Abstract
The t(11;14)(q13;q32) chromosomal translocation, the hallmark of mantle cell lymphoma (MCL), is recurrently found in multiple myelomas (MM) by means of conventional cytogenetics. Unlike MCL, recent molecular studies of MM-derived cell lines with t(11;14) have indicated that the breakpoints are highly dispersed over the 11q13 region; however, the fact that cyclin D1 is generally overexpressed in these cell lines suggests that this gene is the target of the translocation. To evaluate further the involvement of cyclin D1 in MM, we used immunohistochemistry and fluorescence in situ hybridization to investigate cyclin D1 expression and the presence of chromosome 11 abnormalities in a representative panel of 48 MM patients (40 at diagnosis and 8 at relapse). Cyclin D1 overexpression occurred in 12/48 (25%) of cases; combined immunohistochemistry and fluorescence in situ hybridization analyses in 39 patients showed cyclin D1 positivity in all of the cases (7/7) bearing the t(11;14), in two of the 13 cases with trisomy 11, and in one of the 19 cases with no apparent abnormalities of chromosome 11. Our data indicate that the t(11;14) translocation in MM leads to cyclin D1 overexpression and that immunohistochemical analysis may represent a reliable means of identifying this lesion in MM.
Multiple myeloma (MM) is a malignant proliferation of bone marrow plasma cells, which is characterized by a wide spectrum of clinical entities and whose molecular pathogenesis is still largely unknown. 1 In particular, cytogenetic analysis is only informative in approximately 40% of cases because of the limited mitotic activity of malignant plasma cells. However, in about 20 to 40% of the tumors with an abnormal karyotype, a 14q+ marker has been found that is generally considered to be a consequence of translocations involving the immunoglobulin heavy chain locus (IGH) on chromosome 14q32. 2,3 Recent molecular and fluorescence in situ hybridization (FISH) studies have confirmed this evidence by showing that 14q32 translocations represent a very frequent event in MMs involving a variety of chromosome loci, 4-7 mostly the 11q13, 4p16.3, 16q23, and 6p25, where the putative target genes cyclin D1, FGFR3/MMSET, c-MAF, and MUM1/IRF4, respectively, are located. 8-14
The t(11;14)(q13;q32) chromosomal translocation is associated with approximately 70 to 90% of mantle cell lymphomas (MCL) 15-18 and leads to an overexpression of the cyclin D1 gene. 19-22 The 11q13 breakpoints in MCL are prevalently clustered in a 1-kb region, known as the major translocation cluster of the BCL-1 locus, which is located approximately 120 kb centromeric of the cyclin D1 gene. Evidence of this translocation has been reported in almost 30% of the MM cases with cytogenetically detectable 14q+: 2,3 however, rearrangements of the BCL-1/cyclin D1 regions involved in MCL are rarely detected in MM by Southern blotting. 8,9,23 The molecular analysis of 11q13 breakpoints in a limited number of MMs (mostly cell lines) with the t(11;14) indicates that they are highly scattered over a relatively large area encompassing the BCL-1/cyclin D1 loci, but the fact that cyclin D1 was found to be overexpressed in these cases strongly suggests that it is deregulated in MM as a result of the translocation. 8,9,22,24
The detection of the t(11;14) in MM patients must be considered an important step for the evaluation of its prognostic significance, but conventional cytogenetics and Southern blotting are inadequate for this purpose. We and others have recently attempted to provide fluorescence in situ hybridization (FISH) assays for detecting this genetic lesion in MM, 7,9 as has been done in MCL, 17,18 but its functional consequences, particularly the deregulation of cyclin D1, have not yet been investigated in panels of MM tumors with reliable information concerning the translocation. 25-27
The aim of this study was to evaluate a series of primary MMs for structural evidence of BCL-1/cyclin D1 locus involvement by means of FISH, and for cyclin D1 deregulation by means of immunohistochemistry (IHC). Our data indicate that cyclin D1 overexpression occurs in about 25% of cases and closely correlates with the presence of the t(11;14).
Materials and Methods
Patients
Forty-eight cases of multiple myeloma (MM) were selected from 63 patients admitted to the Hematology Service, Ospedale Maggiore IRCCS, Milan, between 1996 and 1999. The selection was based exclusively on the availability of a bone biopsy. The diagnosis of MM was made according to the criteria described by Durie and Salmon. 28 Bone marrow aspirates and biopsies were taken from each patient at the same time and underwent conventional light microscopy examination and immunophenotyping analysis. The clinical and pathological data of the cases are summarized in Table 1 ▶ : 40 were primary tumors at diagnosis (83.3%) and 8 were relapses (16.7%); 22 were males (45.8%) and 26 females (54.2%); median age was 65 (range, 36–85). The monoclonal component was IgG in 26 cases (54.2%), IgA in 16 cases (33.3%), and IgD in one case (2.1%); four cases were λ light chain myelomas (8.3%) and one was IgA/IgG (2.1%). The κ/λ chain ratio was 28/19; one case expressed both the light chains. The Durie and Salmon stage 28 and Bartl’s histological grade and stage 29 are given only for the 40 patients at diagnosis: 23 patients were in clinical stage I (57.5%), six in stage II (15%) and 11 in stage III (27.5%); 22 patients had clinical symptoms (55%). According to Bartl et al, 29 29 (72.5%) were of low-grade malignancy, nine (22.5%) of intermediate-grade malignancy, and two (5%) of high-grade malignancy; 20 cases (50%) showed a low bone marrow infiltration (Bartl’s stage I), 11 (25%) intermediate infiltration (stage II), and nine (22.5%) a high degree of infiltration (stage III). Bone marrow biopsies from ten patients with solid tumors but no bone marrow involvement were used as controls for immunohistochemical analyses.
Table 1.
