Abstract
Neuronal exocytosis is driven by the formation of SNARE complexes between synaptobrevin 2 on synaptic vesicles and SNAP-25/syntaxin 1 on the plasma membrane. It has remained controversial, however, whether SNAREs are constitutively active or whether they are down-regulated until fusion is triggered. We now show that synaptobrevin in proteoliposomes as well as in purified synaptic vesicles is constitutively active. Potential regulators such as calmodulin or synaptophysin do not affect SNARE activity. Substitution or deletion of residues in the linker connecting the SNARE motif and transmembrane region did not alter the kinetics of SNARE complex assembly or of SNARE-mediated fusion of liposomes. Remarkably, deletion of C-terminal residues of the SNARE motif strongly reduced fusion activity, although the overall stability of the complexes was not affected. We conclude that although complete zippering of the SNARE complex is essential for membrane fusion, the structure of the adjacent linker domain is less critical, suggesting that complete SNARE complex assembly not only connects membranes but also drives fusion.
INTRODUCTION
Communication between neurons is mediated by neurotransmitters that are released from presynaptic nerve endings by Ca2+-dependent exocytosis of synaptic vesicles. Exocytotic fusion of the vesicle with the synaptic plasma membrane is mediated by the proteins synaptobrevin 2/VAMP2, SNAP-25, and syntaxin 1 (Jahn and Scheller, 2006; Rizo et al., 2006). These proteins are members of the SNARE protein family that are involved in all fusion events of the secretory pathway. SNAREs are characterized by stretches of 60-70 amino acids arranged in heptad repeats, termed SNARE motifs (Weimbs et al., 1997; Fasshauer et al., 1998b; Bock et al., 2001; Day et al., 2006). Syntaxin and synaptobrevin each contain a single SNARE motif that is located adjacent to a C-terminal transmembrane domain. In contrast, SNAP-25 contains two SNARE motifs connected by a palmitoylated linker region that serves as membrane anchor.
Synaptobrevin resides in synaptic vesicles, whereas SNAP-25 and syntaxin 1 reside in the plasma membrane. The SNARE motifs of syntaxin, SNAP-25, and synaptobrevin readily assemble into quarternary bundles of α-helices (Fasshauer et al., 1997; Sutton et al., 1998). Assembly would thus lead to a tight connection between the membranes. According to this view, assembly is nucleated at the N-terminal ends of the SNARE-motifs and proceeds toward the C-terminal membrane anchor (“zippering”), resulting in a strained “trans”-complex (Hanson et al., 1997). During membrane merger, the trans-complex would relax into a “cis”-complex in which the transmembrane domains are aligned in parallel. To regenerate the SNAREs for another round of fusion, SNARE complexes need to be disassembled by the AAA+-ATPase NEM-sensitive factor (NSF) in conjunction with cofactors termed soluble NSF attachment proteins (SNAPs; Sollner et al., 1993).
Although the “zippering” hypothesis of SNARE function has received a lot of experimental support, it is still unclear how the activity of SNAREs is regulated. In vitro, assembly of SNAREs is essentially irreversible (Fasshauer et al., 2002), suggesting that assembly is associated with a large release of free energy that is used to overcome the fusion barrier. Thus, careful control of this reaction is needed to ensure that membrane fusion occurs only at a defined intracellular location with defined kinetics. Indeed, a variety of in vitro experiments suggest that the reactivity of synaptobrevin is controlled by direct interaction with accessory proteins. Using isolated synaptic vesicles, it was reported that vesicular synaptobrevin is not reactive but requires Ca2+ to interact with syntaxin and SNAP-25, suggesting regulation by a Ca2+-binding protein such as synaptotagmin (Hu et al., 2002). These data were corroborated by observations suggesting that synaptobrevin, once inserted in liposomes, does not bind to its SNARE partners (Hu et al., 2002; Kweon et al., 2003b). The lack of reactivity was attributed to the membrane proximal linker region of synaptobrevin (aa 85–92, Figure 1), which connects the SNARE motif with the C-terminal transmembrane domain. Spin-labeling experiments suggested this region to form an amphipatic helix that is tilted at an angle of 33°, with two highly conserved tryptophan residues (Trp89 and Trp90) dipping into the hydrophobic core of the bilayer (Kweon et al., 2003a). When these Trp-residues were replaced with serine, SNARE binding was restored (Kweon et al., 2003b). It was proposed that the linker region serves to down-regulate synaptobrevin and that an activation step is needed before fusion.
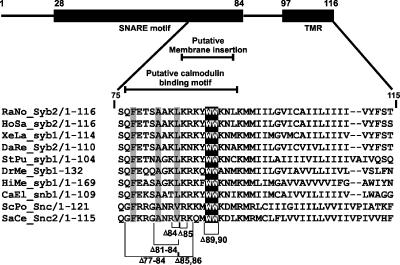
Figure 1.
Schematic drawing of the domain arrangement of synaptobrevin, highlighting the C-terminal end. The SNARE motif (black box) encompasses the region from amino acid (aa) 28 till aa 84 (layer + 8), followed by a short linker (aa 85-96) and a transmembrane region (aa 97-116). The alignment shows that the membrane-proximal tryptophan residues (white letters/black background) are highly conserved across species, irrespective of the proteins being involved in constitutive or regulated exocytosis. Residues that have been deleted in experiments are indicated by brackets. Layers 6, 7, and 8 are highlighted. For the alignment secretory R-SNAREs from the following species were included: Schizosaccharomyces pombe, ScPo; Saccharomyces cerevisiae, SaCe; Hirudo medicinalis, HiMe; Caenorhabditis elegans, CaEl; Strongylocentrotus purpuratus, StPu; Danio rerio, DaRe; Rattus norvegicus, RaNo; Homo sapiens, HoSa; Xenopus laevis, XeLa; Drosophila melanogaster, DrMe.
Several proteins have been invoked in regulating synaptobrevin. For instance, calmodulin was reported to bind to the C-terminal region of synaptobrevin (residues 77-90) in a calcium-dependent manner (Quetglas et al., 2002). Calmodulin was shown to compete with membrane binding of this region, and it was proposed that calmodulin binding inhibits rather than activates synaptobrevin (de Haro et al., 2004). Moreover, synaptobrevin is associated with synaptophysin, a major multispanning membrane protein of synaptic vesicles (Calakos and Scheller, 1994; Edelmann et al., 1995; Washbourne et al., 1995). This interaction appears to be mediated primarily by the transmembrane region of synaptobrevin (Edelmann et al., 1995; Yelamanchili et al., 2005). Interaction of synaptobrevin with synaptophysin and syntaxin 1/SNAP-25 is mutually exclusive (Edelmann et al., 1995), suggesting that release of synaptobrevin from synaptophysin may constitute an activation step.
Together, these reports suggest that the reactivity of synaptobrevin is controlled by protein and lipid interactions of its C-terminal region. However, recent experiments on fusion of synaptobrevin-containing liposomes are difficult to reconcile with the view of an intrinsic inactivation of the protein. Although fusion of synaptobrevin-containing liposomes with liposomes containing SNAP-25 and syntaxin is slow (Weber et al., 1998; Schuette et al., 2004; Tucker et al., 2004), the fusion rate is accelerated dramatically when the syntaxin/SNAP-25 binding site for synaptobrevin is stabilized (Pobbati et al., 2006). Thus it appears that the availability of the acceptor site rather than the intrinsic activity of membrane-anchored synaptobrevin is rate-limiting.
