Abstract
Chromosome ends, known as telomeres, have to be distinguished from DNA breaks that activate DNA damage checkpoint. Two large protein kinases, ataxia-teleangiectasia mutated (ATM) and ATM-Rad3-related (ATR), control not only checkpoint activation but also telomere length. In budding yeast, Mec1 and Tel1 correspond to ATR and ATM, respectively. Here, we show that Cdc13-dependent telomere capping attenuates Mec1 association with DNA ends. The telomeric TG repeat sequence inhibits DNA degradation and decreases Mec1 accumulation at the DNA end. The TG-mediated degradation block requires binding of multiple Cdc13 proteins. The Mre11–Rad50-Xrs2 complex and Exo1 contribute to DNA degradation at DNA ends. Although the TG sequence impedes Exo1 association with DNA ends, it allows Mre11 association. Moreover, the TG sequence does not affect Tel1 association with the DNA end. Our results suggest that the Cdc13 telomere cap coordinates Mec1 and Tel1 accumulation rather than simply covering the DNA ends at telomeres.
INTRODUCTION
DNA double-strand breaks (DSBs) are induced by exogenous DNA-damaging agents and carcinogens or by endogenous by-products, including reactive oxygen species (Friedberg et al., 2006). The repair of DSBs is crucial for maintaining genome stability. All organisms respond to DSBs by promptly launching the DNA damage response. This response involves the recruitment of DNA repair factors and the activation of signal transduction pathways, often termed DNA damage checkpoint pathways (Zhou and Elledge, 2000). Activation of the checkpoint pathway induces cell cycle arrest and expression of genes required for DNA repair.
The checkpoint signals are initiated through two large protein kinases, ataxia-teleangiectasia mutated (ATM) and ATM-Rad3-related (ATR) (Zhou and Elledge, 2000; Abraham, 2001). ATM and ATR are highly conserved among eukaryotes. ATR is closely related to Mec1 in the budding yeast Saccharomyces cerevisiae and Rad3 in the fission yeast Schizosaccharomyces pombe. ATM homologues are termed Tel1 in both budding and fission yeasts. Current evidence indicates that the Mre11–Rad50–Nbs1 (Xrs2 in budding yeast) complex is the primary sensor that recruits ATR/Mec1 and ATM/Tel1 to DSBs (Nakada et al., 2003a, 2004; Falck et al., 2005; You et al., 2005). In budding yeast, there is compelling evidence that the Mre11–Rad50–Xrs2 (MRX) complex collaborates with exonuclease 1 (Exo1) in the generation of single-stranded DNA (ssDNA) at DSB ends (Krogh and Symington, 2004). ATM/Tel1 interacts with the C terminus of Nbs1/Xrs2 to localize to DNA ends (Nakada et al., 2003a; Falck et al., 2005; You et al., 2005). Meanwhile, the ssDNA that is generated at the DSB is covered with replication protein A (RPA) (Wold, 1997; Krogh and Symington, 2004). RPA-covered DNA recruits ATR/Mec1 to a region near the DSB end (Zou and Elledge, 2003; Nakada et al., 2005). ATM and ATR activate the downstream kinases CHK1 and CHK2 with assistance of checkpoint mediators, including 53BP1, BRCA1, and MDC1 (Zhou and Elledge, 2000; Bakkenist and Kastan, 2004). In budding yeast, Tel1 plays a minor role in the response to DSBs, because Mec1 has a predominant role (Morrow et al., 1995; Sanchez et al., 1996). After exposure to DSB-inducing agents, Tel1 checkpoint function is observed only in hypomorphic mec1 mutants (Nakada et al., 2003a,b). Mec1 and Tel1 phosphorylate the Rad9 checkpoint mediator and in turn activate the CHK2-related Rad53 protein (Gilbert et al., 2001; Schwartz et al., 2002; Sweeney et al., 2005). Activated Rad53 becomes hyperphosphorylated and further phosphorylates downstream targets (Sanchez et al., 1996; Sun et al., 1996).
The ends of chromosomes, termed telomeres, contain a double-stranded DNA region of tandem repeats (e.g., human, T2AG3; budding yeast, TG1–3 [TG]) and a 3′-protruding ssDNA region of the G-rich strand (Vega et al., 2003; Smogorzewska and de Lange, 2004). Due to the inability of the DNA replication machinery to copy the most distal telomere sequences, telomeric DNA progressively decreases in length as cells undergo successive cell divisions. To circumvent this problem, cells maintain telomeres by the action of telomerase (Est2 in budding yeast and TERT in human), a reverse transcriptase that uses its associated RNA as a template to elongate the G tail (Lingner et al., 1997; Meyerson et al., 1997; Nakamura et al., 1997). The single-stranded tails on telomeres are covered with the sequence-specific ssDNA-binding proteins, such as Cdc13 in budding yeast and POT1 in mammals (Nugent et al., 1996; Baumann and Cech, 2001). Cdc13 and POT1 act as telomere caps to protect telomeres from degradation (Garvik et al., 1995; Hockemeyer et al., 2005). Cdc13 plays an additional role in recruitment of the telomerase complex to the chromosome ends (Pennock et al., 2001). The maintenance of telomere structure and length is crucial for telomere homeostasis, because dysfunctional or shortened telomeres activate a checkpoint response that is mediated through the ATM and ATR family proteins (d'Adda di Fagagna et al., 2004; de Lange, 2005). However, the ATM and ATR family proteins are also required for proper maintenance of telomeres (Greenwell et al., 1995; Metcalfe et al., 1996; Ritchie et al., 1999). Recent evidence suggests that addition of telomeric TG repeats adjacent to DSBs inhibits the activation of checkpoint response in budding yeast (Michelson et al., 2005). However, it is not clear how telomeres control ATR/Mec1 and ATM/Tel1 to inhibit checkpoint responses.
In this study, we adopted a system that was originally developed to examine de novo telomere synthesis, and we investigated the mechanism by which the TG repeats inhibit checkpoint signaling from the adjacent DNA ends. Cdc13 telomere capping blocks ssDNA accumulation at the DNA end. We show that telomere capping depends on binding of multiple Cdc13 proteins. The Cdc13 telomere cap inhibits Mec1 association with the DNA end, thereby attenuating checkpoint activation. However, the telomere cap does not affect Tel1 association with the DNA end. Our results suggest that the Cdc13 telomere cap does not simply hide the DNA end from the checkpoint machinery; rather, it coordinates localization of Mec1 and Tel1 to telomeres.
MATERIALS AND METHODS
Plasmids and Strains
To prepare the KanMX-marked TG-HO cassette, the KanMX module (Wach et al., 1994) was amplified by polymerase chain reaction (PCR) and then digested with HindIII and XhoI. The resulting DNA fragment was cloned into HindIII-XhoI–digested pSD155 (Diede and Gottschling, 1999) to generate pTG-HO. To create the pTG-HO-CA plasmid, the 81-base pair TG sequence was first added into the TG-HO cassette by PCR. The PCR fragment was digested with EcoRI and NotI and cloned into the EcoRI/NotI sites of pTG-HO. The pTG-HO-CA plasmid was cleaved with EcoRI and XhoI, treated with Klenow, and self-ligated to generate pHO-CA. The EcoRI–XhoI digestion removes the 81-base pair TG sequence and its adjacent 55-base pair sequence. The pHO plasmid was constructed as follows. First, pSD155 was amplified by PCR to create MfeI and SpeI sites deleting the HO site and TG sequence. The PCR fragment was digested with MfeI and SpeI and ligated with an EcoRI–SpeI fragment containing the HO cleavage site (Nakada et al., 2003a). Oligonucleotides containing an 11-base pair TG sequence were inserted into EcoRI-XhoI–treated pTG-HO-CA, generating pTG11-HO-CA. Likewise a 22-base pair TG sequence was cloned to create pTG22-HO-CA. All the aforementioned plasmids were digested with NotI and SalI to integrate the cassettes into the MATa-inc strain (Nakada et al., 2003a). YCp-CDC13-EST1 (pVL1091) (Evans and Lundblad, 1999) was obtained from V. Lundblad (Salk Institute, La Jolla, CA). The EST1 coding sequence was amplified by PCR and cloned into YCplac111 (Gietz and Sugino, 1988) carrying the ADH1 promoter, generating YCp-ADH1-EST1. The CDC13-STN1 in-frame fusion was constructed as follows. The 5′-noncoding and the coding region of CDC13 was amplified by PCR by using YCp-CDC13 (pVL440) (Hughes et al., 2000) as a template. The PCR removed the termination codon of CDC13, creating an NcoI site for fusion. The STN1 coding and 3′-noncoding region was amplified from genomic DNA by PCR. The STN1 gene possesses an NcoI site at the translation initiation codon. The CDC13 fragment was digested with PstI and NcoI, whereas the STN1 fragment was digested with NcoI and SacI. The resulting fragments were cloned into PstI–SacI-treated YCplac111, generating YCp-CDC13-STN1. The CDC13-STN1 construct is functional; it suppresses temperature-sensitive growth defect of both cdc13-1 and stn1-13 mutants. The cdc13-1 and stn1-13 strains were obtained from M. Charbonneau (Ecole Normale Superìeure de Lyon, Lyon, France) (Grandin et al., 2001). Epitope tagging of EXO1 and MRE11 was performed by a PCR-based strategy (Knop et al., 1999; Hirano and Sugimoto, 2006). The CDC13-myc strain was constructed using the integration plasmid as described previously (Taggart et al., 2002). The RFA1-HA strain was generated using YIp-RFA1-HA (obtained from T. Naiki, Nagoya University, Nagoya, Japan) after NheI digestion. Integration of each gene was confirmed by PCR. Other mutations were described previously (Nakada et al., 2003a). All the strains used in this study are isogenic, and they are listed in Table 1. YCpA-GAL-HO and YCpT-RAD53-HA were described previously (Nakada et al., 2003a).