Clinicopathological and Molecular Findings of MM Patients Included in the Study
| Patient no. | Gender/Age | MC | LDH | β2 | Stage* | BG | BS | Cyclin D1 IHC | FISH | |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | M/67 | Aλ | 0 | 1 | IA/N | II | II | – | – | |
| 2 | M/66 | Aκ | 0 | 0 | IA/Y | I | II | – | ND | |
| 3 | M/51 | Aκ | 0 | 1 | IA/N | I | I | – | – | |
| 4 | F/66 | Gκλ | 0 | 1 | IA/Y | I | II | – | +11 | |
| 5 | F/49 | Gκ | 0 | 0 | IA/Y | I | III | + | t(11;14) | |
| 6 | F/46 | Aλ | 0 | 0 | IA/N | II | I | – | – | |
| 7 | F/56 | Gλ | 0 | 1 | IA/N | II | II | – | – | |
| 8 | M/57 | Gκ | 0 | 0 | IA/Y | I | I | + | ND | |
| 9 | F/50 | Gκ | 0 | 0 | IA/Y | II | I | – | – | |
| 10 | F/66 | λ | 1 | 1 | IA/N | I | II | – | – | |
| 11 | M/74 | Aλ | 0 | 1 | IA/N | I | I | – | +11 | |
| 12 | F/55 | Gκ/Aκ | 0 | 1 | IA/Y | I | I | – | – | |
| 13 | F/85 | Aλ | 0 | 1 | IA/N | I | I | – | – | |
| 14 | F/53 | Gκ | 0 | 0 | IA/N | I | I | – | ND | |
| 15 | F/48 | Aλ | 0 | 0 | IA/N | I | I | – | ND | |
| 16 | M/65 | Gκ | 1 | 1 | IA/N | I | I | – | – | |
| 17 | M/54 | Aκ | 0 | 1 | IA/N | II | I | – | +11 | |
| 18 | M/71 | Gλ | 0 | 0 | IA/N | II | I | – | – | |
| 19 | M/48 | Gκ | 0 | 1 | IA/N | I | I | – | – | |
| 20 | F/78 | Gλ | 0 | 1 | IA/N | I | I | – | ND | |
| 21 | F/46 | Gλ | 0 | 0 | IA/N | I | II | – | – | |
| 22 | F/72 | Gκ | 0 | 1 | IA/N | I | II | – | +11 | |
| 23 | F/56 | Gκ | 0 | 0 | IA/N | I | I | – | – | |
| 24 | F/70 | Gκ | 0 | 1 | IIA/Y | I | I | – | – | |
| 25 | F/74 | Aκ | 0 | 1 | IIA/Y | I | I | – | – | |
| 26 | M/36 | Dκ | 1 | 1 | IIA/Y | I | III | + | t(11;14) | |
| 27 | M/77 | Gκ | 0 | 1 | IIA/Y | I | I | – | +11 | |
| 28 | M/44 | Gκ | 0 | 0 | IIA/Y | I | II | – | +11 | |
| 29 | M/67 | Aλ | 0 | 1 | IIA/Y | II | III | + | ND | |
| 30 | M/67 | Gκ | 0 | 1 | IIIA/Y | III | III | – | +11 | |
| 31 | F/61 | Ak | 0 | 1 | IIIA/Y | I | III | – | +11 | |
| 32 | F/65 | λ | 0 | 0 | IIIA/Y | II | I | – | ND | |
| 33 | F/62 | λ | 0 | 1 | IIIA/Y | I | I | + | t(11;14) | |
| 34 | M/53 | Aκ | 0 | 1 | IIIA/Y | I | III | + | t(11;14) | |
| 35 | F/71 | λ | 0 | 1 | IIIA/Y | II | III | – | – | |
| 36 | M/76 | Aλ | 0 | 1 | IIIA/Y | I | II | + | +11 | |
| 37 | F/47 | Gλ | 0 | 0 | IIIA/N | I | II | + | – | |
| 38 | M/57 | Gκ | 1 | 1 | IIIB/Y | III | III | – | +11 | |
| 39 | M/78 | Gκ | 0 | 1 | IIIB/Y | I | III | + | t(11;14) | |
| 40 | F/75 | Gκ | 0 | 1 | IIIB/Y | I | II | – | – | |
| 41 | F/56 | Gκ | 0 | 0 | R | R | R | + | +11 | |
| 42 | F/68 | Gλ | 0 | 0 | R | R | R | – | – | |
| 43 | M/69 | Gκ | 0 | 1 | R | R | R | – | ND | |
| 44 | M/50 | Aλ | 0 | 0 | R | R | R | – | ND | |
| 45 | F/78 | Gκ | 0 | 1 | R | R | R | – | +11 | |
| 46 | M/51 | Aκ | 0 | 1 | R | R | R | – | +11 | |
| 47 | F/68 | Gκ | 0 | 1 | R | R | R | + | t(11;14) | |
| 48 | M/72 | Aλ | 0 | 1 | R | R | R | + | t(11;14) |
MC, monoclonal component; BG, Bartl’s histologic grade; BS, Bartl’s histologic stage; LDH, lactic dehyrogenase; ND, not done.
*Absence (N) or presence (Y) of clinical symptoms; R, relapse.
LDH: 0 ≤ 460 U/l; 1 > 460 U/l; β2: 0 ≤ 2.6 mg/ml; 1 > 2.2 mg/ml.
Immunohistochemical Analysis
All of the 58 bone marrow biopsies (48 MM and 10 controls) were fixed in B5 and decalcified by EDTA (Mielodec, Bio-Optica, Milan). The mean length of the biopsies was 15 mm (range, 7–28 mm). Cyclin D1 protein expression was assayed by means of the avidin-biotin peroxidase complex (ABC) method, using the 3,3′-diaminobenzidine tetrahydrochloride chromogen as previously described. 30 For antigen retrieval, the slides were placed in 0.01 mol/L of EDTA buffer at pH 8.0 and underwent six 5-minute 780W cycles at 90°C in a microwave oven. The sections were immunostained with the DCS-6 monoclonal antibody (Novocastra Laboratories Ltd., Newcastle-on-Tyne, UK) at a working concentration of 1 μg/ml 31 All of the bone marrow tissues were evaluated at 630× magnification and independently analyzed by two of the authors (G. P. and N. C.), who had no knowledge of the genetic and clinicopathological features. Only nuclear staining of cyclin D1 was considered positive. The percentage of cyclin D1-positive plasma cells was expressed as their ratio to the total number of plasma cells. In accordance with Vasef et al, 26 we considered cyclin D1 to be positive if over 10% of plasma cells showed nuclear positivity. Formalin-fixed, paraffin-embedded samples from a laryngeal squamous cell carcinoma bearing a 20-fold BCL-1/cyclin D1 locus amplification, 31 and EDTA-decalcified and B5-fixed samples from a bone marrow biopsy diffusely infiltrated by a mantle cell lymphoma, were used as positive control; the substitution of the primary antibody with nonimmune mouse serum was used as negative control.