In the present study, we have used complementary approaches for probing the reactivity of membrane-anchored synaptobrevin, focusing on the role of its C-terminal region. In particular, we investigated how the interactions with potential regulators including synaptophysin, calmodulin and phospholipid membranes influence synaptobrevin's reactivity. Furthermore, we analyzed the role of the SNARE motif and the linker region between the SNARE motif and the transmembrane domain in SNARE binding and membrane fusion. Our results have important implications in understanding the structure-function relationship of SNARE proteins.
MATERIALS AND METHODS
Antibodies
For immunoprecipitation, the mAb used against synaptobrevin 2 was cl 69.1 (Edelmann et al., 1995) and that used against synaptophysin was cl 7.2 (Jahn et al., 1985). Immunoblots were analyzed using the above antibodies and a rabbit serum against synaptophysin (Jahn et al., 1985) at a 1:1000 dilution.
Protein Constructs
All recombinant proteins were derived from cDNAs encoding for rat proteins and subcloned into the pET28a vector (Novagen, Schwalbach, Germany), which encodes for an amino-terminal His6-tag. SNAP-25 no cysteine, synaptobrevin 1-96, syntaxin (SyxH3, residues 180-262; Fasshauer et al., 1998a), SyxH3 C225, Syx C197, SNAP-25 C84, SNAP-2 C130 (Margittai et al., 2001), full-length synaptobrevin (residues 1-116), SyxH3 with the transmembrane region (residues 183-288; Margittai et al., 1999), and synaptobrevin 49-96 (Pobbati et al., 2006) have been described previously. Single cysteines in the cytsoplasmic portions were introduced at positions 28, 61, and 79 of full-length synaptobrevin by site-directed mutagenesis. Variants of full-length synaptobrevin and of its respective single cysteine mutants in which tryptophan residues at positions 89 and 90 were mutagenized to serine residues were generated (SybW89SW90S). Versions of full-length synaptobrevin carrying small deletions in the C-terminal region of the SNARE motif (SybΔ84, SybΔ81-84, SybΔ77-84) or the linker region between the SNARE motif and the transmembrane domain (SybΔ85, SybΔ85,86, SybΔ89,90) were generated (see Figure 1 for overview). Sequences were checked by DNA sequencing. TeNT light chain was a gift from H. Niemann (Medizinische Hochschule, Hannover, Germany).
Protein Expression and Purification
All proteins were expressed in the Escherichia coli strain BL23 (DE3) and purified by Ni2+-nitrilotriacetic acid affinity chromatography. After elution, His6-tags were removed using thrombin during overnight dialysis in phosphate-buffered saline (PBS, 20 mM Na2HPO4, pH7.4, 150 mM NaCl) or standard buffer (20 mM Tris, pH 7.4, 50 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol). The ternary complex containing SyxH3 with TMR, SNAP-25, and Syb49–96 was purified in the presence of 15 mM CHAPS. Proteins without a transmembrane region were further purified by ion exchange chromatography using a Mono Q or Mono S column on the Ákta system FPLC (GE Healthcare, Waukesha, WI) as described before (Fasshauer et al., 1998a; Margittai et al., 1999, 2001). Calmodulin was purified through hydrophobic interaction chromatography by the use of a phenylsepharose column, after adjusting the calcium concentration to 15 mM. After washing the column with a high-salt containing buffer (0.5 M NaCl), calmodulin was eluted from the column with a buffer containing 5 mM EDTA.
Liposomes Reconstitution
Lipids (Avanti Polar Lipids, Birmingham, AL) phosphatidylcholine, phosphatidyl-ethanolamine, phosphatidylserine, phosphatidylinositol, and cholesterol were mixed in molar ratio 5:2:1:1:1 under argon. It should be noted that this lipid composition is similar to the natural composition reported for synaptic vesicles (Nagy et al., 1976; Takamori et al., 2006). After drying, the lipid mix was resuspended in PBS or HB100 (25 mM HEPES, pH 7.4, 100 mM KCl) containing 5% (wt/vol) sodium cholate at a total lipid concentration of 13.5 mM. Proteins in 1.5% sodium cholate was added to the lipid mix at a lipid-to-protein ratio of 100:1 (100 μl lipid mix and 15 nmol of protein). The protein-lipid mix was incubated at 4°C for 30 min followed by size exclusion chromatography on Sephadex G-50 (superfine) or PC 3.2/10 Fast Desalting column (GE Healthcare) equilibrated in PBS or HB100. For the removal of unincorporated proteins, 500 μl of the liposome fraction was mixed with an equal volume of 80% Nycodenz in PBS, overlaid with 500 μl of 30% Nycodenz and 150 μl of PBS. The gradient was centrifuged at 165,000 × g for 4 h. Liposomes were retrieved from the top of the gradient. To determine protein orientation, proteoliposomes were incubated with Tetanus Toxin light chain (synaptobrevin) or BoNT C light chain (SyxH3) at 37°C for 2 h in the presence or absence of CHAPS. The samples were analyzed on 15% SDS-PAGE.
Fluorescence Measurements
Proteins containing a single cysteine were labeled with the sulfhydryl-reactive fluorophores Oregon Green 488 iodoacetamide (OG) or Texas Red C5 bromoacetamide (TR; Invitrogen, Carlsbad, CA). For labeling, proteins were incubated with 10-fold excess of the fluorophore for 2 h. The reaction was stopped with 10 mM dithiothreitol for 1 h. Unbound dye was removed by gel filtration on a Sephadex G-25 column followed by extensive dialysis.
Förster resonance energy transfer (FRET) and fluorescence anisotropy experiments were carried out in a Fluoromax-2 spectrometer and Fluorolog-3 (Jobin Yvon, Edison, NJ), respectively. All measurements were carried out at 25°C in 1-cm quartz cuvettes (Hellma, Müllheim, Germany) in PBS. FRET measurements were done by excitation at 488 nm and monitoring donor (OG) fluorescence emission (520 nm) and acceptor (TR) fluorescence emission (610 nm). The slit widths were set to 1–2 nm, and the integration time was set to 1 s.
Fluorescence anisotropy was measured using proteins labeled with Oregon Green using a slit widths of 3 nm. The G factor was calculated according to G = IHV/IHH, where I is the fluorescence intensity, and the first subscript letter indicates the direction of the exciting light and the second subscript letter the direction of emitted light. The intensities of the vertically (V) and horizontally (H) polarized emission light after excitation by vertically polarized light were measured. The anisotropy (r) was determined according to r = (IVV − G IVH)/(IVV + 2 G IVH).
SNARE-mediated Lipid-mixing Assay
Liposome fusion reactions were performed at 30°C, essentially as described in Weber et al. (1998). For reactions 2 μl of labeled and 5 μl unlabeled liposomes were mixed in a total volume of 30 μl, resulting in final protein concentrations of approx. 1 μM SyxH3 and 2.5 μM Syb. The reaction was started by the addition of ≈10 μM SNAP25. Fusion between preassembled SyxH3, SNAP-25, and Syb49-96 in proteoliposomes (final concentration 200 nM) and Syb proteoliposomes (final concentration 200 nM) was carried out in a total volume of 1.2 ml. Fluorescence dequenching was measured using 460 nm for excitation and 538 nm for emission. Fluorescence intensities were normalized to the initial fluorescence intensity.
Preparation of Synaptic Vesicles
Synaptic vesicles were prepared from rat brains by a series of differential centrifugation steps, density gradient centrifugation, and control pore glass (CPG) chromatography as described previously (Huttner et al., 1983; Day et al., 2006; Takamori et al., 2006). It is notable that synaptic vesicles prepared by this method are fully functional (Takamori et al., 2000a,b). If indicated, an enriched but less pure fraction (lysate pellet 2, LP2) was used.
