Abstract
ARNO is a soluble guanine nucleotide exchange factor (GEF) for the Arf family of GTPases. Although in biochemical assays ARNO prefers Arf1 over Arf6 as a substrate, its localization in cells at the plasma membrane (PM) suggests an interaction with Arf6. In this study, we found that ARNO activated Arf1 in HeLa and COS-7 cells resulting in the recruitment of Arf1 on to dynamic PM ruffles. By contrast, Arf6 was activated less by ARNO than EFA6, a canonical Arf6 GEF. Remarkably, Arf6 in its GTP-bound form recruited ARNO to the PM and the two proteins could be immunoprecipitated. ARNO binding to Arf6 was not mediated through the catalytic Sec7 domain, but via the pleckstrin homology (PH) domain. Active Arf6 also bound the PH domain of Grp1, another ARNO family member. This interaction was direct and required both inositol phospholipids and GTP. We propose a model of sequential Arf activation at the PM whereby Arf6-GTP recruits ARNO family GEFs for further activation of other Arf isoforms.
INTRODUCTION
Arfs are a family of small GTPases that regulate membrane traffic and structure within the secretory and endocytic pathways. As for all GTPases, the binding of GTP induces conformational changes in Arfs that allow interactions with effector molecules and GTPase-activating proteins (GAPs). Arfs play important roles in regulating membrane lipid composition and in the recruitment of coat proteins to membranes to facilitate sorting and transport of cargo molecules. Arfs can be divided into three classes based on their amino acid sequence similarities: class I contains Arf1 and Arf3, class II comprises Arf4 and Arf5, and class III contains Arf6 (Tsuchiya et al., 1991). Class I and II Arfs function at the Golgi apparatus and within the secretory pathway, whereas Arf6 functions at the plasma membrane (PM; D'Souza-Schorey and Chavrier, 2006). However, the ability of class I and II Arfs to cycle off the Golgi to the cytosol upon GTP hydrolysis makes them available for recruitment onto other membranes, whereas Arf6 appears to remain associated with membranes throughout its activation cycle (Peters et al., 1995; Radhakrishna et al., 1999).
Arf1 and Arf6 are the most divergent and most thoroughly characterized Arfs (D'Souza-Schorey and Chavrier, 2006). Although the function of Arf1 and Arf6 are usually distinct in cells, their GTP-bound conformations are nearly identical, particularly in the switch I and switch II regions that interact with common effectors (Pasqualato et al., 2001). This is consistent with the observation that in cell-free, biochemical assays, Arf1 and Arf6 share the ability to interact with many effectors. Unlike the GTP-bound forms, the GDP-bound conformations differ from one another, particularly in the regions that interact with their guanine nucleotide exchange factors (GEFs; Menetrey et al., 2000). Thus, the spatial-temporal regulation of Arfs, provided at least in part by their GEFs, may be key to determining the cellular function of each Arf isoform. Because GTP binding is coupled to the accessibility of the amino terminal myristoyl group to biological membranes, the GEFs also facilitate the recruitment of the active Arf onto the membranes on which it will function (Antonny et al., 1997). Thus, GEFs play an important role in activating an Arf at a particular location within the cell.
All Arf GEFs identified to date contain a conserved, catalytic Sec7 domain. There are several subfamilies of Arf GEFs including the BFA-sensitive Gea/GBF and BIG families, which localize to and function at the Golgi apparatus on class I and class II Arfs (Jackson and Casanova, 2000). The three other subfamilies of Arf GEFs are resistant to inhibition by BFA and function in the cell periphery and at the plasma membrane. There is consensus that the EFA6 (Franco et al., 1999) and BRAG/GEP100 (Someya et al., 2001; Dunphy et al., 2006) subfamilies function on Arf6 in cell-free assays and in cells.
The Arf specificity of the ARNO/cytohesin subfamily of GEFs, however, is more ambiguous. In biochemical assays, Arf1 is a better substrate than Arf6 for these GEFs (Franco et al., 1998; Frank et al., 1998; Klarlund et al., 1998; Pacheco-Rodriguez et al., 1998; Macia et al., 2001). However, ARNO can activate Arf6 in cells (Frank et al., 1998; Langille et al., 1999; Santy and Casanova, 2001), and, given that Arf6 is present at the plasma membrane where ARNO is also recruited, it has been generally assumed that ARNO/cytohesin family GEFs activate Arf6. Little is known about the mechanism whereby ARNO family GEFs are recruited to the PM other than the fact that the recruitment is dependent upon their pleckstrin homology (PH) domain and its ability to bind specific phosphoinositides (Santy et al., 1999; Venkateswarlu et al., 1999).
This study examines the relationship between the peripheral exchange factor ARNO and its potential substrates, Arf1 and Arf6. Whereas Arf1 serves as a good substrate for ARNO, the relationship between Arf6 and ARNO is more consistent with that of a GTPase and its effector than of a GTPase and its GEF. We show that GTP-bound Arf6 can recruit ARNO to the plasma membrane through a direct interaction between Arf6 and the ARNO PH domain. This interaction is conserved in other members of the ARNO/cytohesin family and is dependent on the presence of the appropriate phosphoinositide. We propose a novel mechanism for activation of Arf1 or other Golgi-associated Arfs downstream of Arf6 activation, through the recruitment by Arf6 of exchange factors of the ARNO/cytohesin family to the PM.
MATERIALS AND METHODS
Antibodies and Reagents
Mouse monoclonal anti-hemagglutinin (HA) coupled to agarose and anti-flag (M5) antibodies were purchased from Sigma-Aldrich (St. Louis, MO). The mouse monoclonal anti-HA antibody, 16b12, was purchased from Covance (Berkeley, CA). The monoclonal anti-GFP antibody (mGFP), and Fugene6 were purchased from Roche (Nutley, NJ). The rabbit polyclonal anti-GFP antibody (rGFP), and the Alexa-conjugated (488, 594, and 680) fluorescent secondary goat-anti-rabbit and goat-anti-mouse antibodies were purchased from Invitrogen (Carlsbad, CA). Glutathione Sepharose 4B was purchased from GE Healthcare (Piscataway, NJ). The rabbit polyclonal anti-Arf6 was previously described (Song et al., 1998). The infrared secondary antibody goat anti-rabbit 800 was purchased from Rockland Biosciences (Gilbertsville, PA). The cross-linker, Dithiobis[succinimidylpropionate] (DSP), was purchased from Pierce (Rockford, IL). Polyclonal antibody to myc epitope was purchased from Abcam (Cambridge, MA).
DNA Constructs
The HA-tagged Arf1 and Arf6 constructs and untagged Arf6 constructs were in pXS vector as previously described (Peters et al., 1995; Radhakrishna and Donaldson, 1997). Arf1-RFP was created by PCR amplification of Arf1-HA, and cloning the resulting product into the BglII and EcoRI sites of monomeric RFP N1 from Dr. Roger Tsien (UCSD; Campbell et al., 2002), and this construct was confirmed by sequencing. Myc-tagged human PIP 5-kinase type I α was previously described (Rozelle et al., 2000). pEGFP-F encoding GFP with the carboxy terminus of H-Ras was from Clontech (Mountain View, CA). Flag-tagged ARNO wild type (wt), ARNO E156K, and GFP ARNO wt of the “3G” splice versions were obtained from James Casanova (University of Virginia, Charlottesville, VA). GFP ARNO 3G PH domain, GFP ARNO 2G PH domain, and YFP GRP1 PH (residues 267–299) and its mutants: R284C, I307E, and K340L were as previously described (Varnai et al., 2005). GST-Arf6T27N and GST-Arf6Q67L were made by fusing GST to the amino termini of Arf6T27N and Arf6Q67L in pGEX4T1 and expressed in Escherichia coli BL21. The resulting fusion proteins were not myristoylated, but GST-Arf6Q67L was capable of binding the Arf6 effector PIP 5-kinase in vitro (unpublished observations).
Cell Culture and Transient Transfections
COS-7 and HeLa cells were maintained in DMEM supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin, and 100 μg/ml streptomycin at 37°C with 5% CO2. Cells were plated the day before transfection with Fugene 6 according to manufacturer's instructions. Experiments were carried out as described ∼18 h after DNA addition.