FISH
FISH analyses were possible on 39 of the 48 patient samples and were performed as previously described. 11 The percentage of malignant plasma cells, assessed by morphologically analyzing cytospins of cell suspensions from bone marrows, ranged from 13 to 85% (median, 31%). The bone marrow cells were cultured in RPMI 1640 medium supplemented with 20% fetal calf serum without any mitogen for 24 hours at 37°C in 5% CO2. The cultures were treated with colcemid (0.05 mg/ml) for 20 minutes. The cells were harvested using hypotonic potassium chloride, fixed by methanol/glacial acetic acid (3:1, v/v), and then stored at −20°C. Bone marrow cells from three healthy donors and three with thrombocythemia were used as negative controls.
The slides were hybridized in situ with probes labeled with biotin or directly with fluorochrome Cy3 (Amersham, Little Chalfont, UK) by means of nick translation as previously described. 32 Biotin-labeled DNA was detected using fluorescein isothiocyanate (FITC)-conjugated avidin (green signal; Vector Laboratories, Burlingame, CA); direct Cy3 was detected as a red signal. The chromosomes were identified by simultaneous 4′,6′-diaminido-2-phenylindole dihydrochloride (DAPI) staining, which produces a Q-banding pattern. Digital images were obtained using a Leica DMR epifluorescence microscope (Leica Imaging Systems Ltd., Cambridge, UK) equipped with a charge-coupled device camera (Cohu Inc., San Diego, CA). The FITC-avidin, Cy3, and DAPI fluorescence signals were detected using specific filters. The images were recorded, pseudocolored, and merged using QFISH software (Leica Imaging Systems Ltd, Cambridge, UK), and finally edited using Adobe Photoshop, Version 3 (Adobe Systems, Mountain View, CA).
FISH Probes
The 11q13 region was investigated using probes recently described by us. 9 In particular, the centromeric YAC 744 probe and the telomeric cosmid FGF3 probe were used in double-color interphase and metaphase FISH, respectively giving red and green signals. YAC 744 was obtained from the human CEPH2 library (YAC Screening Center, DIBIT, Milan) and was reported to contain an insert of approximately 300 kb and to be positive for the D11S97 and D11S146 markers located approximately 400 kb centromeric to the MTC/BCL-1 locus. 33,34 In addition, we have previously shown that this YAC is retained on the der(11) chromosome in KMS-12 cell line carrying the t(11;14) translocation, 9 the 11q13 breakpoint of which has been localized 215 kb centromeric of the BCL-1 locus. 18 To our knowledge, this breakpoint is the most centromeric so far cloned in MM. The clone representative of the FGF3 locus, which is located approximately 300 kb telomeric to the MTC/BCL-1 region, was isolated from a cosmid library of human placenta (Clontech, San Diego, CA). 9 The two probes therefore bound a region of approximately 600 kb, encompassing the BCL-1 and cyclin D1 loci in which the majority of the 11q13 breakpoints in MM have so far been identified or cloned. 8,9,22,24 In normal chromosomes the two signals are colocalized on 11q13, whereas breakpoints occurring within this region should lead to the monoallelic segregation of the two signals. The cutoff value for dissociated red and green signals (neither associated physically nor yellow) in interphase FISH was established as 10% (mean + 3SD) on the basis of the analysis of 200 nuclei from each negative control (4.67 ± 1.63). Aneuploidy of chromosome 11 was evaluated using the biotin-labeled α-satellite DNA probe specific for chromosome 11 (clone D4Z1; Oncor Inc., Gaithersburg, MD) on the same hybridization: the cutoff value for 11 trisomy was established as 1% (mean + 3SD) on the basis of the analysis of 200 nuclei from each negative control (0.1 ± 0.3). To determine whether the breakpoints were the result of a t(11;14) translocation, the positive cases were analyzed by means of double-color FISH using the FGF3 probe (red signal) and a pool of plasmid clones previously reported by us 11 that are specific for the constant regions of the IGH locus (IGH probe; green signal): the constant regions Cγ1, Cγ2, Cγ3, Cγ4 (BamHI/HindIII fragments of 7, 4, 7, and 6 kb respectively), Cα1 and Cα2 (BamHI fragments of approximately 18 kb), and Cμ (HindIII fragment of 10 kb). In the case of a translocation the two signals colocalized in either interphase or metaphase nuclei.
Statistical Analysis
The correlations between the clinicopathological and immunohistochemical data were evaluated by means of Fischer exact test or χ 2 test with Yates correction when necessary. For the purposes of statistical analysis, we considered two cyclin D1 classes: positive and negative. The investigated clinicopathological parameters were classified as follows: gender (male and female); age at diagnosis (≤60 years and >60 years); clinical stage (I, II, and III); clinical symptoms (presence and absence); Bartl’s histological grade (low and intermediate/high grade malignancy); Bartl’s histological stage (I, <20% of bone marrow infiltration; II, 20 to 50%; and III, >50%); lactic dehydrogenase (LDH) (≤460 and >460 U/L); β2 microglobulin (≤2.6 and >2.6 mg/ml).
