological chronology in the Batanes Islands, Philippines and the regional sequence of Neolithic dispersal. J Austronesian Stud 1:25–44 [Google Scholar]
- 16.Bellwood P, Gillespie R, Thompson GB, Vogel GS, Ardika JW, Datan I (1992) New dates for prehistoric Asian rice. Asian Perspect 31:161–170 [Google Scholar]
- 17.Szabó K, O’Connor S (2004) Migration and complexity in Holocene Island Southeast Asia. World Archaeol 36:621–628 10.1080/0043824042000303809 [DOI] [Google Scholar]
- 18.Meacham W (1984–5) On the improbability of Austronesian origins in South China. Asian Perspect 26:89–106 [Google Scholar]
- 19.Wolff J (1995) The position of the Austronesian languages of Taiwan within the Austronesian group. In: Li P, Tsang J-K, Ho D, Tseng C (eds) Austronesian studies relating to Taiwan: Institute of History and Philology Symposium Series 3. Academica Sinica, Taipei [Google Scholar]
- 20.Greenhill S, Gray RD (2005) Testing dispersal hypotheses: Pacific settlement, phylogenetic trees and Austronesian languages. In: Mace R, Holden C, Shennan S (eds) The evolution of cultural diversity: phylogenetic approaches. UCL Press, London, pp 31–52 [Google Scholar]
- 21.Lum JK, Rickards O, Ching C, Cann RL (1994) Polynesian mitochondrial DNAs reveal three deep maternal lineage clusters. Hum Biol 66:567–590 [PubMed] [Google Scholar]
- 22.Redd AJ, Takezaki N, Sherry ST, McGarvey ST, Sofro ASM, Stoneking M (1995) Evolutionary history of the COII/tRNA(Lys) intergenic 9-base-pair deletion in human mitochondrial DNAs from the Pacific. Mol Biol Evol 12:604–615 [DOI] [PubMed] [Google Scholar]
- 23.Melton T, Peterson R, Redd AJ, Saha N, Sofro ASM, Martinson J, Stoneking M (1995) Polynesian genetic affinities with Southeast Asian populations as identified by mtDNA analysis. Am J Hum Genet 57:403–414 [PMC free article] [PubMed] [Google Scholar]
- 24.Sykes B, Leiboff A, Low-Beer J, Tetzner S, Richards M (1995) The origins of the Polynesians—an interpretation from mitochondrial lineage analysis. Am J Hum Genet 57:1463–1475 [PMC free article] [PubMed] [Google Scholar]
- 25.Lum JK, Cann RL, Martinson JJ, Jorde LB (1998) Mitochondrial and nuclear genetic relationships among Pacific Island and Asian populations. Am J Hum Genet 63:613–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pierson MJ, Martinez-Arias R, Holland BR, Gemmell NJ, Hurles ME, Penny D (2006) Deciphering past human population movements in Oceania: provably optimal trees of 127 mtDNA genomes. Mol Biol Evol 23:1966–1975 10.1093/molbev/msl063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Richards M, Oppenheimer S, Sykes B (1998) mtDNA suggests Polynesian origins in eastern Indonesia. Am J Hum Genet 63:1234–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oppenheimer SJ, Richards M (2001) Polynesian origins: slow boat to Melanesia? Nature 410:166–167 10.1038/35065520 [DOI] [PubMed] [Google Scholar]
- 29.Oppenheimer SJ, Richards M (2001) Fast trains, slow boats, and the ancestry of the Polynesian islanders. Sci Prog 84:157–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Capelli C, Wilson JF, Richards M, Stumpf MPH, Gratrix F, Oppenheimer S, Underhill P, Pascali VL, Ko T-M, Goldstein DB (2001) A predominantly indigenous paternal heritage for the Austronesian-speaking peoples of insular Southeast Asia and Oceania. Am J Hum Genet 68:432–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kayser M, Brauer S, Weiss G, Underhill PA, Roewer L, Schiefenhövel W, Stoneking M (2000) Melanesian origin of Polynesian Y chromosomes. Curr Biol 10:1237–1246 10.1016/S0960-9822(00)00734-X [DOI] [PubMed] [Google Scholar]