Immunofluorescent Staining and Live Cell Imaging
For immunofluorescence staining of HeLa cells, cells were plated on to glass coverslips and transfected the following day. Eighteen hours after transfection, cells were fixed in 2% formaldehyde for 10 min. Cells were washed in phosphate-buffered saline (PBS) containing 10% FBS (PBS/FBS) and incubated with primary antibodies diluted in PBS/FBS containing 0.2% saponin for 1 h. Cells were washed three times in PBS/FBS and incubated with appropriate secondary antibodies in PBS/FBS containing 0.2% saponin for 30 min. Cells were washed three times in PBS/FBS and then mounted on glass slides.
For live cell imaging, HeLa or COS-7 cells were plated onto Lab-Tek coverglass chambers (Nunc, Rochester, NY) and transfected with indicated fluorescent constructs. Eighteen hours after transfection, cells were imaged on a 37°C stage in CO2-independent media.
Images were taken using a Zeiss 510 laser scanning confocal microscope (Thornwood, NY) using a 63× 1.3 NA PlanApo objective. After acquisition, images were handled using Adobe Photoshop and Adobe Illustrator (San Jose, CA). All experiments were confirmed at least three times and a representative image is shown.
Immunoprecipitation
Eighteen hours after transfection, HeLa cells were incubated in a solution containing 1 mM DSP for 30 min at room temperature before lysis in a buffer containing 1% IPEGAL CA-430, 10% glycerol, 100 mM NaCl, and 50 mM Tris, pH 7.5. Lysates were cleared by centrifugation. Lysates were probed with anti HA-agarose beads for 2 h at 4°C and then washed four times in lysis buffer. Bound proteins were eluted from the beads by boiling in SDS PAGE sample buffer and resolved by SDS PAGE, followed by transfer to nitrocellulose membrane. Western blot was carried out using indicated primary antibodies and appropriate infrared secondary antibodies. Blots were visualized using an Odyssey infrared scanner (Li-COR Biosciences, Lincoln, NE) according to manufacturer's instructions. All experiments were confirmed at least two times, and a representative example of each experiment is shown.
GAT Assay for GTP-bound Arfs
A GST fusion protein containing the VHS-GAT domain of GGA3 was purified from bacteria on glutathione Sepharose 4B. Forty micrograms of the VHS-GAT domain was incubated with HeLa cell lysates expressing indicated proteins as described by Santy and Casanova (2001). Bound proteins were eluted from beads by boiling in SDS-PAGE sample buffer and resolved by SDS-PAGE, followed by transfer to nitrocellulose membrane. Western blot was carried out using 16B12 to detect HA-tagged Arf and M5 or Rabbit anti-GFP to detect tagged GEFs and appropriate infrared secondary antibody. The Western blot was visualized and quantified using an Odyssey infrared imager. Data from four independent experiments are shown as the average percent of Arf pulldown on VHS-GAT beads with error represented as ±1 SD. The level of expression of HA-tagged Arfs and Flag-tagged GEFs was comparable in these experiments.
In Vitro Binding of GRP PH Domains
Recombinant proteins were produced in BL-21 cells (Invitrogen, Carlsbad, CA). Overnight cultures were grown to OD600: 0.6–0.9 and induced with 300 μM IPTG for 7 h at room temperature. Proteins were purified from the bacterial lysates by Ni-NTA agarose (Qiagen, Valencia, CA) (GRP1-PH constructs) or glutathione Sepharose 4B (Amersham Biosciences, GE Healthcare, Piscataway, NJ) (Arf 6 constructs) following the manufacturer's instructions. Sepharose-bound GST-Arf 6 proteins (∼20 μg) and the purified YFP-tagged GRP1 PH domains (∼20 μg) were incubated for 3 h on ice in 0.4 ml binding buffer (PBS, pH 7.2, 1 mM MgCl2, 1 mM DTT, 0.2% Triton, 0.1% Tween 20) in the presence or in the absence of 30 nM Ins(1,3,4,5)P4. Beads (50 μl) were washed twice with binding buffer (1 ml), and the resulting protein complexes were analyzed by SDS-PAGE. To preserve fluorescence, samples were incubated at room temperature for 30 min instead of boiling. PH domains were visualized using a phosphorimager, and the gels were stained with Coomassie blue.
RESULTS
ARNO Activates Arf1 in Cells
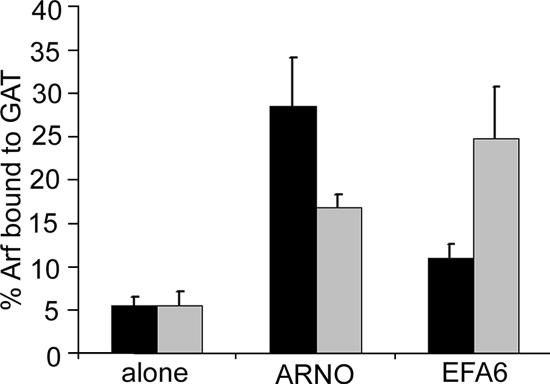
ARNO is one of four related proteins in the ARNO/Cytohesin subfamily of Arf GEFs (Cytohesin-1, ARNO/Cytohesin-2, Grp1/Cytohesin-3, and Cytohesin-4) that are ∼70% identical to one another and share the same domain topography consisting of an amino-terminal coiled-coil domain, a central Sec7 domain, and a carboxyl terminal PH domain and poly basic region (Jackson and Casanova, 2000). To examine ARNO's substrate preference in HeLa cells, we transfected cells with either Arf1 or Arf6, alone or together with ARNO, and determined the extent of activation of Arf1 and Arf6. ARNO expression resulted in a fivefold increase in the activation of Arf1 and only a threefold increase in the activation of Arf6 (Figure 1). We next compared ARNO's activation to EFA6's activation of Arf1 or Arf6 and found the opposite specificity. EFA6 expression caused a fivefold increase in active Arf6 and only a twofold increase in the activation of Arf1 (Figure 1). These results suggest that when the two exogenous Arfs are present at similar levels within a cell, ARNO expression promotes Arf1 activation more than it does Arf6 activation.
Figure 1.
Activation of Arf1 and Arf6 by ARNO and EFA6 in cells. Arf1-HA or Arf6-HA were expressed in HeLa cells either alone or in the presence of Flag-ARNO or Flag-EFA6. The fraction of Arf1 (■) or Arf6 (▩) that was GTP-bound was determined using a GGA3-GAT pulldown assay as described in Materials and Methods. Shown are the means and SD of triplicate experiments.
Having shown that ARNO expression leads to an increase in the amount of Arf1-GTP in cells, we next examined the effect of ARNO expression on the distribution of Arf1 in cells. The lack of good immunological reagents capable of detecting endogenous Arf1 led us to examine the distribution of Arf-RFP in cells because Arf1-GFP has been shown to behave like endogenous Arf1 in cells (Presley et al., 2002; Honda et al., 2005; Liu et al., 2005). Arf1-RFP was associated with the Golgi and was present in the cytosol when expressed alone in HeLa (Figure 2A) and COS-7 cells (data not shown). ARNO alone was largely cytosolic with some labeling along the PM edge (Figures 2A). Coexpression of ARNO with Arf1 led to the recruitment of Arf1 to the PM where it colocalized with ARNO on dynamic membrane ruffles (Figure 2, A and B). Coexpression of ARNO and Arf1 also led to a stimulation of the formation of macropinosomes, which formed from the regions of membrane ruffling. Live cell imaging revealed that ARNO and Arf1 colocalized on the incoming macropinosomes, and as these macropinosomes traveled toward the center of the cell, both ARNO and Arf1 were released from the surface of the macropinosomes within the same 15-s window (Figure 2B and Supplementary Movie 1). This suggests that the loss of ARNO from the macropinosome surface is coupled to the rapid inactivation and subsequent loss of Arf1 from the membrane. In contrast to ARNO's ability to recruit Arf1 to the PM, EFA6 expression did not appreciably recruit ARF1 to PM ruffles or onto the macropinosomes formed and delineated by EFA6 expression (Figure 2C and Supplementary Movie 2). Live cell imaging of HeLa cells showed similar results (data not shown). These data demonstrate that ARNO can recruit and activate Arf1 at the PM.