Table 1.
Strains used in this study
| KSC2214 | VII-L::KanMX-TG-HO |
| KSC2215 | VII-L::KanMX-HO-CA |
| KSC2216 | VII-L::KanMX-TG-HO-CA |
| KSC2217 | VII-L::KanMX-HO |
| KSC2218 | VII-L::KanMX-HO-CA sml1Δ::LEU2 |
| KSC2219 | VII-L::KanMX-HO-CA sml1Δ::LEU2 mec1Δ::LEU2 |
| KSC2221 | VII-L::KanMX-HO-CA MEC1-HA::TRP1 |
| KSC2222 | VII-L::KanMX-TG-HO-CA MEC1-HA::TRP1 |
| KSC2224 | VII-L::KanMX-HO-CA CDC13-myc::TRP1 RFA1-HA::LEU2 |
| KSC2225 | VII-L::KanMX-TG-HO-CA CDC13-myc::TRP1 RFA1-HA::LEU2 |
| KSC2226 | VII -L::KanMX-TG-HO-CA cdc13-1 |
| KSC2227 | VII-L::KanMX-TG-HO-CA cdc13-1-myc::TRP1 RFA1-HA::LEU2 |
| KSC2228 | VII-L::KanMX-TG-HO-CA cdc13-1 MEC1-HA::TRP1 |
| KSC2229 | VII-L::KanMX-TG-HO-CA |
| KSC2230 | VII-L::KanMX-TG-HO-CA |
| KSC2231 | VII-L::KanMX-TG-HO-CA CDC13-myc::TRP1 RFA1-HA::LEU2 |
| KSC2238 | VII-L::KanMX-HO-CA MRE11-myc::TRP1 TEL1-HA::TRP1 |
| KSC2239 | VII-L::KanMX-TG-HO-CA MRE11-myc:: TRP1 TEL1-HA::TRP1 |
| KSC2240 | VII-L::KanMX-HO-CA EXO1-HA::TRP1 |
| KSC2241 | VII-L::KanMX-TG-HO-CA EXO1-HA::TRP1 |
| KSC2242 | VII-L::KanMX-TG-HO-CA EXO1-HA::TRP1 cdc13-1 |
| KSC2258 | VII-L::KanMX-TG-HO-CA CDC13-myc::TRP1 RFA1-HA::LEU2 |
All the strains are isogenic to KSC1514 (MATa-inc ade1 his2 leu2 trp1 ura3) (Nakada et al., 2003a).
Southern Blot Assays
Purified genomic DNA was digested with BsaBI and HindIII, separated on 2.0% agarose gel and transferred to a Hybond nylon membrane (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom). Electrophoresis was performed in TAE buffer (Tris-acetate-EDTA). Hybridization DNA probes were prepared as follows. To generate the probe A, a 500-base pair DNA fragment, corresponding to the region proximal to the HO site, was amplified by PCR by using pTG-HO as a template. The resulting DNA fragment and a 310-base pair DNA fragment derived from the SMC2 locus were fused by PCR. A 510-base pair DNA fragment distal to the HO site was amplified by PCR and similarly fused to the DNA fragment from the SMC2 locus, creating the probe B. DNA probes were labeled with DIG-High prime (Roche Diagnostics, Indianapolis, IN) and detected with anti-Digoxigenin-POD (Roche Diagnostics). Hybridization was performed according to the manufacturer's instructions.
Chromatin Immunoprecipitation (ChIP) Assay
Chromatin immunoprecipitation was performed using anti-hemagglutinin (HA) (16B2) or anti-myc (9E10) antibodies as described previously (Hirano and Sugimoto, 2006). The PCR reaction was performed under nonsaturating conditions, in which the rate of PCR amplification was proportional to substrate concentration and cycling. Quantification of immunoprecipitated DNAs was achieved by using a real-time PCR detection system (Bio-Rad, Hercules, CA). For quantitation, signals from near the DSB (HO) were normalized to a control signal from a region in SMC2. The signals were also normalized to the input signal for each primer set. Finally, the PCR amplification ratio was normalized using the values before HO induction. The sequences of primers for the HO set were 5′-GTTGTTTCTGAAACATGGCAAAGG-3′ and 5′-CAACCAAACCGTTATTCATTCGTG-3′, and those for the SMC2 locus were 5′-AAGAGAAACTTTAGTCAAAACATGGG-3′ and 5′-CCATCACATTATACTAACTACGG-3′.
Other Methods
Degradation of DNA ends and kinase activity of Rad53 were measured as described previously (Nakada et al., 2003a). Immunoblotting analysis and viability assay after HO-induced DNA breaks were performed as described previously (Wakayama et al., 2001; Hirano and Sugimoto, 2006).
RESULTS
The 81-Base Pair TG Repeat Inhibits Checkpoint Signaling from the Adjacent DNA End
To investigate the mechanism by which telomeres control the checkpoint response, we used a system that was originally developed to examine de novo telomere synthesis (Diede and Gottschling, 1999, 2001). This system contains an 81-base pair TG repeat sequence (TG81) placed adjacent to a 30-base pair HO cleavage site inserted at the ADH4 locus. The ADH4 locus is located 15 kb from the left telomere of chromosome VII. The original TG-HO cassette is marked with the ADE2 gene (Diede and Gottschling, 1999, 2001). We have modified the de novo telomere synthesis system as follows. To facilitate detecting specific signals in hybridization analysis and ChIP assay, we marked the ADH4 locus with the KanMX gene (Wach et al., 1994) (Figure 1A). DSB ends are degraded by 5′-to-3′ exonuclease activities (White and Haber, 1990), and it is estimated that the 5′-to-3′ degradation occurs at the rate of 4 kb/h (Vaze et al., 2002). Because the ADH4 locus is only 15 kb away from the telomere, we considered the possibility that the completion of the 5′-to-3′ degradation of the 15-kb DNA fragment might attenuate the checkpoint activation. We therefore constructed two additional cassettes containing TG81; one cassette containing TG distal to the HO cleavage site (HO-CA), and another cassette containing TG on both sides of the HO cleavage site (TG-HO-CA). As a control, we also prepared a cassette containing no TG repeat sequence (HO). This HO cassette contains a 300-base pair fragment containing the HO cleavage site of the MATa locus, because HO does not induce a DSB at the ADH4 locus containing the 30-base pair HO cleavage site (data not shown). All the cassettes were integrated into the ADH4 locus of the strain in which the MATa locus is replaced with the noncleavable MATa-inc allele (Nakada et al., 2003a) (Figure 1A).
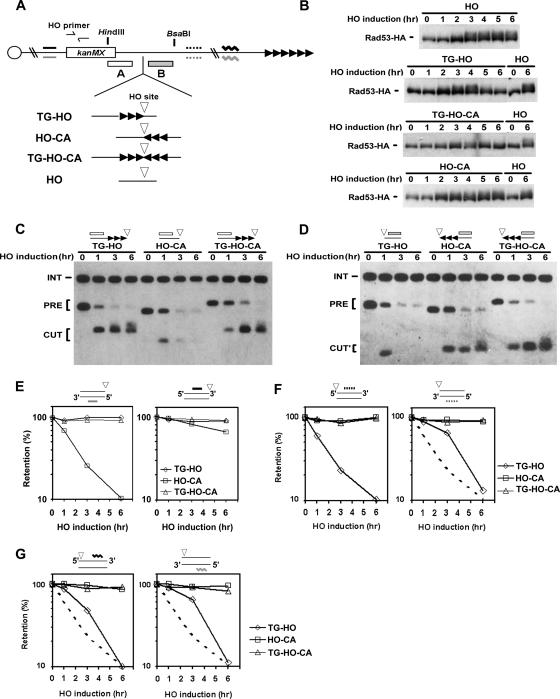
Figure 1.