Results
IHC Analysis
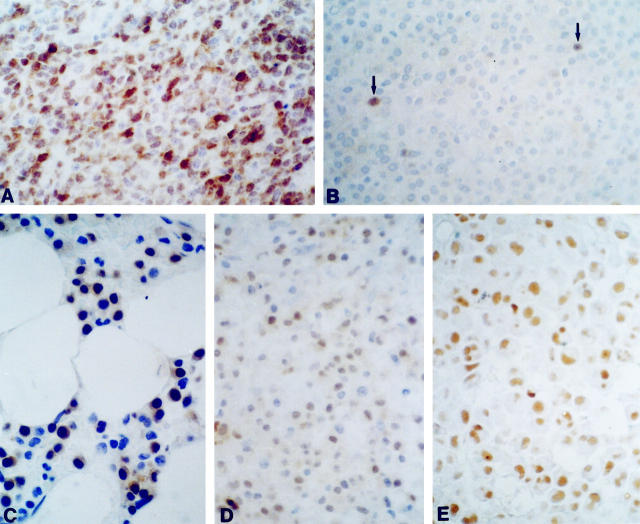
Forty-eight MM patients were investigated, including 40 at clinical presentation and 8 at relapse. The specificity of our IHC assay for cyclin D1 expression was tested using ten normal bone marrow biopsies as negative controls, and a bone marrow biopsy from a MCL case as a positive control. Hematopoietic precursors and plasma cells from negative controls were unreactive to cyclin D1 antibody (data not shown), whereas intense nuclear staining was observed in the MCL case (Figure 1A) ▶ . Cyclin D1 expression was found positive in the plasma cells of 12 (25%) MM patients (see Table 1 ▶ ). The hematopoietic cells in the MM biopsies were also invariably unreactive. Only three cases showed fewer than 10% positive plasma cells, which appeared as scattered cells in the biopsy (a representative case is shown in Figure 1B ▶ ); immunostaining was localized to the nuclei of plasma cells and was variable in intensity (see representative cases in Figure 1 ▶ , C–E).
Figure 1.
IHC analysis of MMs. A: Positive control (diffusely infiltrated bone marrow biopsy by a MCL case). B: A representative case (no. 35) of a cyclin D1-negative MM showing only a very few scattered positive plasma cells (below the 10% threshold; see text) in the biopsy specimen. C−E: Neoplastic plasma cell nuclei from positive cases (nos. 8, 26, and 39, respectively). Hematoxylin counterstain; original magnification, ×630.
FISH Analysis
FISH analysis was possible in 39 of the 48 MMs. Red (YAC 744) and green (FGF3) signals were found to be associated (yellow signal) in the normal interphase and metaphase nuclei (Figure 2 ▶ , A and B), and the green signal from the α-satellite DNA probe specific for chromosome 11, which was used on the same hybridization, could be easily distinguished from the FGF3 signal in the interphase nuclei.
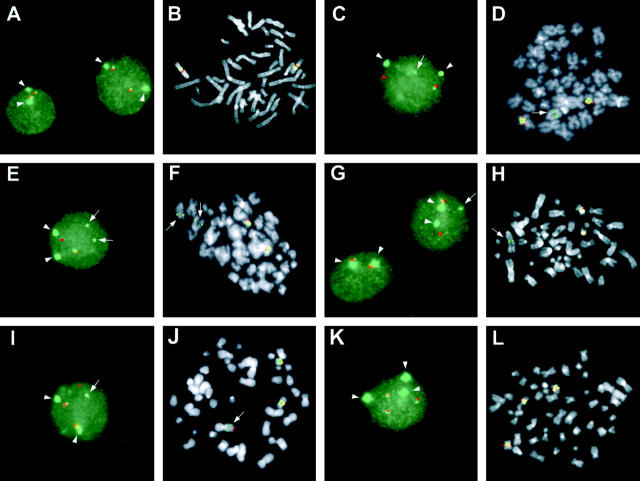
Figure 2.
FISH analysis of 11q13 breakpoints in MM. Red signal: YAC 744; green signal: FGF3; dissociated signals specific for FGF3 (green) are indicated by arrows; the signal (green) from the α-satellite DNA probe specific for chromosome 11 is indicated by arrowhead. A and B: Interphase and metaphase nuclei from a normal control. C and D: Interphase and metaphase nuclei with an 11q13 breakpoint in case no. 26. E and F: Interphase and metaphase nuclei with an 11q13 breakpoint in case no. 34; two dissociated green signals are found due to a duplication of the putative derivative chromosome. G and H: Interphase and metaphase nuclei with an 11q13 breakpoint in case no. 39. I and J: Interphase and metaphase nuclei with an 11q13 breakpoint in case no. 47; there was evidence that the breakpoint is within YAC 744 as the signal was split and associated with the FGF3 in a derivative chromosome. K and L: Interphase and metaphase nuclei of case no. 23 with trisomy 11.
Dissociated red (YAC 744) and green (FGF3) signals, suggestive of an 11q13 breakpoint were detected in the interphase nuclei from seven (18%) of the 39 cases (see Table 1 ▶ ). In all of these cases, associated red and green signals were also found, thus suggesting the presence of a normal chromosome 11; furthermore, no extra copies of chromosome 11 were found in any of the translocated cases as evaluated by the α-satellite DNA probe (see representative cases in Figure 2 ▶ , C and G). Distinct patterns were detected in interphase nuclei from cases 34 and 47, which showed an additional green (FGF3) and red (Yac 744) signal, respectively (see Figure 2 ▶ , E and I; see below). Furthermore, FISH analysis demonstrated trisomy 11 in 13 cases (see Table 1 ▶ ), in all of which a third centromeric signal (green) was found, together with a third series of associated red (Yac 744) and green (FGF3) signals (see representative case in Figure 2K ▶ ).
FISH metaphases with dissociated YAC 744 and FGF3 signals were found in five of seven cases with split signals in interphase nuclei (cases 26, 34, 39, 47, and 48). In particular, case no. 34 showed a duplication of the reciprocal derivative chromosome, which accounts for the third green (FGF3) signal observed in interphase nuclei (Figure 2 ▶ , E and F). In case no. 47, dissociated red (YAC 744) and green (FGF3) signals were detected on one unidentified putative derivative chromosome, whereas a red signal was still present on the putative der (11) chromosome (Figure 2 ▶ I and J); these findings may suggest the occurrence in this case of a breakpoint within YAC 744 with the split of this region being followed by a complex chromosomal rearrangement. In case no. 39, the putative derivative chromosome containing the FGF3 signal showed a large amount of extra telomeric material, again suggesting the occurrence of a complex rearrangement (Figure 2H) ▶ . The metaphases with dissociated signals in case no. 48 (data not shown) had a pattern apparently similar to that shown in Figure 2D ▶ for case no. 26. Finally, metaphases with three copies of chromosome 11 were found in five of the 13 cases with trisomy detected in interphase nuclei (see representative case in Figure 2L ▶ ).