Immunoprecipitation
LP2 (preincubated or not with soluble syntaxin and SNAP-25 or for 1.5 h) was dissolved in ice-cold extraction buffer (20 mM HEPES-KOH, pH 7.3, 140 mM KCl, 2 mM EDTA, 1–1.5% Triton X-100). The protein amount was adjusted to 1–1.5 mg/ml, and the insoluble material was removed by centrifugation for 30 min at 100,000 × g. Ascites fluid, 7.5 μl, corresponding to ∼8–25 μg specific IgG per ml solution was added, and incubation was carried out for 8–10 h at 4°C. Protein G-Sepharose suspension, 75 μl (GE Healthcare), was added and incubated for 1–1.5 h. The beads were collected by centrifugation and washed three times in extraction buffer before elution with sample buffer containing SDS and β-mercaptoethanol.
Cross-linking Experiment
For cross-linking, the LP2 fraction was resuspended at a protein concentration of 1–1.5 mg/ml in Krebs-Ringer buffer (1.2 mM Na2HPO4, 5 mM NaHCO3, 140 mM NaCl, 1 mM MgCl2,) and prewarmed for 10 min at room temperature (RT). Disuccinimidyl suberate (DSS) was dissolved in DMSO at a final concentration of 5 mM. Cross-linking was carried out at RT for 45 min. The reaction was quenched with Tris-Cl, pH 7.4 (100 mM final concentration), for 30 min at RT. The membranes were pelleted in a microfuge at 5000 rpm for 3 min and then resuspended in 1 ml ice-cold 20 mM Tris-Cl, pH 7.4, 150 mM NaCl, containing 1% Triton X-100, and incubated at 4°C for 1 h. Insoluble material was removed by centrifugation at 40,000 rpm (100,000 × g) for 20 min in TLA 55 rotor. Aliquots, 20 μl, of the supernatants were analyzed by SDS-PAGE and immunoblotting.
Kinetic Simulation and Global Fitting
The experimental data were donor fluorescence intensity measured at two wavelength in dependence of time and the concentration of the SNAP 25 ligand. We used ProKineticistII (Applied Photophysics, Leatherhead, Surrey, United Kingdom) to model the binding kinetics in dependence of five rate constants (Supplementary Figure S4, top). This software simulates the corresponding differential equation system using the Newton-Raphson method and numerical integration and minimizes the sum of squares in the residuals by iterative updates of the rates based on the derivatives of the residuals matrix. The stated rate constants resulted from fitting in a global fitting mode (only one set of rate constants to fit all SNAP 25 concentrations).
Other Methods
SDS-PAGE was performed according to Laemmli (1970) and stained with Coomassie Blue. Protein determination was done according to Bradford (1976).
RESULTS
The SNARE Motif of Synaptobrevin But Not the Linker Region Determines Fusion Activity
To investigate whether membrane-bound synaptobrevin is capable of forming SNARE complexes, we reconstituted full-length synaptobrevin in liposomes and added its soluble SNARE partners, full-length SNAP-25 and the SNARE motif of syntaxin 1a (SyxH3). About 50% of the synaptobrevin engaged in SNARE interactions, forming an SDS-resistant band (Figure 2A). The remaining free synaptobrevin was resistant to proteolysis by the light chain of tetanus toxin (which only cleaves free synaptobrevin), suggesting that this pool is inaccessible and thus respresents inversely oriented protein. Thus, the entire pool of surface-exposed synaptobrevin was recruited into SNARE complexes. As typical for SNARE complexes, they dissociated into their individual components when heated to 95°C (Supplementary Figure S1). Similar results were obtained when the entire cytoplasmic region of syntaxin was used (Supplementary Figure S2).
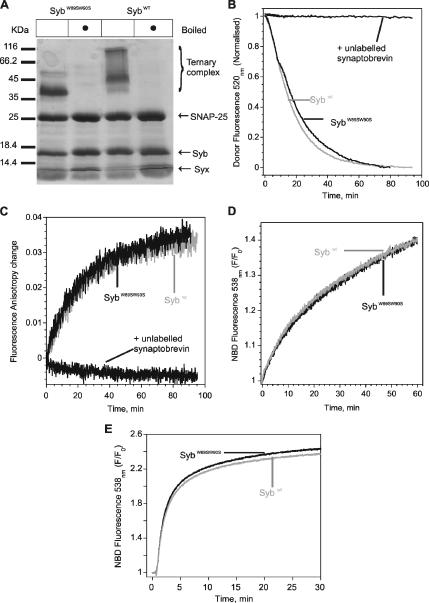
Figure 2.
Substitution of the membrane proximal tryptophans 89 and 90 with serine does not alter the efficiency of synaptobrevin to enter SNARE complexes or mediate fusion of liposomes. (A) Complex formation, monitored by the appearance of heat-resistant bands after SDS-PAGE and Coomassie Blue staining. Liposomes were reconstituted with either mutant or wild-type synaptobrevin (with approximately 200, 100, and 200 pmol of synaptobrevin, SyxH3 and SNAP-25, respectively). Note that the complex containing SynaptobrevinW89S W90S migrates somewhat faster in SDS-PAGE, probably indicating a difference in the amount of bound SDS. (B) Complex formation, monitored by FRET. Synaptobrevin (both wild-type and W89S W90S) labeled at position 61 with Oregon Green was reconstituted in liposomes (∼100 nM final conc.) and incubated with SyxH3 labeled at position 225 with Texas Red (SyxH3225TR; final conc. ∼300 nM). On addition of SNAP-25 (1.28 μM) donor fluorescence decreased, indicating complex formation. Addition of SNAP-25 in the absence of SyxH3 does not cause a measurable change in the FRET signal (not shown). Addition of soluble unlabeled synaptobrevin (2.5 μM) effectively competed with the labeled synaptobrevin for complex formation. For normalization, the minimum value of each trace was subtracted from its respective trace, and every data point of the trace was divided by its starting value. Similar results were obtained with the FRET pairs Syb28OG/SyxH3197TR, Syb28OG/SNAP-25130TR, and Syb79OG/SNAP-2584TR (data not shown). (C) Complex formation monitored by fluorescence anisotropy, using Syb61OG (both wild-type and W89S W90S) containing liposomes as in B. Anisotropy increased when unlabeled soluble syntaxin and SNAP-25 were added. Again, excess soluble synaptobrevin (5 μM) blocks the increase in anisotropy. SNAP-25, alone, does not show any measurable increase in anisotropy (not shown). For normalization, the minimum value of each trace was subtracted from its respective trace. (D) SNARE-mediated lipid mixing monitored by the lipid-dequenching assay as previously described (Schuette et al., 2004). Liposomes reconstituted with both SybW89S W90S fused as effectively as Sybwt with syntaxin 1 liposomes in the presence of SNAP-25. The fusion efficiency was not different when syntaxin 1/SNAP-25 binary complex was stabilized on the liposome (not shown). [NBD: N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl]. (E) SNARE mediates lipid mixing monitored by the lipid-dequenching assay when the SyxH3/SNAP-25 complex was stabilized by Syb49–96. Note that SybW89S W90S and Sybwt reconstituted liposomes fused with equal efficiency.