Checkpoint activation in response to HO-induced DSBs. (A) Schematic of the HO cleavage site and TG repeats at the ADH4 locus on chromosome VII-L. The ADH4 locus was replaced with the KanMX gene and cassettes containing an HO cleavage site (inverted triangle) and 81 base pairs of TG repeat sequence (3 repetitive arrowheads) in different combinations. The TG sequence was inserted centromere-proximal to the HO site in TG-HO cells, distal to the HO site in HO-CA cells, and on both sides of the HO site in TG-HO-CA cells. The HO strain does not contain the TG sequence. The arrowheads indicate the direction of telomere elongation. Centromere is shown as a circle on the left. The HO site is located 15 kb from the endogenous telomere of VII-L (6 repetitive arrowheads). The probe A and B were used to monitor telomere synthesis by Southern blot. Strand-specific oligonucleotides, sequences of which are 2 kb away from the HO site (solid and dotted bars) and 5 kb away from the endogenous telomere (zigzag lines), were used to examine DNA degradation. The HO primer pair was designed to amplify a region 1 kb apart from the HO site. (B) Effect of the 81-base pair TG repeat sequence on Rad53 phosphorylation after DSB induction. HO, HO-CA, TG-HO, and TG-HO-CA cells carrying YCpT-RAD53-HA were transformed with the GAL-HO plasmid. Transformed cells were grown in sucrose and synchronized at G2/M with nocodazole. After arrest, the culture was incubated with galactose to induce HO expression. Aliquots of cells were collected at the indicated times after HO expression and cell extracts were analyzed by immunoblotting analysis with anti-HA antibodies. (C) Telomere addition at the centromere-proximal end of HO-induced DSBs. HO-CA, TG-HO, and TG-HO-CA cells were transformed with the GAL-HO plasmid and cultured as described in B. Cells were harvested at the indicated times after HO expression. Genomic DNA was digested with HindIII and BsaBI and then analyzed by Southern blot by using the probe A. The band labeled PRE indicates the 1-kbp HindIII–BsaBI fragment containing the HO site. After cleavage with HO, this band is converted into a new band (0.47 or 0.60 kbp, marked CUT). Telomere addition retards migration of the CUT fragment. The probe A also detects a 1.8-kbp HindIII fragment from the SMC2 locus on chromosome VI. This band is marked INT and serves as a loading control. (D) Telomere addition at the centromere-distal end of HO-induced DSBs. The same DNAs used for C were analyzed by Southern blot by using the probe B. The band labeled PRE indicates the 1-kbp HindIII–BsaBI fragment containing the HO site. After cleavage with HO, this band is converted into a new one (0.40 or 0.48 kbp, marked CUT'). Telomere addition retards migration of the CUT' fragment. The probe B also detects a 1.8-kbp HindIII fragment (INT) from the SMC2 locus as a loading control. (E) DNA degradation of the centromere-proximal DSB end. Genomic DNA prepared in C was fixed on a membrane and probed with 32P-labeled oligonucleotides each complementary to a 5′-to-3′–degrading strand (left) or 3′-to-5′–degrading strand (right) of a region 2 kb proximal to the HO site. (F) DNA degradation of the centromere-distal DSB end. Genomic DNA was analyzed as in E by using 32P-labeled oligonucleotides each complementary to a 5′-to-3′–dagrading strand (left) or 3′-to-5′–degrading strand (right) of a region 2 kb distal to the HO site. To facilitate comparison, the retention curve of the 5′-to-3′–degrading strand of a region 2kb distal to the HO site in TG-HO cells is included (a broken line). (G) DNA degradation of a region near the endogenous telomere. Genomic DNA was analyzed as in E using 32P-labeled oligonucleotides each complementary to a 5′-to-3′–degrading strand (left) or 3′-to-5′–degrading strand (right) of a region 5 kb apart from the endogenous telomere. The retention curve of the 5′-to-3′–degrading strand of a region 2 kb distal to the HO site in TG-HO cells is included as in F.
We first examined the Rad53 phosphorylation after HO expression in HO, HO-CA, TG-HO and TG-HO-CA cells (Figure 1B). Cells expressing HA-tagged Rad53 (Rad53-HA) were transformed with the GAL-HO plasmid. Transformed cells were grown in sucrose to maintain the GAL promoter inactive and arrested with nocodazole at G2/M. After arrest, cells were incubated with galactose to induce HO expression. Cells were collected at various times and subjected to immunoblotting analysis with anti-HA antibodies. Rad53 phosphorylation can be detected as a slowly migrating form on immunoblots (Sanchez et al., 1996; Sun et al., 1996). In HO cells, phosphorylation became detectable 2 h after HO expression and persisted for at least 6 h. Rad53 was fully phosphorylated in TG-HO cells, but phosphorylation became less evident at 5 h and nearly undetectable at 6 h. Interestingly, phosphorylation was significantly inhibited in TG-HO-CA cells. However, no phosphorylation inhibition was observed in HO-CA cells; Rad53 was phosphorylated similarly in HO and HO-CA cells.
To confirm that the TG81 sequence promotes telomere addition, we monitored telomere addition at the DNA ends in these cells (Figure 1, C and D). Cells transformed with the GAL-HO plasmid were treated as described above to induce HO expression. DNA samples were collected at various times and examined by Southern blot analysis. DSBs were generated within 1 h after HO expression, as evidenced by the appearance of the cleaved fragments (CUT and CUT') and the disappearance of the intact DNA fragment (PRE). The CUT and CUT' fragments were elongated if the DNA contained the TG81 sequence at the end, consistent with the observation that the TG81 sequence acts as a telomere seed and promotes telomere addition at the DNA end (Diede and Gottschling, 1999). However, detection of the CUT or CUT' fragment became weaker over the course of the experiment if the DNA contained no TG sequence at the end. Thus, telomeres were added at DNA ends containing TG81 (TG81 ends), but not at those containing no TG sequence (TG(−) ends), in TG-HO, HO-CA and TG-HO-CA cells.
We next investigated DNA degradation at DNA ends of HO-induced DSBs in TG-HO, HO-CA, and TG-HO-CA cells (Figure 1, E and F). We first monitored DNA degradation of the centromere-proximal DSB ends (Figure 1E). Both strands of the TG81 ends were well protected in TG-HO and TG-HO-CA cells. Unprotected DSB ends are degraded by 5′-to-3′exonuclease activities, thereby generating 3′-ended ssDNA tracts (White and Haber, 1990). Consistently, the 5′ termini of the TG(−) ends in HO-CA cells were immediately degraded, whereas the 3′ termini were protected from DNA degradation. We next monitored DNA degradation of the centromere-distal DSB ends (Figure 1F). Again, both strands of the TG81 end were preserved in HO-CA and TG-HO-CA cells, consistent with the telomere addition. However, the TG(−) end in TG-HO cells behaved differently from unprotected DSB ends, such as the centromere-proximal TG(−) end in HO-CA cells (Figure 1E). The 5′ termini of the TG(−) ends in TG-HO cells were largely degraded 3 h after HO expression. Curiously, degradation of the 3′ termini also proceeded with slow kinetics; its destruction was unclear at 3 h but became apparent at 6 h after HO expression. These results raised a possibility that the distal DNA fragment largely disappears in TG-HO cells within 6 h. To address the possibility, we further monitored DNA degradation near the endogenous telomere (Figure 1G). Given that the 5′-to-3′ degradation occurs at the rate of 4 kb/h (Vaze et al., 2002), the distal DNA fragment that is generated by HO cleavage could be converted to ssDNA within 4 h. Indeed, retention of the 5′-to-3′-degraded strand was quite low at 6 h after HO expression in TG-HO cells, suggesting that the 5′-to-3′ degradation was nearly completed. Again, the other strand was largely degraded at 6 h after HO expression. Thus, both strands of the distal DNA fragments disappeared in TG-HO cells after DSB induction during the time course, indicating that completion of 5′-to-3′ degradation or subsequent loss of the distal DNA fragment results in decreased Rad53 phosphorylation in TG-HO cells. Together, these results suggest that the presence of the TG sequence inhibits checkpoint signaling only from its own DNA end.
The TG Sequence Inhibits the Mec1 Checkpoint Pathway
Mec1 plays a central role in the cellular response to HO-induced DSBs, whereas Tel1 plays a minor role (Nakada et al., 2003a). Consistently, Rad53 phosphorylation in HO-CA cells was largely dependent on Mec1 function; phosphorylation was undetectable in HO-CA cells carrying a mec1Δ mutation (Figure 2A). Moreover, Rad53 kinase activity after HO expression was significantly decreased in HO-CA mec1Δ cells (Figure 2B). Weak Rad53 activation was detectable in TG-HO-CA cells (Figure 2B). However, activation of Rad53 kinase in TG-HO-CA cells was similar to that in HO-CA mec1Δ cells. These results indicate that the TG sequence attenuates Mec1-mediated checkpoint signaling. Interestingly, some Rad53 activation was still observed in TG-HO-CA mec1Δ cells (Figure 2B), suggesting that Tel1 responds to DSBs containing the TG81 end (see below).
Figure 2.