Figure 2.
ARNO recruits Arf1 to the PM to form ruffles and macropinosomes. (A) HeLa cells expressing Arf1-RFP or GFP-ARNO were expressed by themselves or together and imaged live 24 h after transfection. (B) COS-7 cells coexpressing Arf1-RFP and GFP-ARNO (3G) were imaged at 15-s intervals for 30 min at 37°C. (C) COS-7 cells expressing Arf1-RFP and GFP-EFA6 were imaged as above. Shown in B and C are images from a selected 150-s interval from Supplementary Videos 1 and 2, respectively. Bars, 10 μm.
Arf6-GTP Recruits ARNO and Grp1 to the PM
Our data suggests that ARNO functions as an exchange factor for Arf1 in cells. On the other hand, in previous studies expression of the dominant negative, GTP-binding defective mutant of Arf6, T27N, inhibited many events promoted by ARNO expression (Santy and Casanova, 2001; Hernandez-Deviez et al., 2002; Shmuel et al., 2006). To examine the relationship between ARNO and Arf6, we examined the localization of ARNO in cells expressing Arf6. When expressed alone, ARNO was mostly cytosolic with a milky appearance and limited association with the PM (Figure 3A). Coexpression of wt Arf6 with ARNO led to a dramatic shift in ARNO localization from the cytosol to the plasma membrane and onto discrete scattered endocytic structures where it colocalized with Arf6. The ability of Arf6 to recruit ARNO onto membranes was also observed with the constitutively active mutant of Arf6, Q67L, but not with the inactive mutant Arf6T27N (Figure 3A), indicating that it is the GTP-bound form of Arf6 that can recruit ARNO to the membrane. In cells coexpressing Arf6, ARNO, and Arf1, Arf1 localized to the vesicular structures onto which ARNO was recruited (Figure 3B). ARNO belongs to a family of four related proteins that share a common domain structure. We examined whether another family member, Grp1, would also be recruited to membranes by expression of Arf6Q67L. Grp1 alone was partially associated with the PM and also cytosolic but in the presence of Arf6Q67L, Grp1 distribution was shifted onto membranes (Figure 3C). Similar observations were also made for Cytohesin 1 (data not shown). The clearance of these GEFs (both ARNO and Grp1) from the diffuse, milky, cytosolic distribution onto membranes was very distinctive and was observed regardless of the level of expression of these exchange factors. The number of cells exhibiting this shift in distribution was quantified in three separate experiments (see Supplementary Figure 1).
Figure 3.
ARNO is recruited to the PM by GTP-bound Arf6. (A) HeLa cells expressing Flag-ARNO alone, or coexpressing Flag-ARNO and either Arf6, Arf6T27N, or Arf6Q67L, were processed for immunofluorescence as described and labeled with antibodies to Flag and Arf6. (B) Arf1-RFP, GFP-ARNO, and Arf6 were all expressed in HeLa cells. Twenty-four hours after transfection, cells were imaged live for ARNO and Arf1, which both localized to vesicle structures that form with overexpression of Arf6. (C) Flag-GRP1 was expressed alone or with Arf6Q67L and cells were immunolabeled with antibodies to Flag and Arf6. (D) Flag-ARNO was expressed with myc-PIP 5-kinase Iα, and cells were immunolabeled with antibodies to Flag and myc. (E) GFP-ARNO was expressed alone or with HA-tagged Arf6, Arf6T27N, or Arf6Q67L. Lysates from these cells were probed with anti-HA agarose to precipitate the HA-tagged Arfs, and the pulldowns were examined by Western blot against HA (Arf6) and GFP (ARNO) as described in Materials and Methods. Bars, 10 μm.
These localization findings suggest that Arf6 is not acting as a substrate for ARNO or Grp1, but rather as a mechanism for recruitment of these GEFs onto the PM for activation of Arf1. ARNO recruitment onto membranes is dependent on its PH domain, which recognizes phosphatidylinositol 4,5 bisphosphate (PIP2) and phosphatidylinositol 3,4,5 trisphosphate (PIP3; Klarlund et al., 2000). Because Arf6 activation at the plasma membrane stimulates localized PIP2 production through the activation of phosphatidylinositol 4-phosphate 5-kinase (PIP5-kinase; Honda et al., 1999; Brown et al., 2001), it is possible that ARNO is recruited onto the Arf6Q67L structures through excessive PIP2 production on the vacuoles. To see if PIP2 production is sufficient to recruit ARNO on to vacuole structures, we overexpressed PIP 5-kinase I α, which causes the formation of vacuole structures enriched in PIP2 that are similar to those produced by Arf6Q67L (Brown et al., 2001). However, as shown in Figure 3D, ARNO was not recruited on to these structures, suggesting that PIP2 production alone is not sufficient to recruit ARNO onto the Arf6Q67L vacuoles.
To examine more directly whether ARNO interacts with GTP-bound forms of Arf6, we looked to see whether ARNO could be immunoprecipitated with active forms of Arf6 in lysates of transfected cells. Similar to our immunofluorescence results (Figure 3A), we found that ARNO coprecipitated specifically with Arf6 and Arf6Q67L, but not with Arf6T27N (see Figure 3E). Because a fraction of wt Arf6 will be in the active, GTP-bound state but none of the T27N mutant will be in that state, this implies that it is the GTP-bound form of Arf6 that binds to ARNO. By contrast, we observed the opposite nucleotide dependence for EFA6. It coimmunoprecipitated with Arf6T27N and not with wt or Arf6Q67L (data not shown). It is curious that we did not see increased association of ARNO with Arf6Q67L, but rather both wt and Q67L were equally able to coimmunoprecipitate ARNO. We do not know the reason for this, but one possible explanation is that much of the Q67L may already be in association with other effectors and thus not be available for interaction with ARNO. Taken together, these observations suggest that ARNO interacts with Arf6 in a manner more consistent with that of an Arf6 effector than of an Arf6 GEF and, combined with the data in Figure 1, suggests that activation of Arf6 could enhance the activation of Arf1 at the PM by recruiting ARNO/Cytohesin family GEFs.
Active Arf6 Leads to Increased Activation of Arf1
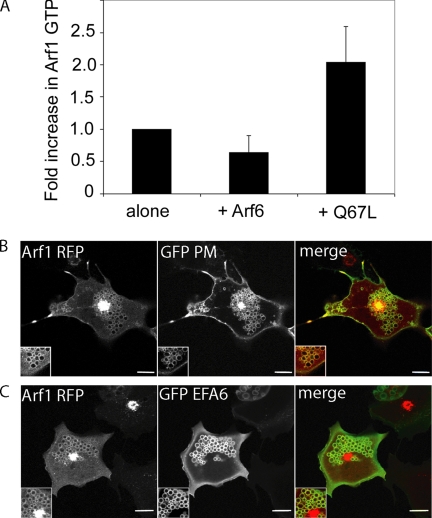
To examine whether activation of Arf6 would result in an increase in activation of Arf1, we measured the activation of Arf1 using the GGA GAT pulldown assay. We found a twofold increase in activation of Arf1 in cells that were coexpressing Arf6Q67L compared with those coexpressing Arf6 wt or expressing Arf1 alone (Figure 4A). The effect of expression of Arf6Q67L on Arf1 distribution was also observed in live cells coexpressing Arf6Q67L, a PM marker that labels the vacuoles (GFP-PM) and Arf1-RFP (Figure 4B). Arf1-RFP was associated with many of the Q67L-induced vacuolar structures, in addition to being localized to the Golgi complex (Figure 4B). Another means of causing activation of Arf6 that mimics the Arf6Q67L phenotype, coexpression of Arf6 and GFP-EFA6 (Brown et al., 2001) also resulted in localization of Arf1-RFP to vacuolar structures (Figure 4C). Taken together these observations provide evidence that the activation of Arf6 leads to the activation and localization of Arf1 in the periphery.
Figure 4.