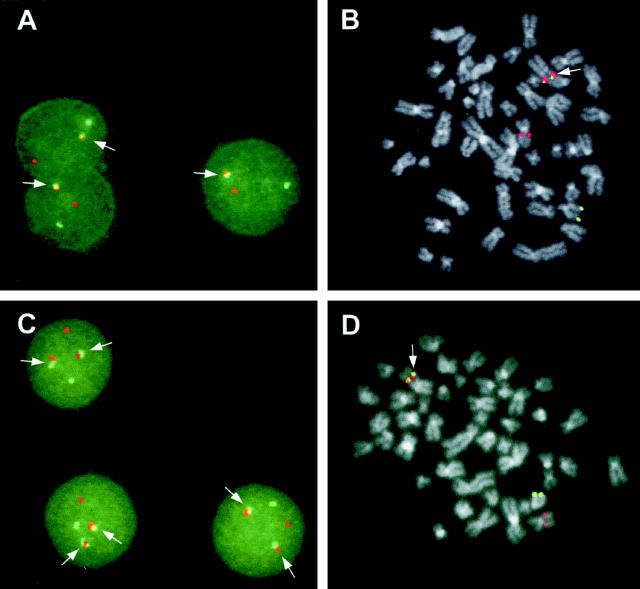
To determine whether these breakpoints were the result of a t(11;14)(q13;q32) chromosomal translocation, we performed double-color FISH using the FGF3 clone (red signal) and a pool of the constant IGH regions (green signal). In all of the seven cases with evidence of 11q13 breakpoints, we found associated red and green signals in the interphase nuclei (see Figure 3A ▶ for representative case). In case no. 34 (Figure 3C) ▶ , the interphase nuclei showed the presence of two series of colocalized green (IGH) and red (FGF3) signals. The IGH-FGF3 colocalization was confirmed in four cases (nos. 26, 39, 47, and 48) on metaphases (see Figure 3 ▶ , B and D; data not shown).
Figure 3.
FISH analysis of the t(11;14) translocation in MM with 11q13 breakpoints. A and B: Interphase and metaphase FISH in case no. 39. C: Interphase FISH in case no. 34. D: Metaphase FISH in case no. 47. Red signal (FGF3); green signal (IGH-constant μ, γ1–4, α1–2 regions). The arrows indicate the FGF3-IGH association on the putative 14 (der).
The t(11;14) translocation was detected in 1/18 (5.5%) cases in clinical stage I, 1/5 (20%) in stage II, 3/10 (30%) in stage III, and 2/6 (33%) in relapse. Trisomy 11 was found in 4/18 (22%) in stage I, 2/5 (40%) in stage II, 4/10 (40%) in stage III, and 3/6 (50%) in relapse. Finally, our combined analysis showed cyclin D1 expression in all of the seven cases bearing the t(11;14) translocation, in two (15%) of the 13 cases with trisomy 11 and in one (5%) of the 19 cases with no apparent abnormalities of chromosome 11 (P < 0.001).
Clinicopathological Parameters and Cyclin D1 Expression
On the basis of the correlation between cyclin D1 overexpression and t(11;14) translocation, we attempted a preliminary analysis of the relationship between cyclin D1 positivity and a series of clinicopathological variables available in our series. Cyclin D1 was expressed in 9 of the 40 MMs at diagnosis (22.5%) and in 3 of the 8 MM at relapse (37.5%; P = 0.654). In the patients at diagnosis, cyclin D1 positivity was more frequent in advanced than in early clinical stages: two of the 23 cases in stage I (8.7%); two of the six cases in stage II (33.3%); and five of the 11 cases in stage III (45.4%; P = 0.044). Moreover, cyclin D1 expression was more frequent in cases with clinical symptoms (8/22; 36.4%) than in those without (1/18; 5.5%; P = 0.026) and progressively increased with the extent of bone marrow infiltration, being detectable in two (10%) of the 20 cases in histological stage I (<20% bone marrow infiltration), two (18.2%) of the 11 cases in stage II (20 to 50% infiltration), and five (55.5%) of the nine cases in stage III (more than 50% infiltration; P = 0.022). No significant association was found in our series between cyclin D1 expression and histological grade (P = 0.399), although it was more frequently detected in cases of low grade (8/29; 27.6%) than in those of intermediate and high grade malignancy (1/11; 9.1%). Finally, no significant association was found between cyclin D1 expression and the variables of age, gender, LDH, or β2 microglobulin serum levels.
Discussion
In the present study we comprehensively investigated the involvement of the cyclin D1 gene in MM. To this end, the overexpression of cyclin D1 in malignant plasma cells was evaluated by means of immunohistochemical analysis of bone marrow biopsies, and then correlated with the presence of 11q13 breakpoints detected by means of double-color interphase FISH. To the best of our knowledge, this is the first report addressing this issue in MM patients. We found that the overexpression of cyclin D1 closely correlated with the presence of the t(11;14) and appeared to be associated with advanced clinical stages. These results may contribute toward improving our understanding of the pathogenesis and clinical evaluation of MM.
Breakpoints at 11q13 were detected in seven (18%) of the 39 cases investigated using interphase FISH; metaphases with dissociated signals were detected in only five of the positive cases. All of the 11q13 breakpoints were the result of a t(11;14) chromosomal translocation. The incidence of t(11;14) found in this study is similar to that (17%) recently reported by Avet-Loiseau et al, 7 who based their FISH analysis on the colocalization of the IGH and 11q13 probes; these data therefore indicate that the t(11;14) occurs in approximately 20% of primary MM tumors. A further finding of our FISH analyses was the presence of trisomy 11 in 13 (33%) of the 39 investigated cases. Like two previous studies in which trisomy 11 was detected by means of interphase FISH in, respectively, 50% and 33% of cases, 35,36 our study indicated that this numerical chromosome abnormality is more frequently associated with patients in advanced clinical stages. Finally, our data further support the use of interphase FISH for the detection of cytogenetic lesions in MM.