- 32.Kayser M, Brauer S, Weiss G, Schiefenhövel W, Underhill PA, Stoneking M (2001) Independent histories of human Y chromosomes from Melanesia and Australia. Am J Hum Genet 68:173–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kayser M, Brauer S, Weiss G, Schiefenhövel W, Underhill P, Shen PD, Oefner P, Tommaseo-Ponzetta M, Stoneking M (2003) Reduced Y-chromosome, but not mitochondrial DNA, diversity in human populations from West New Guinea. Am J Hum Genet 72:281–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Su B, Jin L, Underhill P, Martinson J, Milmani S, McGarvey ST, Shriver MD, Chu J, Oefner P, Chakraborty R, et al (2000) Polynesian origins: insights from the Y chromosome. Proc Natl Acad Sci USA 97:8225–8228 10.1073/pnas.97.15.8225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hill C, Soares P, Mormina M, Macaulay V, Meehan W, Blackburn J, Clarke D, Raja JM, Ismail P, Bulbeck D, et al (2006) Phylogeography and ethnogenesis of aboriginal Southeast Asians. Mol Biol Evol 23:2480–2491 10.1093/molbev/msl124 [DOI] [PubMed] [Google Scholar]
- 36.Melton T, Clifford S, Martinson J, Batzer M, Stoneking M (1998) Genetic evidence for the proto-Austronesian homeland in Asia: mtDNA and nuclear DNA variation in Taiwanese aboriginal tribes. Am J Hum Genet 63:1807–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yao Y-G, Kong Q-P, Bandelt H-J, Kivisild T, Zhang Y-P (2002) Phylogeographic differentiation of mitochondrial DNA in Han Chinese. Am J Hum Genet 70:635–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yao Y-G, Nie L, Harpending H, Fu Y-X, Yuan Z-G, Zhang Y-P (2002) Genetic relationship of Chinese ethnic populations revealed by mtDNA sequence diversity. Am J Phys Anthropol 118:63–76 10.1002/ajpa.10052 [DOI] [PubMed] [Google Scholar]
- 39.Yao Y-G, Zhang Y-P (2002) Phylogeographic analysis of mtDNA variation in four ethnic populations from Yunnan Province: new data and a reappraisal. J Hum Genet 47:311–318 10.1007/s100380200042 [DOI] [PubMed] [Google Scholar]
- 40.Kivisild T, Tolk H-V, Parik J, Wang Y, Papiha SS, Bandelt H-J, Villems R (2002) The emerging limbs and twigs of the east Asian mtDNA tree. Mol Biol Evol 19:1737–1751 (erratum 20:162) [DOI] [PubMed] [Google Scholar]
- 41.Trejaut JA, Kivisild T, Loo JH, Lee CL, He CL, Hsu CJ, Li ZY, Lin M (2005) Traces of archaic mitochondrial lineages persist in Austronesian speaking Formosan populations. PLoS Biol 3:e247 10.1371/journal.pbio.0030247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Betty DJ, Chin-Atkins AN, Croft L, Straml M, Esteal S (1996) Multiple independent origins of the COII/tRNALys intergenic 9-bp mtDNA deletion in aboriginal Australians. Am J Hum Genet 58:428–433 [PMC free article] [PubMed] [Google Scholar]
- 43.Comas D, Calafell F, Mateu E, Pérez-Lezaun A, Bosch E, Martínez-Arias R, Clarimon J, Facchini F, Fiori G, Luiselli D, et al (1998) Trading genes along the Silk Road: mtDNA sequences and the origin of central Asian populations. Am J Hum Genet 63:1824–1838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fucharoen G, Fucharoen S, Horai S (2001) Mitochondrial DNA polymorphisms in Thailand. J Hum Genet 46:115–125 10.1007/s100380170098 [DOI] [PubMed] [Google Scholar]
- 45.Hertzberg M, Mickleson KNP, Serjeanston S, Prior JF, Trent RJ (1989) An Asian-specific 9-bp deletion of mitochondrial DNA is frequently found in Polynesians. Am J Hum Genet 44:510–540 [PMC free article] [PubMed] [Google Scholar]
- 46.Horai S, Hayasaka K (1990) Intraspecific nucleotide sequence differences in the major noncoding region of human mitochondrial DNA. Am J Hum Genet 46:828–842 [PMC free article] [PubMed] [Google Scholar]
- 47.Horai S, Murayama K, Hayasaka K, Matsubayashi S, Hattori Y, Fucharoen G, Harihara S, Park KS, Omoto K, Pan I-H (1996) mtDNA polymorphism in East Asian populations, with special reference to the peopling of Japan. Am J Hum Genet 59:579–590 [PMC free article] [PubMed] [Google Scholar]