Active Arf6 leads to activation of Arf1. (A) Arf1-HA was expressed in HeLa cells either alone or in the presence of Arf6 or Arf6Q67L. The fraction of Arf1 that was GTP-bound was determined using a GGA3-GAT pulldown assay. Shown is the average fold-increase in Arf1-GTP from four independent experiments ±1 SD. (B) Arf1-RFP, Arf6Q67L, and GFP-PM were coexpressed in Cos-7 cells, and cells were imaged live 24 h after transfection. (C) Arf1-RFP, GFP-EFA6, and Arf6 were coexpressed in Cos-7 cells and imaged 18 h after transfection. Bars, 10 μm.
Arf6-GTP Binds ARNO and Grp1 PH Domains
To examine which domains in ARNO are responsible for its interaction with GTP-bound Arf6, we tested several mutants of ARNO for their recruitment to the PM upon Arf6 expression. A point mutation in ARNO, E156K, renders the GEF catalytically inactive and unable to bind Arf1 under physiological conditions (Beraud-Dufour et al., 1998; Cherfils et al., 1998). We found that the ARNO E156K mutant was recruited on to the PM by Arf6 expression in a manner similar to that of wt ARNO, except that it did not form internal vesicles (see Figure 5A). This suggests that the catalytic activity of ARNO, and its ability to interact with Arfs through its Sec7 domain, is not important for Arf6-dependent recruitment of ARNO onto the PM. By contrast, a mutant of ARNO lacking the carboxyl terminal PH domain (ΔPH) remained cytosolic when it was coexpressed with Arf6 (Figure 5A). This suggests that the PH domain is necessary for the Arf6-mediated recruitment of ARNO onto the PM. Remarkably, a chimera of GFP fused to the ARNO PH domain [Figure 5A, PH(3G)] was recruited on to the PM in an Arf6-dependent manner, suggesting that the PH domain is both necessary and sufficient to recruit ARNO onto the PM. We confirmed that the interaction between the full-length ARNO and Arf6-GTP was mediated through the PH domain by coimmunoprecipitation studies. As shown in Figure 5B, like the full-length ARNO, the PH domain coimmunoprecipitated with both Arf6 and Arf6Q67L, and not with Arf6T27N. The coimmunoprecipitation shown was in the presence of cross-linking but could be observed in the absence of cross-linking (data not shown). In addition we found that the PH domain did not coimmunoprecipitate with wt Arf1, Arf1T31N, or Arf1Q71L (Figure 5B), suggesting that the Arf interaction with the ARNO PH domain is specific to Arf6.
Figure 5.
The PH domain of ARNO interacts with GTP-bound Arf6. (A) A flag-tagged catalytically inactive mutant of ARNO (ARNO EK), ARNO lacking the PH domain (ARNO ΔPH), GFP-tagged PH domain of ARNO triglycine variant (PH(3G)), PH domain of ARNO diglycine variant (PH(2G)) or GFP-tagged PH domain of Btk (Btk PH) were expressed alone or coexpressed with Arf6 in HeLa cells and stained with rabbit anti-Arf6 and either anti-Flag antibody (for ARNO EK and ARNO ΔPH) or mouse anti-GFP (for ARNO PH domains and Btk PH domain). For quantification of recruitment onto membranes see Supplementary Figure 1. (B) GFP-ARNO PH(3G) was expressed either alone or coexpressed with wt Arf1-HA or its mutants, or wt Arf6-HA or its mutants. Lysates from these cells were probed with anti-HA agarose, and immunoprecipitations were examined by Western blot using antibodies to HA (Arfs) and GFP (ARNO PH(3G)). (C) GFP-ARNO PH(2G) was expressed either alone or coexpressed with wt Arf6-HA or its mutants and examined with an anti-HA agarose pulldown as described. Bars, 10 μm.
Interestingly, each ARNO/Cytohesin member can generate two splice variants that differ in the number of glycine residues in the PH domain at a site near to the β1/β2 loop region that is involved in phosphoinositide binding (Klarlund et al., 2000; Ogasawara et al., 2000; Cronin et al., 2004). The triglycine (3G) variant used above, which is the predominant form of expressed ARNO (Ogasawara et al., 2000), recognizes both PIP2 and PIP3 with similar affinities, whereas the di-glycine (2G) variant has reduced affinity for PIP2 and preferentially binds PIP3 (Klarlund et al., 2000). We wondered whether the PIP specificity would alter the PH domain's ability to interact with Arf6. When we coexpressed the 2G variant of ARNO with Arf6, the PH domain was recruited onto the plasma membrane [Figure 5A, PH(2G)]. This recruitment was serum-dependent and not observed in cells treated with wortmannin, an inhibitor of PI3-kinase (not shown), indicating that the presence of PM PIP3 was required. We also confirmed an interaction between the 2G ARNO PH domain and Arf6 GTP in coimmunoprecipitation experiments (Figure 5C). Finally, the recruitment of the PH domains by Arf6 to the plasma membrane was specific for PH domains of ARNO/Cytohesin family GEFs, as the PH domain of Btk, which also recognizes PIP3, was not recruited to the plasma membrane by Arf6 expression (Figure 5A).
As shown earlier, Grp1 was also recruited onto the PM when coexpressed with active Arf6, raising the possibility that this recruitment might also be mediated by its PH domain because the ARNO family PH domains are highly conserved. We tested whether Arf6 could recruit Grp1 PH domains to the PM. The Grp1 PH domain is predominantly expressed in the 2G form (Ogasawara et al., 2000) and binds PIP3 (Klarlund et al., 2000). We found that Arf6 expression was capable of recruiting the Grp1 PH domain onto the PM (Figure 6A), and this recruitment was dependent on serum and was blocked by wortmannin treatment (data not shown) indicating a requirement for PIP3. The Grp1 PH domain also coprecipitated with Arf6 (Figure 6B), providing further evidence that Arf6-GTP can bind to PH domains of other ARNO family members.
Figure 6.
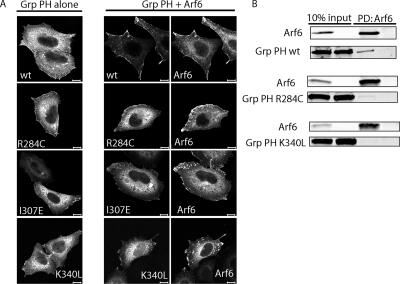
Recruitment of the PH domain of Grp1 to the PM requires an interaction with Arf6 and the ability to bind phospholipids. (A) YFP-Grp1 PH domain or its mutants were expressed alone or coexpressed with Arf6 in HeLa cells and stained with antibodies to Arf6 and GFP (Grp1 PH domains). For quantification of recruitment onto membranes see Supplementary Figure 1. (B) YFP-Grp1 PH domain or its mutants were expressed alone or coexpressed with Arf6-HA in HeLa cells. Lysates from these cells were probed with anti-HA agarose to precipitate Arf6-HA, and pulldowns were examined by Western blot with antibodies to HA (Arf6) and GFP (Grp1 PH domain). Bars, 10 μm.
Although PH domains of either phosphoinositide specificity could be recruited to the membrane by Arf6-GTP, we wondered whether phosphoinositide binding itself was required for this recruitment in cells. The R281C (Venkateswarlu et al., 1999) and R284C (Varnai et al., 2005) mutations within the PH domains of Cytohesin-1 and Grp1, respectively, have been shown to disrupt PM localization. This mutation in the Grp1 PH domain eliminates IP4 binding in vitro (Varnai et al., 2005). We tested whether the R284C mutant of the Grp1 PH domain could be recruited onto the plasma membrane and found that this mutant was not recruited onto the plasma membrane by Arf6 expression (Figure 6A). Additionally, the R284C mutant only weakly coprecipitated with Arf6 (Figure 6B), suggesting that phosphoinositide binding is critical to both Arf6-mediated membrane recruitment of the PH domain and its ability to interact with Arf6.