As stated in the Introduction, synaptobrevin contains two highly conserved tryptophan residues in the linker domain connecting the C-terminal end of the SNARE motif with the transmembrane domain (Figure 1). These tryptophans were previously reported to cause membrane binding of the linker domain and as a consequence, synaptobrevin was proposed to be prevented from entering SNARE complexes (Kweon et al., 2003a,b), but binding activity was restored when the tryptophans were replaced by serines (Kweon et al., 2003b). We therefore tested whether the reactivity of synaptobrevin containing these point mutations (SybW89S, W90S) was increased. Addition of SNAP-25 and syntaxin to membrane-bound SybW89S, W90S again led to the formation of an SDS-resistant SNARE complex (Figure 2A). To compare the rate of SNARE complex formation of wild-type synaptobrevin (Sybwt) and SybW89S, W90S, we monitored SNARE complex assembly by measuring FRET. For this purpose, single cysteines were introduced into SyxH3 and into the synaptobrevin variants (Sybwt and SybW89S, W90S) at positions 225 and 61, respectively (Margittai et al., 2001). Synaptobrevin was labeled with OG as donor (Syb61OG), and SyxH3 with TR as acceptor fluorophore (SyxH3225TR). In the SNARE complex these positions are adjacent to each other on the outside of the four-helix bundle (Sutton et al., 1998). The labeled synaptobrevin variants were reconstituted into liposomes and then mixed with soluble SyxH3225TR. On addition of SNAP-25, donor quenching due to FRET was observed, indicating complex formation. No difference in the rates of complex formation was observed between Sybwt and SybW89S, W90S (Figure 2B).
Complex formation was also monitored by fluorescence anisotropy of Syb61OG, reporting local conformational flexibility of the dye. Again, similar rates for ternary complex formation were observed for reconstituted Sybwt and SybW89S, W90S (Figure 2C). Thus, the exchange of the two C-terminal tryptophan residues did not alter synaptobrevin's capacity to engage in SNARE complexes. As reported for the soluble form of synaptobrevin (Fasshauer and Margittai, 2004), the reaction rate was accelerated by increasing the concentration of SNAP-25 (Supplementary Figure S4). Conversely, the rate is reduced when the entire cytoplasmic region of syntaxin is used (Supplementary Figure S2), in agreement with the notion that the N-terminal regulatory Habc-domain slows down SNARE assembly (Margittai et al., 2003). Furthermore, addition of detergent to synaptobrevin-containing liposomes led to only moderate acceleration of the reaction rate (Supplementary Figure S5), suggesting that there is no significant difference in reactivity regardless of whether synaptobrevin is anchored to bilayers or inserted in detergent micelles.
Next we tested whether the point mutations alter the ability of reconstituted synaptobrevin to mediate fusion with liposomes containing syntaxin and SNAP-25. Liposomes containing Sybwt or SybW89S, W90S were combined with liposomes containing truncated syntaxin (Syx183-288, encompassing the SNARE motif and the transmembrane domain). Fusion was initiated by addition of soluble SNAP-25, and lipid mixing was monitored with a standard fluorescence-dequenching assay (Struck et al., 1981; Schuette et al., 2004). Sybwt or SybW89S,W90S containing liposomes fused with similar efficiency (Figure 2D). Furthermore, we observed a markedly increased rate of liposome fusion when a preformed acceptor complex was used in which the syntaxin/SNAP-25 dimer was stabilized by a short C-terminal peptide of synaptobrevin, in agreement with our previous report (Pobbati et al., 2006). Yet even under these conditions, liposomes containing Sybwt and SybW89S, W90S fused with equal efficiency and at comparable rates (Figure 2E). Thus, substitution of the conserved tryptophans in the linker region neither affects the ability of membrane-anchored synaptobrevin to form complexes with syntaxin and SNAP-25, nor does it affect the rate of liposome fusion.
To study the role of the linker region in more detail, we created mutants containing small deletions in this region (Δ85, Δ85, 86, and Δ89, 90; see Figure 1 for details) and analyzed their effect on the rates of SNARE assembly and membrane fusion. All deletion mutants were purified as full-length proteins and reconstituted into proteoliposomes. First, we added SNAP-25 and fluorescent syntaxin (SyxH3225TR) and monitored the appearance of an SDS-resistant band over time as a readout for complex formation (Supplementary Figure S6). No significant difference in rate of complex formation was observed between wild type and any of the mutant proteins. Second, we measured the rates of fusion of the liposomes containing the synaptobrevin variants with syntaxin liposomes in the presence of SNAP-25, but again no significant difference was observed (Figure 3A). Thus, not only substitutions but also deletions within the linker region do not cause significant changes in the ability of synaptobrevin to engage in SNARE complexes and to fuse liposomes.
Figure 3.
Effects of small deletions either within the SNARE motif or within the linker between the SNARE motif and the transmembrane region on liposome fusion rates. Fusion was monitored by lipid dequenching as described in Figure 2D. (A) Fusion was unaffected when the residues in the linker (indicated in Figure 1) between the SNARE motif and transmembrane region of synaptobrevin were deleted. (B) Deletion of C-terminal SNARE motif residues (indicated in Figure 1) of synaptobrevin retarded fusion. Traces are representative of at least three independent experiments.
According to the zipper hypothesis, fusion is driven by the progressive assembly of SNARE complexes proceeding from the N-terminus toward the C-terminal membrane anchor. If mutations and small deletions of the linker region did not affect fusion, what about the side chains forming the C-terminal end of the four helix bundle? In the crystal structure of the neuronal SNARE complex, the bundle is stabilized by stacked layers of interacting side chains, with residue 84 of synaptobrevin participating in the most C-terminal (+8) of these layers (Figure 1). We therefore generated synaptobrevin variants with the transmembrane domain and the adjacent linker region (aa 85–116) intact but carrying progressive deletions of the C-terminal part of the SNARE motif: Δ84, Δ81-84, and Δ77-84 (Figure 1). When these mutant proteins were inserted into liposomes, a massive reduction of fusion rates was observed (Figure 3B). In contrast, no difference from wild-type synaptobrevin was observed in the formation of SDS-resistant SNARE complexes (Supplementary Figure S6). Thus, minor perturbation of the C-terminal end of the four-helix bundle reduced fusion, which might best be explained by a reduction in the force exerted on the membrane.
Binding of Ca2+/Calmodulin to Synaptobrevin Does Not Alter Its Fusogenic Properties
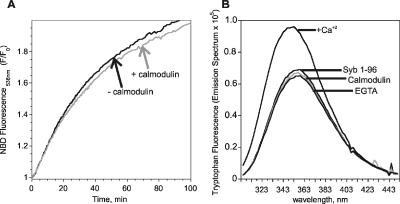
It has been shown previously that the C-terminal portion of the SNARE motif and the adjacent linker region, residues 77-90, constitutes a Ca2+-dependent binding site for calmodulin (Quetglas et al., 2000), raising the possibility that calmodulin controls the activity of synaptobrevin. To investigate this issue, we added Ca2+/calmodulin to a standard liposome fusion reaction, but no change of fusion rates was observed (Figure 4A). We therefore tested whether calmodulin does indeed bind to synaptobrevin in a calcium-dependent manner, and if so, whether binding was preserved when synaptobrevin is reconstituted into liposomes. When calmodulin and synaptobrevin (lacking the transmembrane domain) were mixed, a Ca2+-dependent increase in tryptophan fluorescence emission, associated with a slight blue shift, was observed (Figure 4B), thus confirming the previous findings by Quetglas et al. (2000). In addition, no effect of Ca2+/calmodulin on SNARE complex formation was observed (Supplementary Figure S3). However, no increase in fluorescence was observed when full-length synaptobrevin was used, either in detergent micelles or after reconstitution in liposomes (not shown). Apparently, the binding site for calmodulin is shielded, or alternatively, the conformation is different in the full-length protein, thus challenging a physiological role of calmodulin in regulating synaptobrevin.
