Abstract
The Mas-related genes (Mrgs) comprise a family of >50 G protein-coupled receptors (GPCRs), many of which are expressed in specific subsets of nociceptive sensory neurons in mice. In contrast, humans contain a related but nonorthologous family of genes, called MrgXs or sensory neuron-specific receptors, of which many fewer appear to be expressed in sensory neurons. To determine whether the diversity of murine Mrgs is generic to rodents or is an atypical feature of mice, we characterized MrgA, MrgB, MrgC, and MrgD subfamilies in rat and gerbil. Surprisingly, although mice have ≈22 MrgA and ≈14 MrgC genes, rats and gerbils have just a single MrgA and MrgC gene. This murine-specific expansion likely reflects recent retrotransposon-mediated unequal crossover events. The expression of Mrgs in rat sensory ganglia suggests that the extensive cellular diversity in mice can be simplified to a core subset of approximately four different genes (MrgA, MrgB, MrgC, and MrgD), defining a similar number of neuronal subpopulations. Our results suggest more generally that mouse–human genomic comparisons may sometimes reveal differences atypical of rodents.
In many sensory systems, including taste, olfaction, and vision, primary sensory neurons express diverse families of seven transmembrane domain G protein-coupled receptors (GPCRs) to detect and discriminate among various chemical and visual stimuli (1–3). The expansion of diverse GPCR families is enabled by the fact that functional receptors and their transcriptional controls often reside within small (≈10-kb) segments of DNA present in tandem arrays (4–6). The success of this molecular unit is reflected in the fact that GPCRs constitute the largest single gene family in all metazoan genomes (7–10).
Recent studies have identified a novel family of GPCRs specifically expressed in primary nociceptive sensory neurons in mice and humans (11, 12). In vitro studies suggest that some of these receptors can be activated by neuropeptides that contain C-terminal -RF(Y)amide or -RF(Y)G motifs (11–13). Members of this family have been referred to as Mas-related genes (Mrgs) (11, 14, 15). Alternatively, in humans they have been called sensory neuron-specific receptors (SNSRs) (12). In mice, the Mrg family is comprised of six single-copy genes (MrgD, MrgE, MrgF/RTA, MrgG, MrgH/GPR90, and MAS1), as well as three large clades or subfamilies (MrgA, MrgB, and MrgC) that together comprise ≈50 distinct sequences. The differential expression of various mouse (m)Mrgs defines a surprisingly diverse axis of cellular heterogeneity among murine nociceptive sensory neurons, the functional significance of which is currently unclear (11).
In contrast to the extensive sequence diversity exhibited by mMrgA, mMrgB, and mMrgC subfamilies, in humans there are only four functional hMrgX/SNSR genes. Although some of these genes are specifically expressed in nociceptive sensory neurons like their murine counterparts, none of the human and mouse genes are strictly orthologous (11). This difference raises the question of whether the extensive Mrg sequence diversity characteristic of mice is generic to rodents, perhaps reflecting differences with humans in nociceptive physiology, or rather reflects genomic expansion events unique to mice.
To address this question, we have characterized the complement of Mrg genes in two other rodent species, rat and gerbil.
Our results indicate that the extreme diversity of murine Mrgs is an atypical feature of mice. These findings simplify the problem of understanding the functional significance of Mrg sequence diversity in rodents to a core set of approximately four different genes (MrgA, MrgB, MrgC, and MrgD), defining a similar number of neuronal cell populations.
Methods
Distance Calculations. Representative nucleotide sequences from the coding regions of MrgA (mMrgA1-A8, rMrgA), MrgB (mMrgB1-B5, mMrgB7-B8, rMrgB1, rMrgB2, rMrgB5-B6, rMrgB8), and MrgC (mMrgC1, mMrgC2, mMrgC7, mMrgC11, rMrgC) subfamilies were aligned with clustalw and then manually aligned on a codon-by-codon basis. Nucleotides that introduced gaps within a codon were removed from the analysis (complete-deletion option). The program diverge was then used to calculate the number of pairwise synonymous (Ks) and nonsynonymous (Ka) nucleotide substitutions between mMrgAs, mMrgBs, rMrgBs, and mMrgCs by using the method described by Li et al. (16–18) with recent modifications. A neutral substitution rate of 4.5 = 10–9 substitutions per synonymous site (Ks) per year was used to calculate evolutionary distance between each pair of sequences (10). This rate was based on the assumption that humans and rodents last shared a common ancestor 75 million years ago (MYA), and it is similar to the neutral substitution rate for rodents calculated by others (19).