Although MM-derived cell lines have provided substantial evidence to suggest that the cyclin D1 gene may be deregulated in MM, 8,9,22,24 information concerning its expression in representative series of primary MM tumors is limited and has not been correlated with the presence of t(11;14) translocation. 25-27 We found cyclin D1 positivity by IHC in 25% of the tumors in our series, a frequency similar to the 26% reported by Vasef et al 26 using the same method. Furthermore, we provide evidence that the t(11;14) translocation is strictly associated with gene deregulation; our combined analysis showed cyclin D1 expression in all of the seven cases bearing the t(11;14) translocation, in two of the 13 cases with trisomy 11 and in one of the 19 cases without apparent abnormalities of chromosome 11. Concerning the three cases showing cyclin D1 positivity but apparently negative for translocation, the most plausible explanation is that the breakpoints were outside the region investigated by our FISH. Furthermore, structural alterations other than translocation may be involved, as it has recently been reported by us and others that the breakpoint in the U266 MM-derived cell line occurs without any split of the 11q13 region but with the insertion of a part of the IGH locus. 9,37 However, we did not observe any colocalization of the IGH-FGF3 probes in further FISH analyses of these three cases (data not shown). Alternatively, it is possible that currently unknown mechanisms may induce cyclin D1 expression in MM; in this regard, it has recently been shown by RT-PCR that cyclin D1 is expressed (albeit at lower levels) in non-Hodgkin’s lymphomas other than MCL and negative for the t(11;14). 38
Overall, our study provides the first evidence that, as in MCL, the immunoreactivity to cyclin D1 can be considered a marker of the presence of t(11;14) in native MMs. Our data confirm previous evidence that normal plasma cells and hematopoietic precursors in bone marrow do not react to cyclin D1 antibody, 26,38-40 thus further suggesting the specificity of this analysis. The immunohistochemical analysis of cyclin D1 may therefore be a reliable method of identifying patients with the t(11;14) and, in general, of demonstrating the involvement of this gene in MM. An important factor is that this approach can be adopted easily during routine diagnostic procedures and therefore seems to be superior to the double-color FISH insofar as it generally requires fresh material, cannot be used on archived material, and is beyond the scope of most pathology laboratories for technical reasons. In addition, it can be considered superior to the RT-PCR evaluation of cyclin D1 expression, which requires fresh or frozen material and is sometimes difficult to interpret because of its high degree of sensitivity and the variable percentage of myeloma cells in bone marrow. Taken together, these considerations suggest that IHC is the method of choice for the analysis of cyclin D1 involvement in MM.
Finally, the present study suggests that cyclin D1 overexpression may have prognostic significance in MM. Although we are aware that our series is relatively small, we observed that cyclin D1 expression significantly correlated with the degree of bone marrow involvement in primary tumors, because its incidence progressively increased with the extent of bone marrow infiltration, thus suggesting that it may give in vivo myeloma cells a growth advantage. In keeping with this, our analysis further indicated that cyclin D1 expression was significantly associated with an advanced clinical stage and the presence of clinical symptoms. The association with an advanced clinical stage is in agreement with reports concerning MM cases with t(11;14) and investigated by means of conventional cytogenetics alone, 41,42 but not with those of the FISH studies recently described by Avet-Loiseau et al. 7 Interestingly, a poor prognosis in MM was found to be significantly associated with abnormalities involving chromosome 11q. 43 Further studies involving larger series of patients and an adequate followup are required to demonstrate whether cyclin D1 expression can be considered a prognostic marker in MM.
Footnotes
Address reprint requests to Antonio Neri, M.D., Servizio Ematologia, Istituto di Scienze, Mediche, Università di Milano, Ospedale Maggiore di Milano, IRCCS, Via Francesco Sforza 35, 20122 Milano, Italy. E-mail: filobus@mailserver.unimi.it.
Supported by grants from the Associazione Italiana sul Cancro (to A. N. and M. R.) and from the Ministero Italiano della Sanità (to Ospedale Maggiore IRCCS).
G. P. and S. F. contributed equally to this work.
G. P.’s present address: Department of Pathology and Laboratory Medicine, European Institute of Oncology, Milano, Italy.
References
- 1.Hallek M, Bergsagel PL, Anderson KC: Multiple myeloma: increasing evidence for a multistep transformation process. Blood 1998, 91:3-21 [PMC free article] [PubMed] [Google Scholar]
- 2.Heim S, Mitelman F: Cancer Cytogenetics. 1995. Wiley-Liss, New York
- 3.Dewald GW, Jenkins RB: Cytogenetic studies of patients with monoclonal gammopathies. Wiernik PH Canellos GP Dutcher JP Kyle RA eds. Neoplastic Diseases of the Blood. 1996, :pp 515-523 Churchill Livingstone, New York [Google Scholar]