Figure 4.
Calmodulin binds to soluble synaptobrevin in a calcium-dependent manner but does not influence SNARE-mediated lipid mixing. Calmodulin binding to synaptobrevin was monitored by fluorescence emission of membrane-proximal tryptophan residues of synaptobrevin. (A) SNARE-mediated lipid mixing assay monitored by the lipid dequenching assay, as in Figure 2D. Ca2+/Calmodulin (1 μM) did not influence the rate of SNARE-mediated lipid mixing. Calcium (100 μM) alone had no effect on fusion (not shown). (B) Ca2+/Calmodulin binds synaptobrevin (Syb1–96). Tryptophan fluorescence emission derived from Syb 1–96 (1 μM) remained unchanged when calmodulin (1 μM) was added. Fluorescence emission increased and was slightly blue-shifted when calcium (100 μM) was added, which was reversed upon addition of EGTA (1 mM).
The Reactivity of Synaptobrevin in Synaptic Vesicles Is Comparable to That of Reconstituted Synaptobrevin
The results described so far show that synaptobrevin retains its ability to form SNARE complexes after insertion into liposomes. However, it is possible that in its native environment, the reactivity of synaptobrevin is changed by interaction with other proteins. We have therefore investigated whether synaptobrevin residing in synaptic vesicles can also engage in SNARE complexes. It needs to be noted that although native synaptic vesicles contain some syntaxin 1 and SNAP-25 molecules (Otto et al., 1997), their average copy number (≈6 for syntaxin 1 and ≈2 for SNAP-25) is much less than that of synaptobrevin (≈70; Takamori et al., 2006). Therefore, the vast majority of synaptobrevin molecules on the synaptic vesicle are not engaged in cis-SNARE complexes. We noted that in synaptic vesicles purified from rat brain, almost the entire pool of synaptobrevin is sensitive to digestion by tetanus toxin light chain (Figure 5A, left panel). Addition of SNAP-25 and syntaxin quantitatively shifted synaptobrevin into an SDS-resistant band, which, as expected for SNARE core complexes, was insensitive to toxin digestion (Figure 5B, right panel). Again, similar results were obtained when syntaxin with the intact N-terminal domain was used (Supplementary Figure S2C). We also tested whether complex formation is accelerated in the presence of calcium ions. Synaptic vesicles were incubated with SNAP-25 and syntaxin for 30 min, i.e., conditions under which complex formation is not yet complete. No difference was observed in the presence of several different calcium concentrations up to 1 mM (Figure 5B).
Figure 5.
SNARE complex formation on synaptic vesicles, monitored by SDS-PAGE and immunoblotting for synaptobrevin. SyxH3 (40 pmol) and SNAP-25 (100 pmol) were incubated with 5.6 μg of purified synaptic vesicles for 4 h in 50 μl volume. The samples were analyzed for the presence of SDS-resistant SNARE complexes. (A) Complex formation on synaptic vesicles resulted in an almost complete shift of synaptobrevin to an SDS-resistant band of higher molecular weight (Ternary complex). Treatment of vesicles with the light chain of tetanus toxin (TeNT) before addition of SNAP-25 and SyxH3 cleaved nearly the entire pool of synaptobrevin (left lanes), documenting that it is completely accessible. (B) Efficiency of SNARE complex formation on synaptic vesicles does not change in the presence of increasing amounts of calcium. Synaptic vesicles were incubated with SyxH3 and SNAP-25 for 30 min in the presence of the indicated Ca2+ concentrations. Note that under these conditions, complex formation is not completed (see also Figure 6); thus the differences in calcium concentrations also did not alter the assembly rate.
Next we asked whether the reactivity of synaptobrevin in proteoliposomes differs from that in synaptic vesicles. To make the reaction conditions comparable, we matched the amounts of synaptobrevin, using approximately twice as much for the generation of liposomes, in order to account for random orientation of synaptobrevin in liposomes Because endogenous synaptobrevin cannot be labeled, the reaction was again monitored by the appearance of SDS-resistant SNARE complexes. Addition of SNAP-25 and syntaxin to synaptobrevin-containing liposomes or synaptic vesicles, respectively, resulted in complex formation at comparable rates (Figure 6).
Figure 6.
The rates of SNARE complex (TC) assembly on synaptic vesicles and on synaptobrevin liposomes are comparable. Syx225OG and SNAP-25 were incubated with synaptobrevin liposomes or synaptic vesicles (see Figure 5 legend). SNARE complex formation, measured by the appearance of SDS-resistant bands, was monitored by immunoblotting for synaptobrevin (left panels) or by measuring fluorescence derived from syntaxin (right panels). To ensure that the reactions are completely arrested at the end of the incubation, SDS-containing sample buffer was added, and the samples were immediately shock-frozen and thawed only immediately before SDS-PAGE. For quantitation, the intensity of the bands was determined, corrected for background, and plotted against incubation time. LAU, luminescence arbitrary units.
Synaptobrevin Dissociates from Synaptophysin upon SNARE Complex Formation
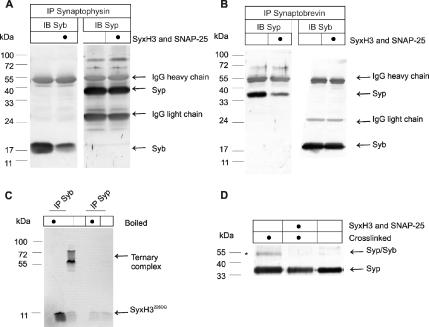
In synaptic vesicles, synaptobrevin is complexed with synaptophysin in a manner that is mutually exclusive with its interaction with SNAREs (Edelmann et al., 1995; Pennuto et al., 2002; Yelamanchili et al., 2005), but it is unclear whether SNARE complex formation leads to a dissociation of synaptobrevin from synaptophysin. Enriched synaptic vesicles were incubated with SNAP-25 and syntaxin, followed by detergent solubilization and immunoprecipitation of either synaptobrevin or synaptophysin. As shown in Figure 7A, addition of the SNAREs caused a massive reduction in the amount of synaptobrevin coprecipitating with synaptophysin. Conversely, upon SNARE addition, a similarly strong reduction was observed in the amount of synaptophysin that coprecipitated with synaptobrevin (Figure 7B). These results indicate that SNARE complex formation effectively dissociates synaptobrevin from synaptophysin. To confirm that synaptophysin does not interact with assembled SNARE complexes, complex formation was carried out using fluorescently labeled syntaxin, followed by solubilization and immunoprecipitation with synaptobrevin- or synaptophysin-specific antibodies, respectively. As shown in Figure 7C, labeled syntaxin is only detectable in the synaptobrevin immunoprecipitates. As an independent readout for the synaptophysin-synaptobrevin complex, which avoids detergent solubilization, we used cross-linking using the bifunctional cross-linker DSS (Edelmann et al., 1995). Cross-linking resulted in the appearance of an additional band of ∼55 kDa that was positive for both synaptophysin (Figure 7D) and synaptobrevin (not shown). Incubation of the vesicles with SNAP-25 and syntaxin before cross-linking prevented the formation of the adduct (Figure 7D).
Figure 7.