The Grp1 PH domain was recently shown to inhibit cell spreading when overexpressed in COS-7 cells (Varnai et al., 2005). In that study, two additional Grp1 PH domain mutants, I307E and K340L, were described that are capable of binding IP4 in vitro like the wt PH domain, but are unable to inhibit cell spreading in COS-7 cells, suggesting that they are deficient in binding to a putative protein partner necessary for the dominant negative activity of the Grp1 PH domain (Varnai et al., 2005). Given our findings that Arf6 can recruit the ARNO/Grp1 PH domains to the PM, we wanted to determine whether Arf6 might be the putative protein postulated in that study. Therefore, we tested whether the Grp1 PH domain mutants, I307E and K340L, were capable of being recruited to the PM by Arf6 and found that they remained cytosolic in cells when Arf6 was overexpressed (Figure 6A). Likewise, we found that the Grp1 PH K340L mutant did not interact with Arf6 in a coprecipitation experiment (Figure 6B). We were unable to ascertain whether the I307E mutant interacted with Arf6 because it nonspecifically interacted with the HA agarose (data not shown). Because the Grp1 PH K340L mutant does not inhibit cell spreading (Varnai et al., 2005) and does not bind to Arf6, these results suggest that the dominant negative effect of the wt PH domain is dependent on its ability to interact with and sequester endogenous Arf6-GTP. Consistent with this, expression of Arf6T27N, which inhibits activation of endogenous Arf6, also inhibits cell spreading (Song et al., 1998). These results suggest that the Grp1 PH domain requires both an interaction with PIP3 and an interaction with Arf6 GTP to be recruited onto the PM. The conservation of these critical residues within the ARNO PH domain (and other family members) suggests that these residues are also critical for its interaction with Arf6 and phosphoinositides.
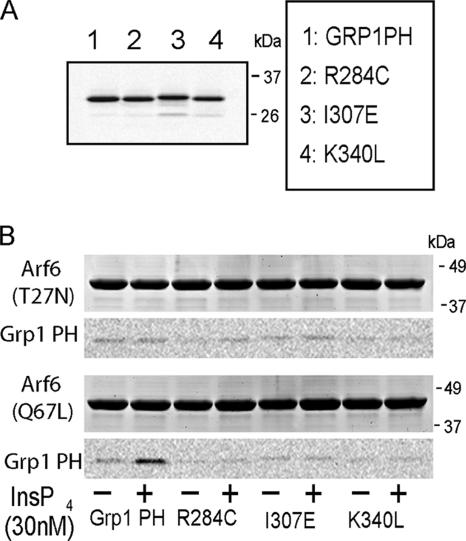
The ability of PH domains from ARNO and Grp1 to interact with Arf6 GTP could be due to a direct interaction or the formation of an indirect complex. To examine whether the interaction was direct, we tested the ability of purified recombinant Grp1 PH domain and several PH domain mutants to interact with Arf 6Q67L and Arf6T27N immobilized on a column. We found that the wt Grp1 PH domain could bind to immobilized Arf6Q67L, but only in the presence of IP4 (Figure 7), suggesting that the efficient and direct interaction of the PH domains with Arf6-GTP requires phosphoinositides. The wt Grp1 did not interact with immobilized Arf6T27N, confirming that the PH domain interaction requires a GTP-bound conformation (Figure 7). The Grp1 R284C PH domain mutant that does not interact with lipids and the Grp1 K340L or I307E mutants that do not function as dominant negatives in cells were unable to bind to Arf6Q67L regardless of the presence of IP4 (Figure 7). This demonstrates that the binding of these PH domains to the PM involves dual recognition of specific phosphoinositides and Arf6-GTP.
Figure 7.
The interaction between Arf6-GTP and the Grp PH domain is direct and requires phosphoinositide. (A) Fluorescence scan of 20 μg purified YFP Grp1 PH domain or its mutants R284C, I307E, or K340L (B) Twenty micrograms of purified YFP Grp1 PH domain or its mutants was incubated with 20 μg purified GST-Arf6 T27N or GST-Arf6 Q67L immobilized on glutathione Sepharose beads in the presence (+) or absence of 30 nM Ins(1,3,4,5)P4. Bound proteins were resolved by SDS-PAGE, scanned for fluorescence of the YFP PH domains, and subsequently stained with Coomassie to visualize the GST-Arf6. The wt YFP Grp1 PH domain bound to GST-Arf6Q67L only in the presence of Ins (1,3,4,5)P4.
DISCUSSION
Defining Arf specificity for each of the Arf GEFs is critical for understanding the function of Arf proteins in cells (Jackson and Casanova, 2000; D'Souza-Schorey and Chavrier, 2006). Here, we set out to explore the relationship between ARNO/Cytohesin family GEFs and their potential substrates Arf1 and Arf6. We found that ARNO prefers Arf1 over Arf6 as a substrate in cells and that ARNO can activate Arf1 at the PM to form membrane ruffles and dynamic macropinosomes. Arf6, instead of acting as a substrate for ARNO, acts upstream of ARNO family GEFs, recruiting them to the PM through a direct interaction with their PH domains. The ability of Arf6-GTP in the presence of either PIP2 or PIP3 to bind directly to PH domains of ARNO family GEFs implies that active Arf6 serves as a PM adaptor for GEF recruitment and signaling. Consistent with this, we observed an increase in Arf1 activation and movement of Arf1 to the PM in cells expressing active Arf6. We propose a new model for Arf function in the periphery where Arf6-GTP acts to recruit ARNO/Cytohesin family GEFs to the PM allowing the subsequent activation of additional Arfs, such as Arf1. This cascade of Arf activation allows the cell to fine-tune and regulate Arf activation during cell signaling.
ARNO GEFs Activate Golgi-associated Arfs at the PM
The ambiguity of assigning Arf specificity for ARNO family GEFs (Jackson and Casanova, 2000; D'Souza-Schorey and Chavrier, 2006) led us to test directly in cells whether ARNO would prefer Arf6 or Arf1 as a substrate. We showed that ARNO clearly preferred Arf1 as a substrate, and in living cells Arf1 was recruited onto ARNO-stimulated PM ruffles and resulting macropinosomes (Figures 1 and 2). This observation is in complete agreement with most biochemical assays performed with recombinant ARNO (Frank et al., 1998; Macia et al., 2001), Cytohesin-1 (Pacheco-Rodriguez et al., 1998), and Grp1 (Franco et al., 1998) that clearly show better exchange activity for Arf1 than for Arf6. In contrast, both EFA6 and Brags 1 and 2, bona fide Arf6 GEFs, activate Arf6 better than Arf1 in biochemical assays (Franco et al., 1998; Someya et al., 2001). Although we cannot exclude the possibility that ARNO family GEFs may activate Arf6 in some instances, the preponderance of evidence suggests that ARNO family GEFs activate Arf1 or other “Golgi-associated” Arfs in the cell periphery. Previous studies implicating Arf6 in ARNO activities might be due to the ability of Arf6-GTP to recruit ARNO to the membrane.
The idea that Arfs 1, 3, 4, and 5 could function at the PM has been largely ignored because these Arfs are observed localizing to the Golgi complex (Volpicelli-Daley et al., 2005). Yet these Arfs, especially Arf1, which are much more abundant than Arf6 (Cavenagh et al., 1996), cycle through the cytoplasm after inactivation and are thus available to be activated on other membrane compartments in the periphery. A lack of specific antibodies that could detect localization of these Arfs to peripheral structures or the PM has hampered our ability to consider extra-Golgi roles for these Arfs. However, Arf1 has been observed associated with apical endosomes in kidney proximal tubules (Maranda et al., 2001) and Arf1-GFP is transiently associated with the PM during insulin stimulation (Li et al., 2003). As we show here, expression of ARNO in mammalian cells can dramatically recruit and activate Arf1 at the PM (see Figure 2B). In support of this observation, several years ago two studies reported that overexpression of ARNO (Monier et al., 1998) or Grp1 (Franco et al., 1998) led to a BFA-like disassembly of the Golgi complex. At the time, these observations were attributed to ARNO-mediated activation of Arf1 at the Golgi causing vesiculation of the Golgi complex. In light of our findings, this observation can now be explained by the ability of ARNO and Grp1 to compete with Golgi GEFs for endogenous “Golgi-associated” Arfs. We also observed that high expression of ARNO or Grp1 can titrate enough of the endogenous Arfs away from the Golgi to cause disassembly of the Golgi apparatus. Consistent with this explanation, we have found that overexpression of Arf1 can rescue the Golgi disassembly in ARNO or Grp1 expressing cells (our unpublished observations).