- 4.Bergsagel PL, Chesi M, Nardini E, Brents LA, Kirby SL, Kuehl WM: Promiscuous translocations into immunoglobulin heavy chain switch regions in multiple myeloma. Proc Natl Acad Sci USA 1996, 93:13931-13936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nishida K, Tamura A, Nakazawa N, Ueda Y, Abe T, Matsuda F, Kashima K, Taniwaki M: The Ig heavy chain gene is frequently involved in chromosomal translocations in multiple myeloma and plasma cell leukemia as detected by in situ hybridization. Blood 1997, 90:526-534 [PubMed] [Google Scholar]
- 6.Richelda R, Ronchetti D, Baldini L, Cro L, Viggiano L, Marzella R, Rocchi M, Otsuki T, Lombardi L, Maiolo AT, Neri A: A novel chromosomal translocation t(4;14)(p16.3;q32) in multiple myeloma involves the fibroblast growth-factor receptor 3 gene. Blood 1997, 90:4062-4069 [PubMed] [Google Scholar]
- 7.Avet-Loiseau H, Li J-Y, Facon T, Brigaudeau C, Morineau N, Maloisel F, Rapp M-J, Talmant P, Trimoreau F, Jaccard A, Harousseau J-L, Bataille R: High incidence of translocations t(11;14)(q13;q32) and t(4;14)(p16;q32) in patients with plasma cell malignancies. Cancer Res 1998, 58:5640-5645 [PubMed] [Google Scholar]
- 8.Chesi M, Bergsagel PL, Brents LA, Smith CM, Gerhard DS, Kuehl WM: Dysregulation of cyclin D1 by translocation into an IgH gamma switch region in two multiple myeloma cell lines. Blood 1996, 88:674-681 [PubMed] [Google Scholar]
- 9.Ronchetti D, Finelli P, Richelda R, Baldini L, Rocchi M, Viggiano L, Cuneo A, Bogni S, Fabris S, Lombardi L, Maiolo AT, Neri A: Molecular analysis of 11q13 breakpoints in multiple myeloma. Blood 1999, 93:1330-1337 [PubMed] [Google Scholar]
- 10.Chesi M, Nardini E, Brents LA, Schroch E, Ried T, Kuehl WM, Bergsagel PL: Frequent translocation t(4;14)(p16.3;q32.3) in multiple myeloma is associated with increased expression and activating mutations of fibroblast growth factor receptor 3. Nat Genet 1997, 16:260-264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Finelli P, Fabris S, Zagano S, Baldini L, Intini D, Nobili L, Lombardi L, Maiolo AT, Neri A: Detection of t(4;14)(p16.3;q32) chromosomal translocation in multiple myeloma by double-color fluorescent in situ hybridization. Blood 1999, 94:724-732 [PubMed] [Google Scholar]
- 12.Chesi M, Nardini E, Lim RSC, Smith KD, Kuehl MW, Bergsagel PL: The t(4;14) translocation in myeloma dysregulates both FGFR3 and a novel gene, MMSET, resulting in IgH/MMSET hybrid transcripts. Blood 1998, 92:3025-3034 [PubMed] [Google Scholar]
- 13.Chesi M, Bergsagel PL, Shonukan OO, Martelli ML, Brents LA, Chen T, Schrock E, Ried T, Kuehl MW: Frequent dysregulation of the c-maf proto-oncogene at 16q23 by translocation to an Ig locus in multiple myeloma. Blood 1998, 91:4457-4463 [PubMed] [Google Scholar]
- 14.Iida S, Rao PH, Butler M, Corradini P, Boccadoro M, Klein B, Chaganti RSK, Dalla-Favera R: Deregulation of MUM1/IRF4 by chromosomal translocation in multiple myeloma. Nat Genet 1997, 17:226-230 [DOI] [PubMed] [Google Scholar]
- 15.Tsujimoto Y, Yunis J, Onorato-Showe L, Erikson J, Nowell PC, Croce CM: Molecular cloning of chromosomal breakpoints of B-cell lymphomas and leukemias with the t(11;14) chromosomal translocation. Science 1984, 224:1403-1406 [DOI] [PubMed] [Google Scholar]
- 16.Williams ME, Meeker TC, Swerdlow SH: Rearrangement of the chromosome 11 bcl-1 locus in centrocytic lymphoma: analysis with multiple breakpoint probes. Blood 1991, 78:493-498 [PubMed] [Google Scholar]
- 17.Coignet LJA, Schuuring E, Kibbelaar RE, Raap TK, Kleiverda KK, Bertheas M-F, Wiegant J, Beverstock G, Kluin PM: Detection of 11q13 rearrangements in hematologic neoplasias by double-color fluorescence in situ hybridization. Blood 1996, 87:1512-1519 [PubMed] [Google Scholar]
- 18.Vaandrager JW, Schuuring E, Zwikstra E, de Boer CJ, Kleiverda K, van Krieken JHJM, Kluin-Nelemans HC, van Ommen GJB, Raap AK, Kluin PM: Direct visualization of dispersed 11q13 chromosomal translocations in mantle cell lymphoma by multicolor DNA fiber fluorescence in situ hybridization. Blood 1996, 88:1177-1182 [PubMed] [Google Scholar]
- 19.Motokura T, Bloom T, Kim HG, Juppner H, Ruderman JV, Kronenberg HM, Arnold A: A novel cyclin encoded by a bcl-1 linked candidate oncogene. Nature 1991, 350:512-515 [DOI] [PubMed] [Google Scholar]
- 20.Seto M, Yamamoto K, Iida S, Akao Y, Utsumi KR, Kubonishi I, Myoshi I, Ohtsuki T, Yawata Y, Namba M, Motokura T, Arnold A, Takahashi T, Ueda R: Gene rearrangement and overexpression of PRAD1 in lymphoid malignancy with t(11;14)(q13;q32) translocation. Oncogene 1992, 7:1401-1406 [PubMed] [Google Scholar]
- 21.DeBoer CJ, Van Krieken JHJM, Kluin-Nelemans JC, Kluin PM, Schuuring E: Cyclin D1 messenger RNA overexpression as a marker for mantle cell lymphoma. Oncogene 1995, 10:1833-1840 [PubMed] [Google Scholar]
- 22.Raynaud SD, Bekri S, Leroux D, Grosgeorge J, Klein B, Bastard C, Gaudray P, Simon M-P: Expanded range of 11 breakpoints with differing patterns of cyclin D1 expression in B-cell malignancies. Genes Chromosomes Cancer 1993, 8:80-87 [DOI] [PubMed] [Google Scholar]
- 23.Fiedler W, Weh HJ, Hossfeld DK: Comparison of chromosome analysis and BCL-1 rearrangement in a series of patients with multiple myeloma. Br J Haematol 1992, 81:58-61 [DOI] [PubMed] [Google Scholar]