Synaptobrevin is displaced from synaptophysin upon formation of SNARE complexes. (A–C) An enriched vesicle fraction (LP2, 50 μg of protein) was incubated or not (as indicated) with 50 μg fluorescently labeled Syx225TR, 200 μg unlabeled SyxH3 and 500 μg SNAP-25 for 2 h, followed by solubilization in Triton X-100 and immunoprecipitation for either synaptophysin (Syp, A) or synaptobrevin (B). All samples were analyzed by SDS-PAGE. LP2 fraction instead of purified synaptic vesicles was used because of the larger yield suitable for this experiment. (A) Immunoblotting for synaptobrevin shows that the amount of synaptobrevin coprecipitating with synaptophysin is reduced in the presence of SyxH3 and SNAP-25. (B) Conversely, immunoblotting for synaptophysin shows that the amount of synaptophysin coprecipitating with synaptobrevin is reduced in the presence of SNAREs. Note that in both cases the efficiency of antigen immunoprecipitation is comparable. (C) SNARE complexes (visualized by fluorescence of Syx225TR) coprecipitated with synaptobrevin but not with synaptophysin. (D) Disappearance of the synaptophysin–synaptobrevin complex in the presence of unlabeled SyxH3 and SNAP-25, monitored by cross-linking with DSS, a bifunctional reagent. In the absence of the SNAREs, cross-linking results in the appearance of a band of ∼55 kDa (*) that is recognized by both synaptophysin- (D) and synaptobrevin-specific (not shown) antibodies and thus represents a heterodimer (Edelmann et al., 1995).
We conclude that the endogenous synaptobrevin of synaptic vesicles is fully active with respect to SNARE complex formation and that it dissociates from synaptophysin when entering SNARE complexes.
DISCUSSION
In the present study we have shown that membrane-anchored synaptobrevin is constitutively active regardless of whether it is reconstituted into proteoliposomes or whether it is in its native environment within synaptic vesicles. Neither association with the proteins calmodulin and synaptophysin nor substitutions in the linker domain had any effect on the ability of synaptobrevin to interact with its SNARE partners and to mediate fusion of proteoliposomes. In contrast, perturbations of the C-terminal end of the SNARE bundle resulted in an impairment of fusion activity. We conclude that although the complete zippering of the helical bundle is essential for fusion, the linker domain appears to be less critical. Possibly, the linker domain merely serves as a force-transducing connection between the helix bundle and the membranes rather than as a separate domain operating by means of specific protein–protein or protein–lipid interactions.
The SNARE-binding properties of synaptobrevin anchored to native or artificial membranes are very similar to those of the soluble SNARE motif of synaptobrevin (Fasshauer and Margittai, 2004; Pobbati et al., 2006). Both the rates of SNARE complex formation and of liposome fusion are solely dependent on the availability of the acceptor site, with so far no evidence for a conformational regulation of synaptobrevin. The binding site for synaptobrevin is formed by a highly unstable dimer of SNAP-25 and syntaxin in a 1:1 stoichiometry, yet is in agreement with our previous reports (Pobbati et al., 2006) that synaptobrevin readily engaged in SNARE interactions when the acceptor site was stabilized. Furthermore, neither calmodulin nor synaptophysin exerted any measurable effect on the rate of SNARE complex formation. Thus, the energy that is stored in these interactions (if any) and that must be overcome during SNARE assembly is relatively small. We cannot exclude, however, that these proteins exert a subtle control of synaptobrevin in a physiological context.
What may be the reasons for the differences between our observations and that of other laboratories that had reported that synaptobrevin in artificial membranes (Kweon et al., 2003b) and in synaptic vesicles (Hu et al., 2002) is largely inhibited? Although the phospholipid composition and protein-lipid ratio of the liposomes used by Shin and colleagues (Kweon et al., 2003b) was somewhat different from the conditions used in our study, we noticed no change when lipid compositions, and other physical parameters, such as temperature, chaotropicity, and dominant counterions in the medium, were altered. Furthermore, their reconstitution procedure was different, using insertion of protein into preformed liposomes instead of simultaneous reconstitution from micellar solutions (Kweon et al., 2003b). Yet, using a similar protocol, we again found no evidence for inactivation of synaptobrevin. We noted, however, that synaptobrevin became refractory to SNARE complex formation after prolonged storage or after repeated freeze–thaw cycles due to the formation of large liposome clusters, thus rendering synaptobrevin inaccessible. In a different study, Davletov and colleagues reported that membrane-anchored synaptobrevin, either reconstituted in liposomes or in purified synaptic vesicles was unable to form complexes with exogenous syntaxin and SNAP-25 unless detergent was added (Hu et al., 2002). Again, we were unable to reproduce these findings because in our hands synaptobrevin both in liposomes and in synaptic vesicles quantitatively formed SNARE complexes. Although it is conceivable that differences in the protein and vesicle purification protocols may account for some of the differences (for instance, Hu et al. used proteins purified by preparative denaturing SDS-PAGE for reconstitution), we have no obvious explanation for these discrepancies. Indeed, several laboratories reported that synaptobrevin-containing liposomes readily fuse with liposomes containing SNAP-25 and syntaxin, a reaction that clearly requires active synaptobrevin (Weber et al., 1998; Schuette et al., 2004; Tucker et al., 2004). Furthermore, clostridial neurotoxins readily cleave membrane-bound synaptobrevin both in liposomes and in synaptic vesicles, with toxin action requiring access to most of the cytoplasmic domain of synaptobrevin (Montecucco et al., 2005). Our data now provide a convenient explanation for these findings. It should be noted that our approach did not allow us to directly assess the conformation of the membrane proximal region of synaptobrevin. A recent study, however, has shown that the transmembrane region of synaptobrevin is tilted at an angle, though it is noteworthy that this study did not propose an interaction of the membrane-proximal region with the membrane (Bowen and Brunger, 2006).
Because the crystal structure of the SNARE complex became available (Sutton et al., 1998), both structure and precise function of the linker region (for which no high resolution structure is available) has been intensely debated (Jahn and Grubmuller, 2002; Rizo et al., 2006). According to the zipper hypothesis, the linker serves to transmit force to the membranes. It is unclear, however, whether it does so as a conformationally flexible rope or whether the connection is stiff (e.g., a contiguous α-helix), thus resulting not only in a pulling but also in a bending force exerted on the membranes. Our data indicate that substitutions of conserved tryptophans and small deletions do not cause any measurable inhibition of SNARE assembly or of liposome fusion. Although these data do not exclude that these tryptophans or other sequence and structural features of the linker are important for function (see e.g., Deak et al., 2006), they clearly rule out a scenario in which these tryptophans control the reactivity of synaptobrevin by membrane insertion as suggested by Kweon et al. (2003b). The tryptophans are conserved between many homologues including the yeast protein Snc2p (Figure 1) for which no tryptophan-based inhibition was observed (Chen et al., 2004). In fact, tryptophan residues are found in many single-spanning membrane proteins near the membrane–water interface where they belong to characteristic belts containing both basic and aromatic residues (Killian and von Heijne, 2000). The positively charged residues are known to contact the phospholipids head groups, whereas the polar-aromatic residues penetrate into a region near the lipid carbonyl chain. The function of the polar-aromatic residues is not entirely understood, but it has been suggested that they maintain the vertical position of the transmembrane helix relative to the membrane–water interface (Ridder et al., 2000). Because synaptobrevin and syntaxin represent a class of proteins called “tail-anchored” proteins that are inserted into the membrane posttranslationally (Kutay et al., 1993, 1995), it is conceivable that the membrane proximal region of synaptobrevin may be critical for its correct localization, as has recently been shown for syntaxin (Ge et al., 2006).