Effect of mec1Δ mutation on checkpoint activation after HO-induced DSBs. (A) Effect on Rad53 phosphorylation. HO-CA and HO-CA mec1Δ cells carrying YCpT-RAD53-HA were transformed with the GAL-HO plasmid and analyzed by immunoblotting analysis as described in Figure 1B. The strains contain an sml1Δ mutation to rescue the viability loss associated with mec1Δ mutation. (B) Effect on Rad53 kinase activity. HO-CA, HO-CA mec1Δ, TG- HO-CA, and TG-HO-CA mec1Δ cells carrying YCpT-RAD53-HA and YCpA-GAL-HO were treated as described in Figure 1B to induce HO expression. Extracts were prepared from cells and subjected to immunoprecipitation with anti-HA antibodies. Kinase activities of the immunoprecipitated Rad53 proteins were analyzed using histone H1 as a substrate. Autoradiogram shows 32P incorporation into histone H1 (top). The amount of Rad53 proteins used for the kinase assay was determined by immunoblotting with anti-HA antibodies (bottom). The strains contain an sml1Δ mutation to rescue the viability loss associated with mec1Δ mutation.
Because accumulation of Mec1 at sites of DNA damage correlates with activation of the Mec1 pathway (Nakada et al., 2004), we asked whether the TG81 sequence decreases Mec1 accumulation at the adjacent DSB end. We monitored Mec1 association with TG(−) or TG81 ends in HO-CA or TG-HO-CA cells, respectively, by ChIP assay (Figure 3A). Cells expressing Mec1-HA were transformed with the GAL-HO plasmid. Transformants were grown initially in sucrose to inhibit HO expression and incubated with nocodazole to arrest at G2/M. After arrest, galactose was added to induce HO expression. Cells were collected at various times, and extracts were prepared after formaldehyde cross-linking were subjected to immunoprecipitation with anti-HA antibodies. Coprecipitated DNA was amplified by PCR by using the HO primer set for a region near the HO cleavage site at the ADH4 locus on chromosome VII (Figure 1A) and the primer set for the SMC2 locus, which contains no HO cleavage site on chromosome VI. PCR amplification with the HO primer set was detected at the TG(−) end after HO expression (Figure 3A). In contrast, there was no increase in the PCR product amplified from the SMC2 locus (Figure 3A). No PCR amplification was detected with cells expressing untagged Mec1 proteins (data not shown). Thus, Mec1 associates efficiently with the TG(−) end. However, the TG sequence reduced Mec1 association with the adjacent DNA end; Mec1 associated poorly with the TG81 end (Figure 3A). We then monitored association of RPA and Cdc13 with TG(−) or TG81 ends (Figure 3B). RPA is a heterotrimeric ssDNA binding protein, which consists of Rfa1, Rfa2, and Rfa3 in budding yeast (Wold, 1997). At telomeres, another ssDNA binding protein, Cdc13, covers single-stranded TG regions (Nugent et al., 1996). Cells expressing HA-tagged Rfa1 and myc-tagged Cdc13 were transformed with the GAL-HO plasmid and analyzed as described above by ChIP assay by using anti-HA and anti-myc antibodies. Similar to Mec1 association, Rfa1 association was readily detected at TG(−) ends but its association was decreased at TG81 ends. In sharp contrast, Cdc13 association was clearly detected at TG81 ends whereas only weak association was observed at TG(−) ends. No association was detected with untagged cells (data not shown). Association of Cdc13 with TG81 ends is consistent with the fact that telomere addition occurs at the DNA end. These results are consistent with the current model in which RPA covers long ssDNA tracts and recruits Mec1 to the ssDNA regions (Zou and Elledge, 2003; Nakada et al., 2005).
Figure 3.
Association of Mec1, Rfa1, and Cdc13 with the TG81 end in wild-type and cdc13-1 cells. (A) Association of Mec1 with a region near the TG(−) or TG81 end. HO-CA (TG(−)) and TG-HO-CA (TG81) cells expressing Mec1-HA were transformed with the GAL-HO plasmid and cultured as described in Figure 1B. Aliquots of cells were collected at the indicated times after HO expression and subjected to ChIP assay. PCR was done with the HO primer pair (see Figure 1A) and the SMC2 primer pair for the control SMC2 locus on chromosome VI. PCR products from the respective input extracts are shown in a bottom panel. (B) Association of Rfa1 and Cdc13 with the TG(−) or TG81 end. HO-CA (TG(−)) and TG-HO-CA (TG81) cells expressing Rfa1-HA and Cdc13-myc were analyzed by ChIP assay as described in A. (C) Effect of cdc13-1 mutation on DNA degradation at TG81 ends. HO-CA (TG(−)), TG-HO-CA (TG81), and TG-HO-CA (TG81) cdc13-1 cells were transformed with the GAL-HO plasmid. Transformed cells were grown in sucrose at 23°C and synchronized at G2/M with nocodazole. After synchronization, the culture was incubated with galactose and concomitantly shifted to 32°C. Aliquots of cells were collected at the indicated times after HO expression and analyzed as described in Figure 1E to examine the rate of degradation of the centromere-proximal DSB end. (D) Rfa1 association with the TG81 end in cdc13-1 mutants. HO-CA (TG(−)), TG-HO-CA (TG81), and TG-HO-CA (TG81) cdc13-1 cells expressing Rfa1-HA were transformed with the GAL-HO plasmid. Cells were treated as in C and subjected to ChIP assay as described in A. (E) Mec1 association with the TG81 end in cdc13-1 mutants. HO-CA (TG(−)), TG-HO-CA (TG81), and TG-HO-CA (TG81) cdc13-1 cells expressing Mec1-HA were analyzed by ChIP assay as described in D.
Cdc13 Is Required for Inhibition of Mec1 Association with the TG81 DNA End
We next asked whether checkpoint inhibition at the TG81 end requires Cdc13 function. When cdc13-1 mutants are shifted to the restrictive temperature, cells generate long tracts of ssDNA at telomere ends (Garvik et al., 1995). Cells containing the CDC13 wild-type or cdc13-1 mutant gene were transformed with the GAL-HO plasmid. Transformants were grown in sucrose at the permissive temperature (23°C) and arrested with nocodazole at G2/M. After arrest, cells were incubated with galactose at the restrictive temperature (32°C). Consistent with the role of Cdc13 in DNA end protection, the 5′-to-3′ degradation of the TG81 end was observed in cells carrying the cdc13-1 mutation (Figure 3C). Accordingly, association of Rfa1 and Mec1 with the TG81 end was increased in cdc13-1 mutants (Figure 3, D and E). Previous studies showed that Mec1 localizes to telomeric regions in cdc13-1 mutants at the restrictive temperature (Rouse and Jackson, 2002). Together, these results indicate that Cdc13 is critical for inhibition of Mec1 association with the TG81 end, and suggest that the TG81 end behaves like an endogenous telomere to attenuate checkpoint signaling.
Binding of Multiple Cdc13 Proteins Is Required for DNA End Protection and Telomere Addition
Cdc13 binds to a minimum 11-base telomeric substrate in vitro (Hughes et al., 2000). We therefore examined whether a shorter TG sequence supports telomere addition or DNA end protection. To this end, we constructed TG11-HO-CA and TG22-HO-CA cells, in which the 81-base pair TG sequence at the centromere-proximal side of the HO site was replaced with 11- or 22-base pair TG sequence, respectively. We first determined viability of HO-CA, TG-HO-CA, TG11-HO-CA, and TG22-HO-CA cells after HO expression (Figure 4A). More than 90% of TG-HO-CA cells retained viability, whereas only 0.02% of HO-CA cells survived. The 22-base pair TG sequence fully rescued the viability loss; TG22-HO-CA cells were as viable as TG-HO-CA cells. In contrast, only 5% of TG11-HO-CA cells were viable after HO expression. We then examined Cdc13 binding to the adjacent DNA end in these cells by ChIP assay (Figure 4B and Supplemental Figure 1). Cdc13 was found to associate with the TG11 end; the TG11 end was more enriched than the TG(−) end. However, Cdc13 associated more efficiently with the TG22 and the TG81 end than the TG11 end (1.5-fold and 3-fold enrichment of the TG22 and the TG81 end, respectively). We monitored telomere addition at the DNA ends in TG11-HO-CA and TG22-HO-CA cells (Figure 4C). Although the TG11 construct supports Cdc13 binding in vivo, no apparent telomere addition was detected at the TG11 end. In contrast, telomere addition occurred efficiently at the TG22 end; telomere extension at TG22 ends was more prominent than that at TG81 ends. We next investigated whether the TG11 and TG22 constructs protect DNA ends and inhibit DNA checkpoint activation (Figure 4, D and E). The 11-base pair TG sequence did not protect the DNA end; the 5′-to-3′ degradation occurred at the TG11 end as fast as at the TG(−) end (Figure 4D). Moreover, no checkpoint inhibition was observed with the 11-base pair TG sequence; Rad53 was phosphorylated after HO expression in TG11-HO-CA cells as observed in HO-CA cells (Figure 4E). The TG22 construct partially suppressed the 5′-to-3′ DNA degradation; 3′-ended ssDNA was still accumulated at the TG22 end (Figure 4D). Correspondingly, Rad53 was phosphorylated after HO expression in TG22-HO-CA cells (Figure 4E). Biochemical experiments have shown that multiple Cdc13 proteins bind to longer TG repeat sequence (Hughes et al., 2000). Together, these observations indicate that binding of multiple Cdc13 proteins is required for both telomere addition and end protection.