ARNO PH Domains Bind Arf6-GTP and Phosphoinositides
The recruitment of ARNO to the PM by Arf6 was due to a specific and direct interaction between Arf6-GTP and the PH domains of ARNO family GEFs. The dual requirement of phosphoinositide and Arf6-GTP for ARNO PH domain recruitment reveals that PH domains likely engage in protein-protein interactions in the context of specific phosphoinositides to specify their localization within particular membrane compartments (Varnai et al., 2002; Balla, 2005). Each of the ARNO family GEFs target to unique membrane environments through alternative splicing of the PH domain. The splice forms generated depend on the ARNO isoform. For example, for ARNO and cytohesin 1, 80–90% of the transcripts are of the 3G form, which can bind to either PIP2 or PIP3, whereas for Grp1 80% is of the 2G form that preferentially binds PIP3 (Klarlund et al., 2000; Ogasawara et al., 2000). Arf6-GTP was able to bind the PH domains of Grp1 (Figure 6) and either the 2G or 3G splice variant of the PH domain of ARNO (Figure 3).
In addition to Arf6-GTP recruiting ARNO family PH domains to the PM, Arf1-GTP has been shown to recruit PH domains of FAPP1 and OSBP (Godi et al., 2004) and ARHGAP10 (Dubois et al., 2005) to the Golgi. The ability of several PH domains to interact with active Arfs is intriguing given the ability of Arfs to regulate lipid metabolism. This could allow Arfs to regulate cellular events by altering lipid metabolism and coordinately controlling the localized association and activity of downstream PH domain-containing molecules to specific places on biological membranes. Because Arf6 activates PIP 5-kinase at the PM, the PIP2 formed would thus facilitate recruitment of the 3G PH domain to the sites where Arf6 is active. Because PIP2 is a precursor for PIP3, Arf6 in this environment could recruit the 2G PH domains to these areas of the PM. This type of reinforcement mechanism parallels that of the FAPP and OSBP PH domains. Binding to PI4P and Arf1-GTP localizes these PH domains to the Golgi (Godi et al., 2004). Arf1 is known to recruit PI 4-kinase IIIβ to the Golgi (Godi et al., 1999); thus, the FAPP and OSBP PH domains act as coincidence detectors of Arf1 activity and PI4P production in a manner that reinforces their localization to sites at which Arf1 is active. A third protein ARHGAP 10 has been shown to interact with Arf1-GTP through its PH domain and a region just adjacent to its PH domain (Dubois et al., 2005).
Several recent studies including this one have revealed that secondary sites on GEFs, adjacent to but not within the catalytic site, can bind to GTP-bound GTPases and either create a positive feedback mechanism or further downstream signaling by relating activation of one GTPase to that of another. For example, son of sevenless, a Ras GEF, binds to Ras-GTP at a site adjacent to the catalytic core. Binding of Ras-GTP to son of sevenless enhances the exchange activity of this GEF for Ras (Margarit et al., 2003). Dbs, an exchange factor for Rac, binds to GTP-bound Cdc42 through the PH domain adjacent to its catalytic region, creating cross-talk between the two Rho family GEFs (Cheng et al., 2004). Similar to the Dbs study, we show that ARNO interacts with Arf6-GTP through its PH domain adjacent to the catalytic Sec7 domain and this can lead to the activation of “Golgi-associated” Arfs like Arf1 at the PM.
Sequential Activation of Arfs
Our model suggests that Arf6-GTP at the PM might serve as an adaptor for recruitment of ARNO GEFs to sites at the PM leading to Arf1 (or 3, 4, or 5) activation. Although all Arfs possess the ability to activate PIP 5-kinase and phospholipase D, Arf1 recruits various coat proteins and PI 4-kinase IIIβ onto the Golgi, which could represent Arf1-specific activities. We have observed recruitment of PI 5-kinase IIIβ onto endosomes containing ARNO (data not shown), suggesting that Arf1 activation on the endosomes could promote the resynthesis of PI4P after PIP phosphatase activity. Another interesting possibility is that Arf1-GTP recruits a specific GAP to the plasma membrane that sets up a scaffold for further signaling (Nie et al., 2003). ASAP1 is a multidomain peripheral GAP with SH3, ankryn repeats, and proline-rich domains that acts preferentially on Arf1 and Arf5, and it is possible that ARNO activation of Arf1 at the PM sets up signaling through this GAP (Brown et al., 1998; Yoon et al., 2004). Finally, Arf1 and other cytosolic Arfs might be able to function acutely at the PM during a signaling event to amplify, attenuate, or alter the initial signaling coming from Arf6-GTP. Indeed, a recent study on the mechanism of Fc-mediated phagocytosis has found evidence for the sequential activation of Arf6 followed by Arf1 during phagosome formation (Beemiller et al., 2006). It will be interesting to examine whether an ARNO family GEF is recruited by Arf6-GTP to mediate the subsequent activation of Arf1 in this case.
A model for Arfs acting sequentially in a pathway might also be relevant at the Golgi complex. There are multiple Arfs and multiple GEFs associated with the Golgi complex and sorting out which GEFs are working on which Arfs poses a special challenge (Jackson and Casanova, 2000; Donaldson et al., 2005). A recent study that examined the requirement for Arfs 1–5 at the Golgi found that knock down of any one individual Arf had only subtle phenotypes on Golgi structure and transport, whereas knockdown of pairs of Arfs revealed distinct functions of these pairs at specific sites in the ER–Golgi system (Volpicelli-Daley et al., 2005). Our model provides a possible mechanism for this phenomenon and raises the possibility that one Arf in its active state could recruit the GEF to activate another Arf at the Golgi.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jim Casanova (University of Virginia) and Roger Tsien (UCSD) for plasmids and Cathy Jackson and Ed Korn for comments on the manuscript. This research was supported by the Intramural Research Programs of the National Heart, Lung, and Blood Institute and the National Institute of Child Health and Human Development.
Abbreviations used:
- GAP
GTPase-activating protein
- GEF
guanine nucleotide exchange factor
- PH
pleckstrin homology
- PIP2
phosphatidylinositol 4,5 bisphosphate
- PIP3,
phosphatidylinositol 3,4,5 trisphosphate
- PIP5-kinase
phosphatidylinositol 4-phosphate 5-kinase
- PM
plasma membrane.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-11-0998) on April 4, 2007.