In contrast to mutations in the linker, deletions of amino acids involved in the most C-terminal interacting layers of the SNARE complex led to an impairment of fusion. Presently, we cannot exclude that impairment of the C-terminal layer results in a destabilization of the penultimate layers and thus of the entire complex, although no difference in SDS resistance was observed. Whether or not this is the case, our findings strongly support the zipper hypothesis of SNARE-mediated membrane fusion. Progressive assembly of the SNARE complex is expected to result in a progressively increasing strain on the membrane (trans-complex intermediate). The fact that perturbation of the most C-terminal layer (which is expected to bear the highest strain) reduces fusion supports the view that SNARE assembly not only connects membranes but also drives fusion itself by coupling the mechanical energy of SNARE complex formation to the merger of the bilayers.
Supplementary Material
ACKNOWLEDGMENTS
We thank A. Pobbati (Max-Planck-Institute for Biophysical Chemistry) for providing calmodulin. This work was supported by National Institutes of Health Grant P01 GM072694.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-01-0049) on March 14, 2007.
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Bock J. B., Matern H. T., Peden A. A., Scheller R. H. A genomic perspective on membrane compartment organization. Nature. 2001;409:839–841. doi: 10.1038/35057024. [DOI] [PubMed] [Google Scholar]
- Bowen M., Brunger A. T. Conformation of the synaptobrevin transmembrane domain. Proc. Natl. Acad. Sci. USA. 2006;103:8378–8383. doi: 10.1073/pnas.0602644103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Calakos N., Scheller R. H. Vesicle-associated membrane protein and synaptophysin are associated on the synaptic vesicle. J. Biol. Chem. 1994;269:24534–24537. [PubMed] [Google Scholar]
- Chen Y., Xu Y., Zhang F., Shin Y. K. Constitutive versus regulated SNARE assembly: a structural basis. EMBO J. 2004;23:681–689. doi: 10.1038/sj.emboj.7600083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day M., et al. Selective elimination of glutamatergic synapses on striatopallidal neurons in Parkinson disease models. Nat. Neurosci. 2006;9:251–259. doi: 10.1038/nn1632. [DOI] [PubMed] [Google Scholar]
- de Haro L., Ferracci G., Opi S., Iborra C., Quetglas S., Miquelis R., Leveque C., Seagar M. Ca2+/calmodulin transfers the membrane-proximal lipid-binding domain of the v-SNARE synaptobrevin from cis to trans bilayers. Proc. Natl. Acad. Sci. USA. 2004;101:1578–1583. doi: 10.1073/pnas.0303274101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deak F., Shin O.-H., Kavalali E. T., Sudhof T. C. Structural determinants of synaptobrevin 2 function in synaptic vesicle fusion. J. Neurosci. 2006;26:6668–6676. doi: 10.1523/JNEUROSCI.5272-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelmann L., Hanson P. I., Chapman E. R., Jahn R. Synaptobrevin binding to synaptophysin: a potential mechanism for controlling the exocytotic fusion machine. EMBO J. 1995;14:224–231. doi: 10.1002/j.1460-2075.1995.tb06995.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasshauer D., Antonin W., Subramaniam V., Jahn R. SNARE assembly and disassembly exhibit a pronounced hysteresis. Nat. Struct. Biol. 2002;9:144–151. doi: 10.1038/nsb750. [DOI] [PubMed] [Google Scholar]
- Fasshauer D., Eliason W. K., Brunger A. T., Jahn R. Identification of a minimal core of the synaptic SNARE complex sufficient for reversible assembly and disassembly. Biochemistry. 1998a;37:10354–10362. doi: 10.1021/bi980542h. [DOI] [PubMed] [Google Scholar]
- Fasshauer D., Margittai M. A transient N-terminal interaction of SNAP-25 and syntaxin nucleates SNARE assembly. J. Biol. Chem. 2004;279:7613–7621. doi: 10.1074/jbc.M312064200. [DOI] [PubMed] [Google Scholar]
- Fasshauer D., Otto H., Eliason W. K., Jahn R., Brunger A. T. Structural changes are associated with soluble N-ethylmaleimide-sensitive fusion protein attachment protein receptor complex formation. J. Biol. Chem. 1997;272:28036–28041. doi: 10.1074/jbc.272.44.28036. [DOI] [PubMed] [Google Scholar]
- Fasshauer D., Sutton R. B., Brunger A. T., Jahn R. Conserved structural features of the synaptic fusion complex: SNARE proteins reclassified as Q- and R-SNAREs. Proc. Natl. Acad. Sci. USA. 1998b;95:15781–15786. doi: 10.1073/pnas.95.26.15781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge W.-P., Yang X.-J., Zhang Z., Wang H.-K., Shen W., Deng Q.-D., Duan S. Long-term potentiation of neuron-glia synapses mediated by Ca2+-permeable AMPA receptors. Science. 2006;312:1533–1537. doi: 10.1126/science.1124669. [DOI] [PubMed] [Google Scholar]
- Hanson P. I., Heuser J. E., Jahn R. Neurotransmitter release—four years of SNARE complexes. Curr. Opin. Neurobiol. 1997;7:310–315. doi: 10.1016/s0959-4388(97)80057-8. [DOI] [PubMed] [Google Scholar]
- Hu K., Carroll J., Fedorovich S., Rickman C., Sukhodub A., Davletov B. Vesicular restriction of synaptobrevin suggests a role for calcium in membrane fusion. Nature. 2002;415:646–650. doi: 10.1038/415646a. [DOI] [PubMed] [Google Scholar]
- Huttner W. B., Schiebler W., Greengard P., De Camilli P. Synapsin I (protein I), a nerve terminal-specific phosphoprotein. III. Its association with synaptic vesicles studied in a highly purified synaptic vesicle preparation. J. Cell Biol. 1983;96:1374–1388. doi: 10.1083/jcb.96.5.1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn R., Grubmuller H. Membrane fusion. Curr. Opin. Cell Biol. 2002;14:488–495. doi: 10.1016/s0955-0674(02)00356-3. [DOI] [PubMed] [Google Scholar]
- Jahn R., Scheller R. H. SNAREs—engines for membrane fusion. Nat. Rev. Mol. Cell Biol. 2006;7:631–643. doi: 10.1038/nrm2002. [DOI] [PubMed] [Google Scholar]
- Jahn R., Schiebler W., Ouimet C., Greengard P. A 38,000-dalton membrane protein (p38) present in synaptic vesicles. Proc. Natl. Acad. Sci. USA. 1985;82:4137–4141. doi: 10.1073/pnas.82.12.4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killian J. A., von Heijne G. How proteins adapt to a membrane-water interface. Trends Biochem. Sci. 2000;25:429–434. doi: 10.1016/s0968-0004(00)01626-1. [DOI] [PubMed] [Google Scholar]
- Kutay U., Ahnert-Hilger G., Hartmann E., Wiedenmann B., Rapoport T. A. Transport route for synaptobrevin via a novel pathway of insertion into the endoplasmic reticulum membrane. EMBO J. 1995;14:217–223. doi: 10.1002/j.1460-2075.1995.tb06994.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutay U., Hartmann E., Rapoport T. A. A class of membrane proteins with a C-terminal anchor. Trends Cell Biol. 1993;3:72–75. doi: 10.1016/0962-8924(93)90066-a. [DOI] [PubMed] [Google Scholar]