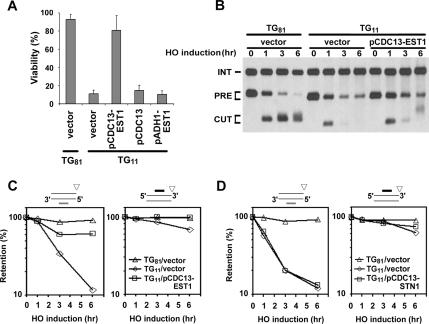
Figure 4.
Effect of the TG repeat length on telomere maintenance. (A) Effect of the TG repeat length on cell viability after DSB induction. TG-HO-CA (TG81), HO-CA (TG(−)), TG11-HO-CA (TG11), and TG22-HO-CA (TG22) cells were transformed with the GAL-HO plasmid. Transformed cells were grown in sucrose and then plated out on either glucose or galactose medium selectable for the plasmid. Viability was estimated from colony formation ability on galactose medium. The bars represent SEs. TG11-HO-CA and TG22-HO-CA cells contain 11- and 22-base pair TG sequences, respectively, on the centromere-proximal side of the HO site. (B) Effect of the TG length on Cdc13 association with DNA ends. HO-CA (TG(−)), TG11-HO-CA (TG11), TG22-HO-CA (TG22), and TG-HO-CA (TG81) cells expressing Cdc13-myc were subjected to ChIP as described in Figure 3A, and precipitated DNA was analyzed by real-time PCR. Cdc13 enrichment was calculated from three independent experiments. The bars represent SEs. (C) Effect of the 11- and 22-base pair TG sequence on telomere addition. HO-CA (TG(−)), TG11-HO-CA (TG11), TG22-HO-CA (TG22), and TG-HO-CA (TG81) cells were analyzed by Southern blot by using the probe A as described in Figure 1C. (D) Effect of the 11- and 22-base pair TG sequence on DNA degradation at DNA ends. HO-CA (TG(−)), TG11-HO-CA (TG11), TG22-HO-CA (TG22), and TG-HO-CA (TG81) cells were transformed with the GAL-HO plasmid and analyzed as described in Figure 1E to determine the DNA degradation rate of the centromere-proximal DSB end. (E) Effect of the 11- and 22-base pair TG sequence on Rad53 phosphorylation after DSB induction. HO-CA (TG(−)), TG11-HO-CA (TG11), TG22-HO-CA (TG22), and TG-HO-CA (TG81) cells carrying YCpT-RAD53-HA were transformed with the GAL-HO plasmid and analyzed by immunoblotting analysis as described in Figure 1B.
Cdc13 has been proposed to recruit the telomerase complex and the end-protection complex to telomeres. Indeed, delivery of telomerase and Stn1 to telomeres, by fusing the DNA binding domain of Cdc13, can rescue the lethality and telomere replication defect of a cdc13 null strain (Pennock et al., 2001). We first addressed whether binding of multiple Cdc13 proteins is required for telomerase recruitment. To this end, we examined the effect of Cdc13-Est1 fusion on survival of TG11-HO-CA cells after HO expression (Figure 5A). Expression of Cdc13–Est1 proteins suppressed the colony-formation defect of TG11-HO-CA cells, whereas high dosage of neither CDC13 nor EST1 rescued. Moreover, Cdc13–Est1 expression promoted telomere addition and end protection at the TG11 end (Figure 5, B and C). Accordingly, recovered TG11-HO-CA cells expressing the Cdc13–Est1 protein contained telomeres (Supplemental Figure 2). Because Cdc13 binding requires an 11-base-long TG repeat, recruitment of a single copy of Cdc13-Est1 seems to promote telomere addition at the TG11 end. These results suggest that binding of multiple Cdc13 proteins promotes telomerase recruitment. However, we cannot rule out a possibility that the Cdc13–Est1 fusion increases associated telomerase activity. The observed end protection is likely to result from telomere addition, which in turn recruits more Cdc13 proteins. To address whether binding of multiple Cdc13 proteins is required for recruitment of the end protection complex, we examined the effect of Cdc13–Stn1 fusion on DNA degradation in TG11-HO-CA cells (Figure 5D). No apparent suppression was observed with the Cdc13–Stn1 protein, suggesting that the Stn1 recruitment itself is not enough to protect TG11 ends from DNA degradation.
Figure 5.
Effect of Cdc13-Est1 or Cdc13-Stn1 fusion on telomere maintenance. (A) Effect of Cdc13-Est1 expression on cell viability after DSB induction. TG-HO-CA (TG81) or TG11-HO-CA (TG11) cells carrying the GAL-HO plasmid were transformed with YCp-CDC13-EST1 (pCDC13-EST1), YCp-CDC13 (pCDC13), YCp-ADH1-EST1 (pADH1-EST1) or the control vector. Cell viabilities after DSB induction were determined as described in Figure 4A. The bars represent SEs. (B) Effect of Cdc13-Est1 expression on telomere addition at the TG11 end. TG-HO-CA (TG81) or TG11-HO-CA (TG11) cells carrying the GAL-HO plasmid were transformed with YCp-CDC13-EST1 (pCDC13-EST1) or the control vector, and analyzed by Southern blot using the probe A as described in Figure 1C. (C) Effect of Cdc13-Est1 expression on DNA degradation at the TG11 end. TG-HO-CA (TG81) or TG11-HO-CA (TG11) cells carrying the GAL- HO plasmid were transformed with YCp-CDC13-EST1 (pCDC13-EST1) or the control vector, and analyzed as described in Figure 1E to determine the DNA degradation rate of the centromere-proximal DSB end. (D) Effect of Cdc13-Stn1 expression on DNA degradation at the TG11 end. TG-HO-CA (TG81) or TG11-HO-CA (TG11) cells carrying the GAL-HO plasmid were transformed with YCp-CDC13-STN1 (pCDC13-STN1) or the control vector, and analyzed as described in Figure 1E to determine the DNA degradation rate of the centromere-proximal DSB end.
Cdc13-mediated Telomere Capping Does Not Inhibit Tel1 Recruitment to the DNA End
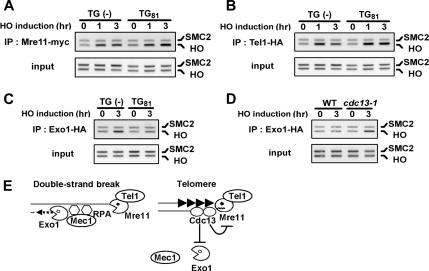
The aforementioned findings have suggested that binding of multiple Cdc13 proteins is required to block DNA degradation at telomeres. Degradation of DNA ends requires functions of the MRX complex (Ivanov et al., 1994; Lee et al., 1998). Because the MRX complex associates with DSB ends, we hypothesized that the Cdc13 telomere cap inhibits association of the MRX complex with DNA ends. We therefore examined Mre11 association with the TG(−) or TG81 end by ChIP assay (Figure 6A). The 81-base pair TG sequence did not decrease Mre11 association with the adjacent DSB end; Mre11 associated similarly with the TG81 and TG(−) ends. Because Tel1 interacts with the C terminus of Xrs2 to localize to DSB ends (Nakada et al., 2003a), we examined whether the TG sequence impedes Tel1 association with the adjacent DNA end (Figure 6B). Again, Tel1 associated efficiently with the TG81 end. Thus, the TG sequence does not affect MRX association or Tel1 association with the adjacent DNA end. Exo1 possesses a 5′-to-3′ exonuclease activity and acts redundantly with the MRX complex in DNA degradation of DSB ends (Moreau et al., 2001; Nakada et al., 2004). We further examined whether Exo1 localizes to the TG81 end by ChIP assay (Figure 6C). Although Exo1 associated with the TG(−) end, no association with the TG81 end was detected. As shown above, the TG81 end is degraded in cdc13-1 mutants. Accordingly, Exo1 association was restored in cdc13-1 mutants at the restrictive temperature (Figure 6D). These results indicate that the Cdc13 telomere cap acts differently on the MRX complex and Exo1, thereby modulating Mec1 and Tel1 recruitment to the DNA end.
Figure 6.