- 48.Kolman C, Sambuughin N, Bermingham E (1996) Mitochondrial DNA analysis of Mongolian populations and implications for the origin of New World founders. Genetics 142:1321–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee SD, Shin CH, Kim KB, Lee YS, Lee JB (1997) Sequence variation of mitochondrial DNA control region in Koreans. Forensic Sci Int 87:99–116 10.1016/S0379-0738(97)02114-2 [DOI] [PubMed] [Google Scholar]
- 50.Nishimaki Y, Sato K, Fang L, Ma M, Hasekura H, Boettcher B (1999) Sequence polymorphism in the HV1 region in Japanese and Chinese. Legal Med 1:238–249 10.1016/S1344-6223(99)80044-3 [DOI] [PubMed] [Google Scholar]
- 51.Oota H, Kurosaki K, Pookajorn S, Ishida T, Ueda S (2001) Genetic study of the Palaeolithic and Neolithic Southeast Asians. Hum Biol 73:225–231 [DOI] [PubMed] [Google Scholar]
- 52.Pfeiffer H, Steighner R, Fisher R, Mörnstad H, Yoon CL, Holland MM (1998) Mitochondrial DNA extraction and typing from isolated dentin—experimental evaluation in a Korean population. Int J Legal Med 111:309–313 10.1007/s004140050177 [DOI] [PubMed] [Google Scholar]
- 53.Prasad BVR, Ricker CE, Watkins WS, Dixon ME, Rao BB, Naidu JM, Jorde LB, Bamshad M (2001) Mitochondrial DNA variation in Nicobarese islanders. Hum Biol 73:715–725 [DOI] [PubMed] [Google Scholar]
- 54.Qian YP, Chu Z-T, Dai Q, Wei C-D, Chu YJ, Tajima A, Horai S (2001) Mitochondrial DNA polymorphism in Yunnan nationalities in China. J Hum Genet 46:211–220 10.1007/s100380170091 [DOI] [PubMed] [Google Scholar]
- 55.Seo Y, Stradmann-Bellinghausen B, Rittner C, Takahama K, Schneider PM (1998) Sequence polymorphism of mitochondrial DNA control region in Japanese. Forensic Sci Int 97:155–164 10.1016/S0379-0738(98)00153-4 [DOI] [PubMed] [Google Scholar]
- 56.Zainuddin Z, Goodwin W (2003) Mitochondrial DNA profiling of modern Malay and Orang Asli populations in peninsular Malaysia. Prog Forensic Genet 10:428–430 [Google Scholar]
- 57.Tanaka M, Cabrera VM, González AM, Larruga JM, Takeyasu T, Fuku N, Guo L-J, Hirose R, Fujita Y, Kurata M, et al (2004) Mitochondrial genome variation in eastern Asia and the peopling of Japan. Genome Res 14:1832–1850 10.1101/gr.2286304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wen B, Li H, Gao S, Mao X, Gao Y, Li F, Zhang F, He Y, Dong Y, Zhang Y, et al (2004) Genetic structure of Hmong-Mien speaking populations in East Asia as revealed by mtDNA lineages. Mol Biol Evol 22:725–734 10.1093/molbev/msi055 [DOI] [PubMed] [Google Scholar]
- 59.Wen B, Li H, Lu D, Song X, Zhang F, He Y, Li F, Gao Y, Mao X, Zhang L, et al (2004) Genetic evidence supports demic diffusion of Han culture. Nature 431:302–305 10.1038/nature02878 [DOI] [PubMed] [Google Scholar]
- 60.Richards M, Macaulay V, Hickey E, Vega E, Sykes B, Guida V, Rengo C, Sellitto D, Cruciani F, Kivisild T, et al (2000) Tracing European founder lineages in the Near Eastern mitochondrial gene pool. Am J Hum Genet 67:1251–1276 [PMC free article] [PubMed] [Google Scholar]
- 61.Andrews RM, Kubacka I, Chinnery PF, Lightowlers R, Turnbull D, Howell N (1999) Reanalysis and revision of the Cambridge Reference Sequence for human mitochondrial DNA. Nat Genet 23:147 10.1038/13779 [DOI] [PubMed] [Google Scholar]
- 62.Bandelt H-J, Quintana-Murci L, Salas A, Macaulay V (2002) The fingerprint of phantom mutations in mitochondrial DNA data. Am J Hum Genet 71:1150–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bandelt H-J, Forster P, Sykes BC, Richards MB (1995) Mitochondrial portraits of human populations using median networks. Genetics 141:743–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Forster P, Harding R, Torroni A, Bandelt H-J (1996) Origin and evolution of Native American mtDNA variation: a reappraisal. Am J Hum Genet 59:935–945 [PMC free article] [PubMed] [Google Scholar]