 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Antonny B., Beraud-Dufour S., Chardin P., Chabre M. N-terminal hydrophobic residues of the G-protein ADP-ribosylation factor-1 insert into membrane phospholipids upon GDP to GTP exchange. Biochemistry. 1997;36:4675–4684. doi: 10.1021/bi962252b. [DOI] [PubMed] [Google Scholar]
- Balla T. Inositol-lipid binding motifs: signal integrators through protein-lipid and protein-protein interactions. J. Cell Sci. 2005;118:2093–2104. doi: 10.1242/jcs.02387. [DOI] [PubMed] [Google Scholar]
- Beemiller P., Hoppe A. D., Swanson J. A. A phosphatidylinositol- 3-kinase-dependent signal transition regulates ARF1 and ARF6 during Fcgamma receptor-mediated phagocytosis. PLoS Biol. 2006;4:e162. doi: 10.1371/journal.pbio.0040162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beraud-Dufour S., Robineau S., Chardin P., Paris S., Chabre M., Cherfils J., Antonny B. A glutamic finger in the guanine nucleotide exchange factor ARNO displaces Mg2+ and the beta-phosphate to destabilize GDP on ARF1. EMBO J. 1998;17:3651–3659. doi: 10.1093/emboj/17.13.3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown F. D., Rozelle A. L., Yin H. L., Balla T., Donaldson J. G. Phosphatidylinositol 4,5-bisphosphate and Arf6-regulated membrane traffic. J. Cell Biol. 2001;154:1007–1017. doi: 10.1083/jcb.200103107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown M. T., Andrade J., Radhakrishna H., Donaldson J. G., Cooper J. A., Randazzo P. A. ASAP1, a phospholipid-dependent arf GTPase-activating protein that associates with and is phosphorylated by Src. Mol. Cell. Biol. 1998;18:7038–7051. doi: 10.1128/mcb.18.12.7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell R. E., Tour O., Palmer A. E., Steinbach P. A., Baird G. S., Zacharias D. A., Tsien R. Y. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA. 2002;99:7877–7882. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavenagh M. M., Whitney J. A., Carroll K., Zhang C., Boman A. L., Rosenwald A. G., Mellman I., Kahn R. A. Intracellular distribution of Arf proteins in mammalian cells. Arf6 is uniquely localized to the plasma membrane. J. Biol. Chem. 1996;271:21767–21774. doi: 10.1074/jbc.271.36.21767. [DOI] [PubMed] [Google Scholar]
- Cheng L., Mahon G. M., Kostenko E. V., Whitehead I. P. Pleckstrin homology domain-mediated activation of the rho-specific guanine nucleotide exchange factor Dbs by Rac1. J. Biol. Chem. 2004;279:12786–12793. doi: 10.1074/jbc.M313099200. [DOI] [PubMed] [Google Scholar]
- Cherfils J., Menetrey J., Mathieu M., Le Bras G., Robineau S., Beraud-Dufour S., Antonny B., Chardin P. Structure of the Sec7 domain of the Arf exchange factor ARNO. Nature. 1998;392:101–105. doi: 10.1038/32210. [DOI] [PubMed] [Google Scholar]
- Cronin T. C., DiNitto J. P., Czech M. P., Lambright D. G. Structural determinants of phosphoinositide selectivity in splice variants of Grp1 family PH domains. EMBO J. 2004;23:3711–3720. doi: 10.1038/sj.emboj.7600388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Souza-Schorey C., Chavrier P. ARF proteins: roles in membrane traffic and beyond. Nat. Rev. Mol. Cell Biol. 2006;7:347–358. doi: 10.1038/nrm1910. [DOI] [PubMed] [Google Scholar]
- Donaldson J. G., Honda A., Weigert R. Multiple activities for Arf1 at the Golgi complex. Biochim. Biophys. Acta. 2005;1744:364–373. doi: 10.1016/j.bbamcr.2005.03.001. [DOI] [PubMed] [Google Scholar]
- Dubois T., Paleotti O., Mironov A. A., Fraisier V., Stradal T. E., De Matteis M. A., Franco M., Chavrier P. Golgi-localized GAP for Cdc42 functions downstream of ARF1 to control Arp2/3 complex and F-actin dynamics. Nat. Cell Biol. 2005;7:353–364. doi: 10.1038/ncb1244. [DOI] [PubMed] [Google Scholar]
- Dunphy J. L., Moravec R., Ly K., Lasell T. K., Melancon P., Casanova J. E. The Arf6 GEF GEP100/BRAG2 regulates cell adhesion by controlling endocytosis of beta1 integrins. Curr. Biol. 2006;16:315–320. doi: 10.1016/j.cub.2005.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco M., Boretto J., Robineau S., Monier S., Goud B., Chardin P., Chavrier P. ARNO3, a Sec7-domain guanine nucleotide exchange factor for ADP ribosylation factor 1, is involved in the control of Golgi structure and function. Proc. Natl. Acad. Sci. USA. 1998;95:9926–9931. doi: 10.1073/pnas.95.17.9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco M., Peters P. J., Boretto J., van Donselaar E., Neri A., D'Souza-Schorey C., Chavrier P. EFA6, a sec7 domain-containing exchange factor for ARF6, coordinates membrane recycling and actin cytoskeleton organization. EMBO J. 1999;18:1480–1491. doi: 10.1093/emboj/18.6.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank S., Upender S., Hansen S. H., Casanova J. E. ARNO is a guanine nucleotide exchange factor for ADP-ribosylation factor 6. J. Biol. Chem. 1998;273:23–27. doi: 10.1074/jbc.273.1.23. [DOI] [PubMed] [Google Scholar]
- Godi A., Di Campli A., Konstantakopoulos A., Di Tullio G., Alessi D. R., Kular G. S., Daniele T., Marra P., Lucocq J. M., De Matteis M. A. FAPPs control Golgi-to-cell-surface membrane traffic by binding to ARF and PtdIns(4)P. Nat. Cell Biol. 2004;6:393–404. doi: 10.1038/ncb1119. [DOI] [PubMed] [Google Scholar]
- Godi A., Santone I., Pertile P., Marra P., Di Tullio G., Luini A., Corda D., De Matteis M. A. ADP-ribosylation factor regulates spectrin skeleton assembly on the Golgi complex by stimulating phosphatidylinositol 4,5-bisphosphate synthesis. Biochem. Soc. Trans. 1999;27:638–642. doi: 10.1042/bst0270638. [DOI] [PubMed] [Google Scholar]
- Hernandez-Deviez D. J., Casanova J. E., Wilson J. M. Regulation of dendritic development by the ARF exchange factor ARNO. Nat. Neurosci. 2002;5:623–624. doi: 10.1038/nn865. [DOI] [PubMed] [Google Scholar]
- Honda A., Al-Awar O. S., Hay J. C., Donaldson J. G. Targeting of Arf-1 to the early Golgi by membrin, an ER-Golgi SNARE. J. Cell Biol. 2005;168:1039–1051. doi: 10.1083/jcb.200409138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda A., et al. Phosphatidylinositol 4-phosphate 5-kinase alpha is a downstream effector of the small G protein ARF6 in membrane ruffle formation. Cell. 1999;99:521–532. doi: 10.1016/s0092-8674(00)81540-8. [DOI] [PubMed] [Google Scholar]
- Jackson C. L., Casanova J. E. Turning on ARF: the Sec7 family of guanine-nucleotide-exchange factors. Trends Cell Biol. 2000;10:60–67. doi: 10.1016/s0962-8924(99)01699-2. [DOI] [PubMed] [Google Scholar]
- Klarlund J. K., Rameh L. E., Cantley L. C., Buxton J. M., Holik J. J., Sakelis C., Patki V., Corvera S., Czech M. P. Regulation of GRP1-catalyzed ADP ribosylation factor guanine nucleotide exchange by phosphatidylinositol 3,4,5-trisphosphate. J. Biol. Chem. 1998;273:1859–1862. doi: 10.1074/jbc.273.4.1859. [DOI] [PubMed] [Google Scholar]
- Klarlund J. K., Tsiaras W., Holik J. J., Chawla A., Czech M. P. Distinct polyphosphoinositide binding selectivities for pleckstrin homology domains of GRP1-like proteins based on diglycine versus triglycine motifs. J. Biol. Chem. 2000;275:32816–32821. doi: 10.1074/jbc.M002435200. [DOI] [PubMed] [Google Scholar]
- Langille S. E., Patki V., Klarlund J. K., Buxton J. M., Holik J. J., Chawla A., Corvera S., Czech M. P. ADP-ribosylation factor 6 as a target of guanine nucleotide exchange factor GRP1. J. Biol. Chem. 1999;274:27099–27104. doi: 10.1074/jbc.274.38.27099. [DOI] [PubMed] [Google Scholar]