Effect of the 81-base pair TG sequence on association of Mre11, Exo1, and Tel1 with the adjacent DNA end. (A) Association of Mre11 with the TG(−) or TG81 end. HO-CA (TG(−)), and TG-HO-CA (TG81) cells expressing Mre11-myc were analyzed by ChIP assay as described in Figure 3A. (B) Association of Tel1 with the TG(−) or TG81 end. HO-CA (TG(−)) and TG-HO-CA (TG81) cells expressing Tel1-HA were analyzed by ChIP assay as described in A. (C) Association of Exo1 with the TG(−) or TG81 end. HO-CA (TG(−)) and TG-HO-CA (TG81) cells expressing Exo1-HA were analyzed by ChIP assay as described in A. (D) Effect of cdc13-1 mutation on Exo1 association with the TG81 end. TG-HO-CA and TG-HO-CA cdc13-1 cells expressing Exo1-HA were analyzed by ChIP assay as described in Figure 3D. (E) Regulation of Mec1 and Tel1 recruitment to DSBs and telomeres. The Cdc13 telomere cap blocks DNA degradation at the DNA end; however, it regulates the MRX complex and Exo1 differently. The Cdc13 telomere cap inhibits Exo1 localization to the DNA end, whereas it allows the MRX complex to associate with the end. Inhibition of DNA degradation decreases ssDNA generation and RPA accumulation at the DNA end, thereby reducing Mec1 association with the DNA ends. However, the Cdc13 telomere cap does not inhibit Tel1 association with the DNA ends. See Discussion for details.
DISCUSSION
DNA ends are degraded by 5′-to-3′ exonuclease activities, generating long ssDNA tracts at the 3′ ends (White and Haber, 1990). In turn, RPA covers the ssDNA region, promoting the recruitment of Mec1 to the DSB ends (Zou and Elledge, 2003). However, the telomeric TG sequence blocks DNA degradation, decreasing RPA and Mec1 association with the adjacent DSB end. As a result, the TG sequence attenuates the Mec1 checkpoint pathway. The TG-mediated DNA degradation block depends on binding of multiple Cdc13 proteins. The TG sequence inhibits Exo1 association with the adjacent DNA end, whereas it allows association of the MRX complex. Moreover, the TG sequence did not affect Tel1 association. Thus, Cdc13-mediated telomere capping decreases Mec1 association, but not Tel1 association, with the adjacent DNA end.
Cdc13-mediated telomere capping does not simply hide DNA ends from the DNA repair and checkpoint machinery, although it protects the DNA ends at telomeres. Degradation of DNA ends depends on the MRX complex and Exo1 (Moreau et al., 2001; Nakada et al., 2004). To inhibit Exo1-dependent degradation, the Cdc13 telomere cap prevents Exo1 from binding to the DNA end. In contrast, the Cdc13 telomere cap allows the MRX complex to localize to DNA ends yet blocks MRX-dependent DNA degradation. At this moment, however, it remains to be determined how the Cdc13 cap inhibits MRX-dependent end processing. Previous studies showed that the MRX complex is essential for de novo telomere synthesis at TG81 ends (Diede and Gottschling, 2001). The MRX complex seems to promote 5′-to-3′ degradation of the DNA ends, generating single-stranded TG tracts. In turn, Cdc13 covers the single-stranded TG tracts. Binding of multiple Cdc13 proteins could establish telomere caps, which in turn attenuate MRX and other nuclease functions. However, the Cdc13 telomere cap may not completely inhibit the nuclease activities. Similar to DSB ends, endogenous telomeres are degraded in a Cdk1 (Cdc28)-dependent manner (Frank et al., 2006; Vodenicharov and Wellinger, 2006), and the degradation process is partially dependent on the MRX complex (Larrivee et al., 2004). DNA degradation occurs even in cells lacking the MRX complex and Exo1; for example, other nucleases such as Fen1/Rad27 are implicated in the DNA degradation (Moreau et al., 2001). Likewise, Fen1/Rad27 has been shown to play a role in telomere maintenance (Parenteau and Wellinger, 2002). The Cdc13 telomere cap could repress functions of other nucleases, such as Fen1/Rad27, by either blocking localization or inhibiting activities.
The MRX complex controls both the Mec1 and Tel1 checkpoint pathways (Usui et al., 2001; Nakada et al., 2004). However, the Cdc13 cap can modulate MRX complex functions to regulate Mec1 and Tel1 differently at telomeres. The Cdc13 telomere cap blocks MRX-dependent DNA degradation and in turn decreases Mec1 accumulation at the DNA end, whereas it permits the MRX complex to recruit Tel1 to the DNA end. Although Tel1 plays a minor role in checkpoint response (Morrow et al., 1995; Sanchez et al., 1996), it plays a predominant role in telomere length regulation (Greenwell et al., 1995; Ritchie et al., 1999). Consistently, telomere synthesis from TG81 ends largely depends on Tel1 (Frank et al., 2006). Cdc13-dependent inhibition of Mec1 association with telomeres could explain why Tel1 plays an important role in telomere maintenance. One group reported that Mec1 associates with endogenous telomeres and stimulates recruitment of telomerase to telomeres (Takata et al., 2004; Takata et al., 2005). However, recent studies demonstrated that telomerase recruitment depends largely on Tel1, suggesting that Tel1 associates with endogenous telomeres (Goudsouzian et al., 2006).
We found that both telomere addition and end protection depend on binding of multiple Cdc13 proteins at the DNA ends. The TG22 construct exhibits full telomere addition but partial telomere protection, whereas the TG11 construct fails to promote both telomere addition and protection. DNA degradation at the TG22 end could be eventually repaired by lagging-strand synthesis, because cells containing the TG22 construct recovered similar to those containing the TG81 construct. Recovered cells containing the TG22 construct acquired telomeres at the DNA end (data not shown). Thus, telomere maintenance seems to depend on multiple copies of Cdc13 at DNA ends. Enhanced telomere addition at TG22 ends could result from the absence of double-stranded TG repeat. Double-stranded regions of telomeres are covered with Rif1 and Rif2, both of which negatively control telomere length (Vega et al., 2003; Smogorzewska and de Lange, 2004). In the absence of Rif2, telomere addition occurs more extensively at TG81 ends (Diede and Gottschling, 2001; Frank et al., 2006).
It has been shown that Cdc13 delivers telomerase and Stn1 to the DNA end for telomere addition and end protection, respectively (Pennock et al., 2001). The Cdc13–Est1 fusion restores telomere addition at TG11 ends, implying that multiple Cdc13 binding stimulates telomerase recruitment. However, the Cdc13–Stn1 fusion fails to suppress DNA degradation at TG11 ends; the Stn1 recruitment by itself does not protect DNA ends containing the 11-base pair TG sequence. One explanation could be that localization of multiple Stn1 proteins to telomeres is important for DNA end protection. Stn1 collaborates with Ten1 in telomere end protection (Grandin et al., 2001). Alternatively, Ten1 might localize to DNA ends that are covered with multiple Cdc13 proteins. Previous findings indicate that telomere addition takes place at a very limited number of chromosome sites in budding yeast (Mangahas et al., 2001). On chromosome VII, telomere addition frequently occurs at only a single location, which corresponds to a 35-base pair TG stretch that is 50 kb from the left telomere end (35 kb away from the ADH4 locus). Chromosome VII has no other TG tract >20 base pairs (Mangahas et al., 2001), consistent with our finding that the 22-base pair TG sequence assists DSB repair by telomere addition, whereas the 11-base pair TG sequence does not. Notably, the haploid genome contains only 14 TG repeats, the length of which is >20 base pairs (Mangahas et al., 2001). We detected weak Cdc13 association with TG(−) ends at the ADH4 locus on chromosome VII (Figure 4B). It is possible that Cdc13 binds to the region containing a 10-base pair single-stranded TG-like repeat (AACCACACCA), sequence that locates 5 kb from the HO cleavage site. Cdc13 shows some affinity to 10 nucleotides of embedded TG sequence in vitro (Lin and Zakian, 1996; Hughes et al., 2000). Alternatively, Cdc13 might interact weakly with more diverged sequences at other locations. Cdc13 has been shown to bind to mammalian telomeric T2AG3 repeat sequences in vivo and in vitro (Lin and Zakian, 1996; Alexander and Zakian, 2003).
The TG sequence seems to attenuate checkpoint signaling only from the adjacent DNA end. In TG-HO cells, Rad53 is phosphorylated after DSB induction but its phosphorylation tapers off at a later time point. We propose that this checkpoint attenuation in TG-HO cells results from completion of 5′-to-3′ degradation or subsequent loss of the distal DNA fragment. The cleavage site is 15 kb away from the chromosome end in TG-HO cells. It is estimated that the 5′-to-3′ degradation of DNA ends occurs at the rate of 4 kb/h (Vaze et al., 2002). Consistently, the 5′-to-3′ degradation of the distal DNA fragment was largely completed at 6 h after HO induction. Completion of the 5′-to-3′ degradation could relocate checkpoint proteins, thereby attenuating checkpoint activation. The PCNA-like checkpoint clamps have been shown to target partial duplex DNA (Majka and Burgers, 2003; Majka et al., 2006). Although retained at early time points, the other strand of the distal DNA fragment disappeared at 6 h after HO induction. It is possible that the 5′-to-3′ degradation disrupts telomeric structures at the chromosome end. Therefore, the resulting ssDNA could be promptly degraded by exonuclease activities. Alternatively, the remaining strand might be digested by endonuclease activities after the completion of the 5′-to-3′ degradation. Currently, it is not clear how ssDNA tracts are processed if they are not used for DSB repair. In any case, loss of the distal DNA fragment could shut off checkpoint signaling. Recently, Michelson et al. (2005) showed that telomeric TG repeat sequence inhibits checkpoint signaling from damage sites, and they proposed a model in which the TG sequence could inhibit checkpoint signaling nearby. In their report, the TG sequence was inserted only into the centromere-proximal side of the HO cleavage site that is 20 kb away from the chromosome end. After DSB induction, degradation of the distal DNA fragment was monitored by Southern blot. They found that one strand of the DNA fragment was retained within the time frame. However, it would be difficult to determine degradation of each strand, because DNA was electrophoresed in nondenaturing conditions. The observed checkpoint attenuation might result from completion of 5′-to-3′ degradation or subsequent loss of the distal 20-kb DNA fragment.