- Kweon D. H., Kim C. S., Shin Y. K. Insertion of the membrane proximal region of the neuronal SNARE coiled coil into the membrane. J. Biol. Chem. 2003a;278:12367–12373. doi: 10.1074/jbc.M211123200. [DOI] [PubMed] [Google Scholar]
- Kweon D. H., Kim C. S., Shin Y. K. Regulation of neuronal SNARE assembly by the membrane. Nat. Struct. Biol. 2003b;10:440–447. doi: 10.1038/nsb928. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Margittai M., Fasshauer D., Pabst S., Jahn R., Langen R. Homo- and heterooligomeric SNARE complexes studied by site-directed spin labeling. J. Biol. Chem. 2001;276:13169–13177. doi: 10.1074/jbc.M010653200. [DOI] [PubMed] [Google Scholar]
- Margittai M., Otto H., Jahn R. A stable interaction between syntaxin 1a and synaptobrevin 2 mediated by their transmembrane domains. FEBS Lett. 1999;446:40–44. doi: 10.1016/s0014-5793(99)00028-9. [DOI] [PubMed] [Google Scholar]
- Margittai M., et al. Single-molecule fluorescence resonance energy transfer reveals a dynamic equilibrium between closed and open conformations of syntaxin 1. Proc. Natl. Acad. Sci. USA. 2003;100:15516–15521. doi: 10.1073/pnas.2331232100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montecucco C., Schiavo G., Pantano S. SNARE complexes and neuroexocytosis: how many, how close? Trends Biochem. Sci. 2005;30:367–372. doi: 10.1016/j.tibs.2005.05.002. [DOI] [PubMed] [Google Scholar]
- Nagy A., Baker R. R., Morris S. J., Whittaker V. P. The preparation and characterization of synaptic vesicles of high purity. Brain Res. 1976;109:285–309. doi: 10.1016/0006-8993(76)90531-x. [DOI] [PubMed] [Google Scholar]
- Otto H., Hanson P. I., Jahn R. Assembly and disassembly of a ternary complex of synaptobrevin, syntaxin, and SNAP-25 in the membrane of synaptic vesicles. Proc. Natl. Acad. Sci. USA. 1997;94:6197–6201. doi: 10.1073/pnas.94.12.6197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennuto M., Dunlap D., Contestabile A., Benfenati F., Valtorta F. Fluorescence resonance energy transfer detection of synaptophysin I and vesicle-associated membrane protein 2 interactions during exocytosis from single live synapses. Mol. Biol. Cell. 2002;13:2706–2717. doi: 10.1091/mbc.E02-01-0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pobbati A. V., Stein A., Fasshauer D. N- to C-terminal SNARE complex assembly promotes rapid membrane fusion. Science. 2006;313:673–676. doi: 10.1126/science.1129486. [DOI] [PubMed] [Google Scholar]
- Quetglas S., Iborra C., Sasakawa N., De Haro L., Kumakura K., Sato K., Leveque C., Seagar M. Calmodulin and lipid binding to synaptobrevin regulates calcium-dependent exocytosis. EMBO J. 2002;21:3970–3979. doi: 10.1093/emboj/cdf404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quetglas S., Leveque C., Miquelis R., Sato K., Seagar M. Ca2+-dependent regulation of synaptic SNARE complex assembly via a calmodulin- and phospholipid-binding domain of synaptobrevin. Proc. Natl. Acad. Sci. USA. 2000;97:9695–9700. doi: 10.1073/pnas.97.17.9695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridder A.N.J.A., Morein S., Stam J. G., Kuhn A., de Kruijff B., Killian J. A. Analysis of the role of interfacial tryptophan residues in controlling the topology of membrane proteins. Biochemistry. 2000;39:6521–6528. doi: 10.1021/bi000073v. [DOI] [PubMed] [Google Scholar]
- Rizo J., Chen X., Arac D. Unraveling the mechanisms of synaptotagmin and SNARE function in neurotransmitter release. Trends Cell Biol. 2006;16:339–350. doi: 10.1016/j.tcb.2006.04.006. [DOI] [PubMed] [Google Scholar]
- Schuette C. G., Hatsuzawa K., Margittai M., Stein A., Riedel D., Kuster P., Konig M., Seidel C., Jahn R. Determinants of liposome fusion mediated by synaptic SNARE proteins. Proc. Natl. Acad. Sci. USA. 2004;101:2858–2863. doi: 10.1073/pnas.0400044101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollner T., Bennett M. K., Whiteheart S. W., Scheller R. H., Rothman J. E. A protein assembly-disassembly pathway in vitro that may correspond to sequential steps of synaptic vesicle docking, activation, and fusion. Cell. 1993;75:409–418. doi: 10.1016/0092-8674(93)90376-2. [DOI] [PubMed] [Google Scholar]
- Struck D. K., Hoekstra D., Pagano R. E. Use of resonance energy transfer to monitor membrane fusion. Biochemistry. 1981;20:4093–4099. doi: 10.1021/bi00517a023. [DOI] [PubMed] [Google Scholar]
- Sutton R. B., Fasshauer D., Jahn R., Brunger A. T. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution. Nature. 1998;395:347–353. doi: 10.1038/26412. [DOI] [PubMed] [Google Scholar]
- Takamori S., et al. Molecular anatomy of a trafficking organelle. Cell. 2006;127:831–846. doi: 10.1016/j.cell.2006.10.030. [DOI] [PubMed] [Google Scholar]
- Takamori S., Rhee J. S., Rosenmund C., Jahn R. Identification of a vesicular glutamate transporter that defines a glutamatergic phenotype in neurons. Nature. 2000a;407:189–194. doi: 10.1038/35025070. [DOI] [PubMed] [Google Scholar]
- Takamori S., Riedel D., Jahn R. Immunoisolation of GABA-specific synaptic vesicles defines a functionally distinct subset of synaptic vesicles. J. Neurosci. 2000b;20:4904–4911. doi: 10.1523/JNEUROSCI.20-13-04904.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker W. C., Weber T., Chapman E. R. Reconstitution of Ca2+-regulated membrane fusion by synaptotagmin and SNAREs. Science. 2004;304:435–438. doi: 10.1126/science.1097196. [DOI] [PubMed] [Google Scholar]
- Washbourne P., Schiavo G., Montecucco C. Vesicle-associated membrane protein-2 (synaptobrevin-2) forms a complex with synaptophysin. Biochem. J. 1995;305(Pt 3):721–724. doi: 10.1042/bj3050721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber T., Zemelman B. V., McNew J. A., Westermann B., Gmachl M., Parlati F., Sollner T. H., Rothman J. E. SNAREpins: minimal machinery for membrane fusion. Cell. 1998;92:759–772. doi: 10.1016/s0092-8674(00)81404-x. [DOI] [PubMed] [Google Scholar]
- Weimbs T., Low S. H., Chapin S. J., Mostov K. E., Bucher P., Hofmann K. A conserved domain is present in different families of vesicular fusion proteins: a new superfamily. Proc. Natl. Acad. Sci. USA. 1997;94:3046–3051. doi: 10.1073/pnas.94.7.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelamanchili S. V., Reisinger C., Becher A., Sikorra S., Bigalke H., Binz T., Ahnert-Hilger G. The C-terminal transmembrane region of synaptobrevin binds synaptophysin from adult synaptic vesicles. Eur. J. Cell Biol. 2005;84:467–475. doi: 10.1016/j.ejcb.2004.11.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.