- Li H. S., Shome K., Rojas R., Rizzo M. A., Vasudevan C., Fluharty E., Santy L. C., Casanova J. E., Romero G. The guanine nucleotide exchange factor ARNO mediates the activation of ARF and phospholipase D by insulin. BMC Cell Biol. 2003;4:13. doi: 10.1186/1471-2121-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Duden R., Phair R. D., Lippincott-Schwartz J. ArfGAP1 dynamics and its role in COPI coat assembly on Golgi membranes of living cells. J. Cell Biol. 2005;168:1053–1063. doi: 10.1083/jcb.200410142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macia E., Chabre M., Franco M. Specificities for the small G proteins ARF1 and ARF6 of the guanine nucleotide exchange factors ARNO and EFA6. J. Biol. Chem. 2001;276:24925–24930. doi: 10.1074/jbc.M103284200. [DOI] [PubMed] [Google Scholar]
- Maranda B., Brown D., Bourgoin S., Casanova J. E., Vinay P., Ausiello D. A., Marshansky V. Intra-endosomal pH-sensitive recruitment of the Arf-nucleotide exchange factor ARNO and Arf6 from cytoplasm to proximal tubule endosomes. J. Biol. Chem. 2001;276:18540–18550. doi: 10.1074/jbc.M011577200. [DOI] [PubMed] [Google Scholar]
- Margarit S. M., Sondermann H., Hall B. E., Nagar B., Hoelz A., Pirruccello M., Bar-Sagi D., Kuriyan J. Structural evidence for feedback activation by Ras.GTP of the Ras-specific nucleotide exchange factor SOS. Cell. 2003;112:685–695. doi: 10.1016/s0092-8674(03)00149-1. [DOI] [PubMed] [Google Scholar]
- Menetrey J., Macia E., Pasqualato S., Franco M., Cherfils J. Structure of Arf6-GDP suggests a basis for guanine nucleotide exchange factors specificity. Nat. Struct. Biol. 2000;7:466–469. doi: 10.1038/75863. [DOI] [PubMed] [Google Scholar]
- Monier S., Chardin P., Robineau S., Goud B. Overexpression of the ARF1 exchange factor ARNO inhibits the early secretory pathway and causes the disassembly of the Golgi complex. J. Cell Sci. 1998;111(Pt 22):3427–3436. doi: 10.1242/jcs.111.22.3427. [DOI] [PubMed] [Google Scholar]
- Nie Z., Hirsch D. S., Randazzo P. A. Arf and its many interactors. Curr. Opin. Cell Biol. 2003;15:396–404. doi: 10.1016/s0955-0674(03)00071-1. [DOI] [PubMed] [Google Scholar]
- Ogasawara M., Kim S. C., Adamik R., Togawa A., Ferrans V. J., Takeda K., Kirby M., Moss J., Vaughan M. Similarities in function and gene structure of cytohesin-4 and cytohesin-1, guanine nucleotide-exchange proteins for ADP-ribosylation factors. J. Biol. Chem. 2000;275:3221–3230. doi: 10.1074/jbc.275.5.3221. [DOI] [PubMed] [Google Scholar]
- Pacheco-Rodriguez G., Meacci E., Vitale N., Moss J., Vaughan M. Guanine nucleotide exchange on ADP-ribosylation factors catalyzed by cytohesin-1 and its Sec7 domain. J. Biol. Chem. 1998;273:26543–26548. doi: 10.1074/jbc.273.41.26543. [DOI] [PubMed] [Google Scholar]
- Pasqualato S., Menetrey J., Franco M., Cherfils J. The structural GDP/GTP cycle of human Arf6. EMBO Rep. 2001;2:234–238. doi: 10.1093/embo-reports/kve043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters P. J., Hsu V. W., Ooi C. E., Finazzi D., Teal S. B., Oorschot V., Donaldson J. G., Klausner R. D. Overexpression of wild-type and mutant ARF1 and ARF6, distinct perturbations of nonoverlapping membrane compartments. J. Cell Biol. 1995;128:1003–1017. doi: 10.1083/jcb.128.6.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presley J. F., Ward T. H., Pfeifer A. C., Siggia E. D., Phair R. D., Lippincott-Schwartz J. Dissection of COPI and Arf1 dynamics in vivo and role in Golgi membrane transport. Nature. 2002;417:187–193. doi: 10.1038/417187a. [DOI] [PubMed] [Google Scholar]
- Radhakrishna H., Al-Awar O., Khachikian Z., Donaldson J. G. ARF6 requirement for Rac ruffling suggests a role for membrane trafficking in cortical actin rearrangements. J. Cell Sci. 1999;112(Pt 6):855–866. doi: 10.1242/jcs.112.6.855. [DOI] [PubMed] [Google Scholar]
- Radhakrishna H., Donaldson J. G. ADP-ribosylation factor 6 regulates a novel plasma membrane recycling pathway. J. Cell Biol. 1997;139:49–61. doi: 10.1083/jcb.139.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozelle A. L., Machesky L. M., Yamamoto M., Driessens M. H., Insall R. H., Roth M. G., Luby-Phelps K., Marriott G., Hall A., Yin H. L. Phosphatidylinositol 4,5-bisphosphate induces actin-based movement of raft-enriched vesicles through WASP-Arp2/3. Curr. Biol. 2000;10:311–320. doi: 10.1016/s0960-9822(00)00384-5. [DOI] [PubMed] [Google Scholar]
- Santy L. C., Casanova J. E. Activation of ARF6 by ARNO stimulates epithelial cell migration through downstream activation of both Rac1 and phospholipase D. J. Cell Biol. 2001;154:599–610. doi: 10.1083/jcb.200104019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santy L. C., Frank S. R., Hatfield J. C., Casanova J. E. Regulation of ARNO nucleotide exchange by a PH domain electrostatic switch. Curr. Biol. 1999;9:1173–1176. doi: 10.1016/S0960-9822(00)80019-6. [DOI] [PubMed] [Google Scholar]
- Shmuel M., Santy L. C., Frank S., Avrahami D., Casanova J. E., Altschuler Y. ARNO through its coiled-coil domain regulates endocytosis at the apical surface of polarized epithelial cells. J. Biol. Chem. 2006;281:13300–13308. doi: 10.1074/jbc.M513723200. [DOI] [PubMed] [Google Scholar]
- Someya A., Sata M., Takeda K., Pacheco-Rodriguez G., Ferrans V. J., Moss J., Vaughan M. ARF-GEP(100), a guanine nucleotide-exchange protein for ADP-ribosylation factor 6. Proc. Natl. Acad. Sci. USA. 2001;98:2413–2418. doi: 10.1073/pnas.051634798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J., Khachikian Z., Radhakrishna H., Donaldson J. G. Localization of endogenous ARF6 to sites of cortical actin rearrangement and involvement of ARF6 in cell spreading. J. Cell Sci. 1998;111(Pt 15):2257–2267. doi: 10.1242/jcs.111.15.2257. [DOI] [PubMed] [Google Scholar]
- Tsuchiya M., Price S. R., Tsai S. C., Moss J., Vaughan M. Molecular identification of ADP-ribosylation factor mRNAs and their expression in mammalian cells. J. Biol. Chem. 1991;266:2772–2777. [PubMed] [Google Scholar]
- Varnai P., Bondeva T., Tamas P., Toth B., Buday L., Hunyady L., Balla T. Selective cellular effects of overexpressed pleckstrin-homology domains that recognize PtdIns(3,4,5)P3 suggest their interaction with protein binding partners. J. Cell Sci. 2005;118:4879–4888. doi: 10.1242/jcs.02606. [DOI] [PubMed] [Google Scholar]
- Varnai P., Lin X., Lee S. B., Tuymetova G., Bondeva T., Spat A., Rhee S. G., Hajnoczky G., Balla T. Inositol lipid binding and membrane localization of isolated pleckstrin homology (PH) domains. Studies on the PH domains of phospholipase C delta 1 and p130. J. Biol. Chem. 2002;277:27412–27422. doi: 10.1074/jbc.M109672200. [DOI] [PubMed] [Google Scholar]
- Venkateswarlu K., Gunn-Moore F., Tavare J. M., Cullen P. J. EGF-and NGF-stimulated translocation of cytohesin-1 to the plasma membrane of PC12 cells requires PI 3-kinase activation and a functional cytohesin-1 PH domain. J. Cell Sci. 1999;112(Pt 12):1957–1965. doi: 10.1242/jcs.112.12.1957. [DOI] [PubMed] [Google Scholar]
- Volpicelli-Daley L. A., Li Y., Zhang C. J., Kahn R. A. Isoform-selective effects of the depletion of ADP-ribosylation factors 1–5 on membrane traffic. Mol. Biol. Cell. 2005;16:4495–4508. doi: 10.1091/mbc.E04-12-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon H. Y., Jacques K., Nealon B., Stauffer S., Premont R. T., Randazzo P. A. Differences between AGAP1, ASAP1 and Arf GAP1 in substrate recognition: interaction with the N-terminus of Arf1. Cell Signal. 2004;16:1033–1044. doi: 10.1016/j.cellsig.2004.02.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.