In summary, we have shown that efficient telomere capping requires binding of multiple Cdc13 proteins. We also have shown that the Cdc13 telomere cap acts differently on the MRX complex and Exo1, thereby modulating Mec1 and Tel1 recruitment to the DNA end. However, it remains to be determined exactly how the Cdc13 telomere cap blocks the MRX-dependent DNA degradation at telomeres. Future work will focus on the regulation of the MRX catalytic activity at TG-containing DNA ends.
Supplementary Material
ACKNOWLEDGMENTS
We thank M. Takami for construction of the KanMX-marked TG-HO cassette; M. Charbonneau, D. Gottschling, V. Lundblad, T. Naiki, D. Nakada, and V. Zakian for materials; and J. Kang and C. Newlon for helpful discussion and critical reading. This work was supported by National Institutes of Health grant GM-073876.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-12-1074) on March 21, 2007.
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Abraham R. T. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 2001;15:2177–2196. doi: 10.1101/gad.914401. [DOI] [PubMed] [Google Scholar]
- Alexander M. K., Zakian V. A. Rap1p telomere association is not required for mitotic stability of a C(3)TA(2) telomere in yeast. EMBO J. 2003;22:1688–1696. doi: 10.1093/emboj/cdg154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakkenist C. J., Kastan M. B. Initiating cellular stress responses. Cell. 2004;118:9–17. doi: 10.1016/j.cell.2004.06.023. [DOI] [PubMed] [Google Scholar]
- Baumann P., Cech T. R. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science. 2001;292:1171–1175. doi: 10.1126/science.1060036. [DOI] [PubMed] [Google Scholar]
- d'Adda di Fagagna F., Teo S. H., Jackson S. P. Functional links between telomeres and proteins of the DNA-damage response. Genes Dev. 2004;18:1781–1799. doi: 10.1101/gad.1214504. [DOI] [PubMed] [Google Scholar]
- de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- Diede S. J., Gottschling D. E. Telomerase-mediated telomere addition in vivo requires DNA primase and DNA polymerases alpha and delta. Cell. 1999;99:723–733. doi: 10.1016/s0092-8674(00)81670-0. [DOI] [PubMed] [Google Scholar]
- Diede S. J., Gottschling D. E. Exonuclease activity is required for sequence addition and Cdc13p loading at a de novo telomere. Curr. Biol. 2001;11:1336–1340. doi: 10.1016/s0960-9822(01)00400-6. [DOI] [PubMed] [Google Scholar]
- Evans S. K., Lundblad V. Est1 and Cdc13 as comediators of telomerase access. Science. 1999;286:117–120. doi: 10.1126/science.286.5437.117. [DOI] [PubMed] [Google Scholar]
- Falck J., Coates J., Jackson S. P. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature. 2005;434:605–611. doi: 10.1038/nature03442. [DOI] [PubMed] [Google Scholar]
- Frank C. J., Hyde M., Greider C. W. Regulation of telomere elongation by the cyclin-dependent kinase CDK1. Mol. Cell. 2006;24:423–432. doi: 10.1016/j.molcel.2006.10.020. [DOI] [PubMed] [Google Scholar]
- Friedberg E. C., Walker G. C., Siede W., Wood R. D., Schultz R. A., Ellenberger T. Washington, DC: ASM Press; 2006. DNA Repair and Mutagenesis. [Google Scholar]
- Garvik B., Carson M., Hartwell L. Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol. Cell Biol. 1995;15:6128–6138. doi: 10.1128/mcb.15.11.6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz R. D., Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Gilbert C. S., Green C. M., Lowndes N. F. Budding yeast Rad9 is an ATP-dependent Rad53 activating machine. Mol. Cell. 2001;8:129–136. doi: 10.1016/s1097-2765(01)00267-2. [DOI] [PubMed] [Google Scholar]
- Goudsouzian L. K., Tuzon C. Z., Zakian V. A. S. cerevisiae Tel1p and Mre11p are required for normal Levels of Est1p and Est2p telomere association. Mol. Cell. 2006;24:603–610. doi: 10.1016/j.molcel.2006.10.005. [DOI] [PubMed] [Google Scholar]
- Grandin N., Damon C., Charbonneau M. Ten1 functions in telomere end protection and length regulation in association with Stn1 and Cdc13. EMBO J. 2001;20:1173–1183. doi: 10.1093/emboj/20.5.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwell P. W., Kronmal S. L., Porter S. E., Gassenhuber J., Obermaier B., Petes T. D. TEL1, a gene involved in controlling telomere length in S. cerevisiae, is homologous to the human ataxia telangiectasia gene. Cell. 1995;82:823–829. doi: 10.1016/0092-8674(95)90479-4. [DOI] [PubMed] [Google Scholar]
- Hirano Y., Sugimoto K. ATR homolog Mec1 controls association of DNA polymerase zeta-Rev1 complex with regions near a double-strand break. Curr. Biol. 2006;16:586–590. doi: 10.1016/j.cub.2006.01.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D., Sfeir A. J., Shay J. W., Wright W. E., de Lange T. POT1 protects telomeres from a transient DNA damage response and determines how human chromosomes end. EMBO J. 2005;24:2667–2678. doi: 10.1038/sj.emboj.7600733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes T. R., Weilbaecher R. G., Walterscheid M., Lundblad V. Identification of the single-strand telomeric DNA binding domain of the Saccharomyces cerevisiae Cdc13 protein. Proc. Natl. Acad. Sci. USA. 2000;97:6457–6462. doi: 10.1073/pnas.97.12.6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov E. L., Sugawara N., White C. I., Fabre F., Haber J. E. Mutations in XRS2 and RAD50 delay but do not prevent mating-type switching in Saccharomyces cerevisiae. Mol. Cell Biol. 1994;14:3414–3425. doi: 10.1128/mcb.14.5.3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M., Siegers K., Pereira G., Zachariae W., Winsor B., Nasmyth K., Schiebel E. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast. 1999;15:963–972. doi: 10.1002/(SICI)1097-0061(199907)15:10B<963::AID-YEA399>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Krogh B. O., Symington L. S. Recombination proteins in yeast. Annu. Rev. Genet. 2004;38:233–271. doi: 10.1146/annurev.genet.38.072902.091500. [DOI] [PubMed] [Google Scholar]
- Larrivee M., LeBel C., Wellinger R. J. The generation of proper constitutive G-tails on yeast telomeres is dependent on the MRX complex. Genes Dev. 2004;18:1391–1396. doi: 10.1101/gad.1199404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. E., Moore J. K., Holmes A., Umezu K., Kolodner R. D., Haber J. E. Saccharomyces Ku70, Mre11/Rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell. 1998;94:399–409. doi: 10.1016/s0092-8674(00)81482-8. [DOI] [PubMed] [Google Scholar]
- Lin J. J., Zakian V. A. The Saccharomyces CDC13 protein is a single-strand TG1–3 telomeric DNA-binding protein in vitro that affects telomere behavior in vivo. Proc. Natl. Acad. Sci. USA. 1996;93:13760–13765. doi: 10.1073/pnas.93.24.13760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J., Hughes T. R., Shevchenko A., Mann M., Lundblad V., Cech T. R. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science. 1997;276:561–567. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- Majka J., Binz S. K., Wold M. S., Burgers P. M. Replication protein A directs loading of the DNA damage checkpoint clamp to 5′-DNA junctions. J. Biol. Chem. 2006;281:27855–27861. doi: 10.1074/jbc.M605176200. [DOI] [PubMed] [Google Scholar]
- Majka J., Burgers P. M. Yeast Rad17/Mec3/Ddc1, a sliding clamp for the DNA damage checkpoint. Proc. Natl. Acad. Sci. USA. 2003;100:2249–2254. doi: 10.1073/pnas.0437148100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangahas J. L., Alexander M. K., Sandell L. L., Zakian V. A. Repair of chromosome ends after telomere loss in Saccharomyces. Mol. Biol. Cell. 2001;12:4078–4089. doi: 10.1091/mbc.12.12.4078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalfe J. A., Parkhill J., Campbell L., Stacey M., Biggs P., Byrd P. J., Taylor A. M. Accelerated telomere shortening in ataxia telangiectasia. Nat. Genet. 1996;13:350–353. doi: 10.1038/ng0796-350. [DOI] [PubMed] [Google Scholar]