- 65.Saillard J, Forster P, Lynnerup N, Bandelt H-J, Nørby SS (2000) mtDNA variation among Greenland Eskimos: the edge of the Beringian expansion. Am J Hum Genet 67:718–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kong Q-P, Bandelt H-J, Sun C, Yao Y-G, Salas A, Achilli A, Wang C-Y, Zhong L, Zhu C-L, Wu S-F, et al (2006) Updating the East Asian mtDNA phylogeny: a prerequisite for the identification of pathogenic mutations. Hum Mol Genet 15:2076–2086 10.1093/hmg/ddl130 [DOI] [PubMed] [Google Scholar]
- 67.Torroni A, Bandelt H-J, Macaulay V, Richards M, Cruciani F, Rengo C, Martinez-Cabrera V, Villems R, Kivisild T, Metspalu E, et al (2001) A signal, from human mtDNA, of post-glacial recolonization in Europe. Am J Hum Genet 69:844–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Richards MB, Macaulay VA, Torroni A, Bandelt H-J (2002) In search of geographical patterns in European mitochondrial DNA. Am J Hum Genet 71:1168–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schneider S, Roessli D, Excoffier L (2000) Arlequin: a software for population genetics data analysis: version 2.000. Genetics and Biometry Laboratory, Department of Anthropology, University of Geneva, Geneva [Google Scholar]
- 70.Trivedi R, Sitalaximi T, Banerjee J, Singh A, Sircar PK, Kashyap VK (2006) Molecular insights into the origins of the Shompen, a declining population of the Nicobar archipelago. J Hum Genet 51:217–226 10.1007/s10038-005-0349-2 [DOI] [PubMed] [Google Scholar]
- 71.Kivisild T, Shen P, Wall DP, Do B, Sung R, Davis K, Passarino G, Underhill PA, Scharfe C, Torroni A, et al (2006) The role of selection in the evolution of human mitochondrial genomes. Genetics 172:373–387 10.1534/genetics.105.043901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Friedlaender J, Schurr T, Gentz F, Koki G, Friedlaender F, Horvat G, Babb P, Cerchio S, Kaestle F, Schanfield M, et al (2005) Expanding Southwest Pacific mitochondrial haplogroups P and Q. Mol Biol Evol 22:1506–1517 10.1093/molbev/msi142 [DOI] [PubMed] [Google Scholar]
- 73.Macaulay V, Hill C, Achilli A, Rengo C, Clarke D, Meehan W, Blackburn J, Semino O, Scozzari R, Cruciani F, et al (2005) Single, rapid coastal settlement of Asia revealed by analysis of complete mitochondrial genomes. Science 308:1034–1036 10.1126/science.1109792 [DOI] [PubMed] [Google Scholar]
- 74.Hurles M (2002) Can the hypothesis of language/agriculture co-dispersal be tested with archaeogenetics? In: Bellwood P, Renfrew C (eds) Examining the farming/language dispersal hypothesis. MacDonald Institute for Archaeological Research, Cambridge, pp 299–309 [Google Scholar]
- 75.Hurles ME, Nicholson J, Bosch E, Renfrew C, Sykes BC, Jobling MA (2002) Y chromosomal evidence for the origins of Oceanic-speaking peoples. Genetics 160:289–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Reesink G (2005) West Papuan languages: roots and development. In: Pawley A, Attenborough R, Golson G, Hide R (eds) Papuan pasts: cultural, linguistic and biological histories of Papuan-speaking peoples. The Australian National University, Canberra, pp 185–215 [Google Scholar]
- 77.Terrell JE, Welsch RL (1997) Lapita and the temporal geography of prehistory. Antiquity 71:548–572 [Google Scholar]
- 78.Détroit F (2006) Homo sapiens in Southeast Asian archipelagos: the Holocene fossil evidence with special reference to funerary practises in East Java. In: Simanjuntak T, Pojoh IHE, Hisyam M (eds) Austronesian diaspora and the ethnogeneses of the people in Indonesian Archipelago. LIPI Press, Jakarta [Google Scholar]
- 79.Bird MI, Taylor D, Hunt C (2005) Palaeoenvironments of insular Southeast Asia during the last glacial period: a savanna corridor in Sundaland? Quaternary Sci Rev 24:2228–2242 10.1016/j.quascirev.2005.04.004 [DOI] [Google Scholar]
- 80.Solheim WG (1994) South-east Asia and Korea from the beginnings of food production to the first states. In: De Laet SJ (ed) The history of humanity. Vol I. Routledge, London, pp 468–481 [Google Scholar]