- 24.Vaandrager JW, Kluin P, Schuuring E: The t(11;14)(q13;q32) in multiple myeloma cell line KMS12 has its 11q13 breakpoint 330 kb centromeric from the cyclin D1 gene. Blood 1997, 89:349-350 [PubMed] [Google Scholar]
- 25.Zukeberg LR, Yang W, Arnold A, Harris NL: Cyclin D1 expression in non-Hodgkin’s lymphomas: detection by immunohistochemistry. Am J Clin Pathol 1995, 103:756-760 [DOI] [PubMed] [Google Scholar]
- 26.Vasef MA, Medeiros LJ, Yospur LS, Sun NCJ, McCourty A, Brynes RK: Cyclin D1 protein in multiple myeloma and plasmacytoma: an immunohistochemical study using fixed, paraffin-embedded tissue sections. Mod Pathol 1997, 10:927-932 [PubMed] [Google Scholar]
- 27.Sonoki T, Hata H, Kuribayashi N, Yoshida M, Harada N, Nagasaki A, Kimura T, Matsuno F, Mitsuya H, Matsuzaki H: Expression of PRAD1/cyclin D1 in plasma cell malignancy: incidence and prognostic aspects. Br J Haematol 1999, 104:614-617 [DOI] [PubMed] [Google Scholar]
- 28.Durie BGM, Salmon S: A clinical staging system for multiple myeloma. Cancer 1975, 36:842-854 [DOI] [PubMed] [Google Scholar]
- 29.Bartl R, Frisch B, Fateh-Moghadam A, Kettner G, Jaeger K, Sommerfeld W: Histologic classification and staging of multiple myeloma: a retrospective and prospective study of 674 cases. Am J Clin Pathol 1987, 87:342-355 [DOI] [PubMed] [Google Scholar]
- 30.Hsu SM, Raine L, Fanger H: Use of avidin-biotin-peroxidase complex (ABC): a comparison between ABC and unlabeled antibody (PAP) procedures. J Histochem Cytochem 1981, 29:577-580 [DOI] [PubMed] [Google Scholar]
- 31.Fracchiolla NS, Pruneri G, Pignataro L, Carboni N, Capaccio P, Boletini A, Buffa R, Neri A: Molecular and immunohistochemical analysis of the bcl-1/cyclin D1 gene in laryngeal squamous cell carcinomas. Cancer 1997, 79:1114-1121 [PubMed] [Google Scholar]
- 32.Lichter P, Tang Chang C-J, Call K, Hermanson G, Evans GA, Housman D, Ward DC: High resolution mapping of human chromosome 11 by in situ hybridization with cosmid clones. Science 1990, 247:64-69 [DOI] [PubMed] [Google Scholar]
- 33.Courseaux A, Szepetowski P, Fernandes M, Serized C, Kawaguchi Y, Grosgeorge J, Perucca-Lostanlen D, Shows TB, Todd JA, Novak NJ, Gaudray P: Framework Yac contig anchored into a 3.2-Mb high-resolution physical map in proximal 11q13. Genomics 1997, 40:13-23 [DOI] [PubMed] [Google Scholar]
- 34.Bekri S, Adélaide J, Merscher S, Grosgeorge J, Caroli-Bosc F, Perucca-Lostanlen D, Kelley PM, Pébusque M-J, Theillet C, Birnbaum D, Gaudray P: Detailed map of a region commonly amplified at 11q13–q14 in human breast carcinoma. Cytogenet Cell Genet 1997, 79:125-131 [DOI] [PubMed] [Google Scholar]
- 35.Drach J, Schuster J, Nowotny H, Angerler J, Rosenthal F, Fiegl M, Rothermundt C, Gsur A, Jager U, Heinz R, Lechner K, Ludwig H, Huber H: Multiple myeloma: high incidence of chromosomal aneuploidy as detected by interphase fluorescence in situ hybridization. Cancer Res 1995, 55:3854-3859 [PubMed] [Google Scholar]
- 36.Tabernero D, San Miguel JF, Garcia-Sanz R, Najera L, Garcia-Isidoro M, Pérez-Simon JA, Gonzales M, Wiegant J, Raap AK, Orfao A: Incidence of chromosome numerical changes in multiple myeloma. Fluorescence in situ hybridization analysis using 15 chromosome-specific probes. Am J Pathol 1996, 149:153-161 [PMC free article] [PubMed] [Google Scholar]
- 37.Gabrea A, Bergsagel PL, Chesi M, Shou Y, Kuehl WM: Insertion of excised IgH switch sequences causes overexpression of cyclin D1 in a myeloma tumor cell. Mol Cell 1999, 3:119-123 [DOI] [PubMed] [Google Scholar]
- 38.Aguilera NSI, Bijwaard KE, Duncan B, Krafft AE, Chu W-S, Abbondanzo SL, Lichy JH, Taubenberger JK: Differential expression of cyclin D1 in mantle cell lymphoma and other non-Hodgkin’s lymphomas. Am J Pathol 1998, 153:1969-1976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang W-I, Zukerberg LR, Motokura T, Arnold A, Harris NL: Cyclin D1 (Bcl-1, PRAD1) protein expression in low-grade B-cell lymphomas and reactive hyperplasia. Am J Pathol 1994, 145:86-96 [PMC free article] [PubMed] [Google Scholar]
- 40.Ott MM, Bartkova J, Bartek J, Durr A, Fischer L, Ott G, Muller-Hermelink HK, Kreipe H: Cyclin D1 expression in mantle cell lymphoma is accompanied by downregulation of cyclin D3 and is not related to proliferation activity. Blood 1997, 90:3154-3159 [PubMed] [Google Scholar]
- 41.Fonseca R, Witzig TE, Gertz MA, Kyle RA, Hoyer JD, Jalal SM, Greipp PR: Multiple myeloma and the translocation t(11;14)(q13;q32): a report on 13 cases. Br J Haematol 1998, 101:296-301 [DOI] [PubMed] [Google Scholar]
- 42.Lai JL, Michaux L, Dastugue N, Vasseur F, Daudignon A, Facon T, Bauters F, Zandecki M: Cytogenetics in multiple myeloma: a multicentric study of 24 patients with t(11;14)(q13;q32) or its variant. Cancer Genet Cytogenet 1998, 104:133-138 [DOI] [PubMed] [Google Scholar]
- 43.Tricot GF, Barlogie B, Jagannath S, Bracy D, Mattox S, Vesole DH, Naucke S, Sawyer JR: Poor prognosis in multiple myeloma is associated only with partial or complete deletions of chromosome 13 or abnormalities involving 11q and not with other karyotype abnormalities. Blood 1995, 86:4250-4256 [PubMed] [Google Scholar]