- Meyerson M., et al. hEST2, the putative human telomerase catalytic subunit gene, is up-regulated in tumor cells and during immortalization. Cell. 1997;90:785–795. doi: 10.1016/s0092-8674(00)80538-3. [DOI] [PubMed] [Google Scholar]
- Michelson R. J., Rosenstein S., Weinert T. A telomeric repeat sequence adjacent to a DNA double-stranded break produces an anticheckpoint. Genes Dev. 2005;19:2546–2559. doi: 10.1101/gad.1293805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau S., Morgan E. A., Symington L. S. Overlapping functions of the Saccharomyces cerevisiae Mre11, Exo1 and Rad27 nucleases in DNA metabolism. Genetics. 2001;159:1423–1433. doi: 10.1093/genetics/159.4.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow D. M., Tagle D. A., Shiloh Y., Collins F. S., Hieter P. TEL1, an S. cerevisiae homolog of the human gene mutated in ataxia telangiectasia, is functionally related to the yeast checkpoint gene MEC1. Cell. 1995;82:831–840. doi: 10.1016/0092-8674(95)90480-8. [DOI] [PubMed] [Google Scholar]
- Nakada D., Hirano Y., Sugimoto K. Requirement of the Mre11 complex and exonuclease 1 for activation of the Mec1 signaling pathway. Mol. Cell Biol. 2004;24:10016–10025. doi: 10.1128/MCB.24.22.10016-10025.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakada D., Hirano Y., Tanaka Y., Sugimoto K. Role of the C terminus of mec1 checkpoint kinase in its localization to sites of DNA damage. Mol. Biol. Cell. 2005;16:5227–5235. doi: 10.1091/mbc.E05-05-0405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakada D., Matsumoto K., Sugimoto K. ATM-related Tel1 associates with double-strand breaks through an Xrs2-dependent mechanism. Genes Dev. 2003a;17:1957–1962. doi: 10.1101/gad.1099003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakada D., Shimomura T., Matsumoto K., Sugimoto K. The ATM-related Tel1 protein of Saccharomyces cerevisiae controls a checkpoint response following phleomycin treatment. Nucleic Acids Res. 2003b;31:1715–1724. doi: 10.1093/nar/gkg252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T. M., Morin G. B., Chapman K. B., Weinrich S. L., Andrews W. H., Lingner J., Harley C. B., Cech T. R. Telomerase catalytic subunit homologs from fission yeast and human. Science. 1997;277:955–959. doi: 10.1126/science.277.5328.955. [DOI] [PubMed] [Google Scholar]
- Nugent C. I., Hughes T. R., Lue N. F., Lundblad V. Cdc13p: a single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science. 1996;274:249–252. doi: 10.1126/science.274.5285.249. [DOI] [PubMed] [Google Scholar]
- Parenteau J., Wellinger R. J. Differential processing of leading- and lagging-strand ends at Saccharomyces cerevisiae telomeres revealed by the absence of Rad27p nuclease. Genetics. 2002;162:1583–1594. doi: 10.1093/genetics/162.4.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennock E., Buckley K., Lundblad V. Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell. 2001;104:387–396. doi: 10.1016/s0092-8674(01)00226-4. [DOI] [PubMed] [Google Scholar]
- Ritchie K. B., Mollory J. C., Petes T. D. Interactions of TLC1 (which encodes the RNA subunit of telomerase), TEL1, and MEC1 in regulating telomere length in the yeast Saccharomyces cerevisiae. Mol. Cell Biol. 1999;19:6065–6075. doi: 10.1128/mcb.19.9.6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouse J., Jackson S. P. Lcd1p recruits Mec1p to DNA lesions in vitro and in vivo. Mol. Cell. 2002;9:857–869. doi: 10.1016/s1097-2765(02)00507-5. [DOI] [PubMed] [Google Scholar]
- Sanchez Y., Desany B. A., Jones W. J., Liu Q., Wang B., Elledge S. J. Regulation of RAD53 by the ATM-like kinase MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science. 1996;271:357–360. doi: 10.1126/science.271.5247.357. [DOI] [PubMed] [Google Scholar]
- Schwartz M. F., Duong J. K., Sun Z., Morrow J. S., Pradhan D., Stern D. F. Rad9 phosphorylation sites couple Rad53 to the Saccharomyces cerevisiae DNA damage checkpoint. Mol. Cell. 2002;9:1055–1065. doi: 10.1016/s1097-2765(02)00532-4. [DOI] [PubMed] [Google Scholar]
- Smogorzewska A., de Lange T. Regulation of telomerase by telomeric proteins. Annu. Rev. Biochem. 2004;73:177–208. doi: 10.1146/annurev.biochem.73.071403.160049. [DOI] [PubMed] [Google Scholar]
- Sun Z., Fay D. S., Marini F., Foiani M., Stern D. F. Spk1/Rad53 is regulated by Mec1-dependent protein phosphorylation in DNA replication and damage checkpoint pathways. Genes Dev. 1996;10:395–406. doi: 10.1101/gad.10.4.395. [DOI] [PubMed] [Google Scholar]
- Sweeney F. D., Yang F., Chi A., Shabanowitz J., Hunt D. F., Durocher D. Saccharomyces cerevisiae Rad9 acts as a Mec1 adaptor to allow Rad53 activation. Curr. Biol. 2005;15:1364–1375. doi: 10.1016/j.cub.2005.06.063. [DOI] [PubMed] [Google Scholar]
- Taggart A. K., Teng S. C., Zakian V. A. Est1p as a cell cycle-regulated activator of telomere-bound telomerase. Science. 2002;297:1023–1026. doi: 10.1126/science.1074968. [DOI] [PubMed] [Google Scholar]
- Takata H., Kanoh Y., Gunge N., Shirahige K., Matsuura A. Reciprocal association of the budding yeast ATM-related proteins Tel1 and Mec1 with telomeres in vivo. Mol. Cell. 2004;14:515–522. doi: 10.1016/s1097-2765(04)00262-x. [DOI] [PubMed] [Google Scholar]
- Takata H., Tanaka Y., Matsuura A. Late S phase-specific recruitment of Mre11 complex triggers hierarchical assembly of telomere replication proteins in Saccharomyces cerevisiae. Mol. Cell. 2005;17:573–583. doi: 10.1016/j.molcel.2005.01.014. [DOI] [PubMed] [Google Scholar]
- Usui T., Ogawa H., Petrini J. H. A DNA damage response pathway controlled by Tel1 and the Mre11 complex. Mol. Cell. 2001;7:1255–1266. doi: 10.1016/s1097-2765(01)00270-2. [DOI] [PubMed] [Google Scholar]
- Vaze M. B., Pellicioli A., Lee S. E., Ira G., Liberi G., Arbel-Eden A., Foiani M., Haber J. E. Recovery from checkpoint-mediated arrest after repair of a double-strand break requires Srs2 helicase. Mol. Cell. 2002;10:373–385. doi: 10.1016/s1097-2765(02)00593-2. [DOI] [PubMed] [Google Scholar]
- Vega L. R., Mateyak M. K., Zakian V. A. Getting to the end: telomerase access in yeast and humans. Nat. Rev. Mol. Cell Biol. 2003;4:948–959. doi: 10.1038/nrm1256. [DOI] [PubMed] [Google Scholar]
- Vodenicharov M. D., Wellinger R. J. DNA degradation at unprotected telomeres in yeast is regulated by the CDK1 (Cdc28/Clb) cell-cycle kinase. Mol. Cell. 2006;24:127–137. doi: 10.1016/j.molcel.2006.07.035. [DOI] [PubMed] [Google Scholar]
- Wach A., Brachat A., Pohlmann R., Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;13:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- Wakayama T., Kondo T., Ando S., Matsumoto K., Sugimoto K. Pie1, a protein interacting with Mec1, controls cell growth and checkpoint responses in Saccharomyces cerevisiae. Mol. Cell Biol. 2001;21:755–764. doi: 10.1128/MCB.21.3.755-764.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White C. I., Haber J. E. Intermediates of recombination during mating type switching in Saccharomyces cerevisiae. EMBO J. 1990;9:663–673. doi: 10.1002/j.1460-2075.1990.tb08158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wold M. S. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- You Z., Chahwan C., Bailis J., Hunter T., Russell P. ATM activation and its recruitment to damaged DNA require binding to the C terminus of Nbs1. Mol. Cell Biol. 2005;25:5363–5379. doi: 10.1128/MCB.25.13.5363-5379.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B.-B.S., Elledge S. J. The DNA damage response: putting checkpoints in perspective. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- Zou L., Elledge S. J. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.