The details describing how rat and gerbil Mrgs were cloned as well as methods for in situ hybridizations and Southern blot hybridizations can be found in Supporting Methods, which is published as supporting information on the PNAS web site, www.pnas.org.
Results
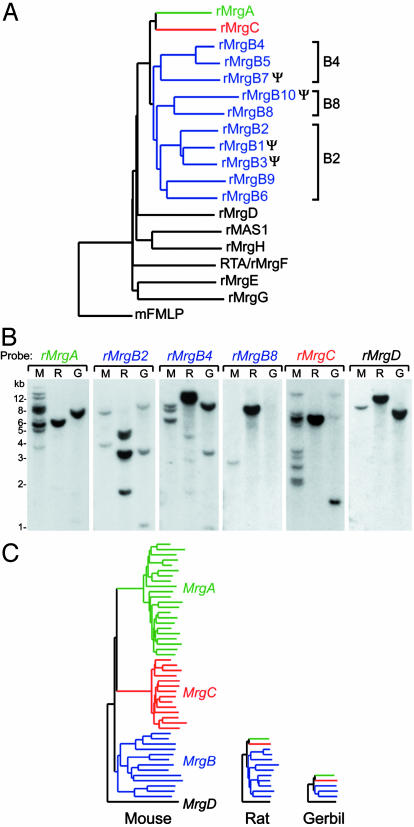
Identification of the Rat Mrg Family. Searches of the January 21, 2003, release of the rat genome using mouse Mrgs as query sequences revealed that the rat has a single copy each of rat (r)MrgA and rMrgC (Fig. 1A). These two rat genes have been previously characterized as an adenine receptor and rSNSR, respectively (12, 20). However, the total number of rat Mrgs was not systematically examined in these previous studies. Our searches also identified 10 rMrgBs, many of which are orthologous to at least one of the mouse MrgB genes (Fig. 6, which is published as supporting information on the PNAS web site). The mouse and rat MrgB subfamily was divided further into phylogenetically defined B2, B4, and B8 subdivisions (Figs. 1 A and 6).
Fig. 1.
Analysis of the rat and gerbil Mrg families. (A) Phylogenetic analysis of the rat Mrg family. The program clustalw was used to align rat MRG protein sequences and assemble them into a dendrogram by using the neighbor-joining method. The mouse formyl peptide receptor 1 (mFMLP) was used as the outgroup. Genes that fall into the B2, B4, and B8 subdivisions are bracketed. Ψ, predicted pseudogenes. (B) Southern blot analysis of rodent Mrgs. Each lane contains 9 μgof BglII digested liver genomic DNA from mouse (M), rat (R), or gerbil (G). Blots were probed and washed under high stringency conditions with the designated rat Mrg probes. For all lanes, no bands were visible below 1 kb. (C) Summary of rodent MrgA, MrgB, MrgC, and MrgD subfamilies based on data obtained from Southern blots, degenerate PCR, and genomic analyses. For mouse and rat, the number of bands detected by Southern blotting is similar to the number of genes predicted from the draft genomic sequences.
The rat genomic dataset also contained complete sequences of rMrgD, rMrgE, rMrgF/RTA, rMrgG, rMrgH/GPR90, and rMAS1. With the exception of MrgH, which has not been identified in humans, these genes all have orthologs in mice and humans. As in mice, we were unable to identify rat genes that are orthologous to the human MrgX subfamily or to hMrg, the first Mas-related gene identified in humans (14). Taken together, these data indicate that rats and mice contain orthologous sets of the MrgA, MrgB, MrgC, and MrgD genes, albeit in different numbers, and that neither species contains genes orthologous to human MrgX/SNSRs.
Because our bioinformatic analysis was based on draft genomic sequence, we performed Southern blot experiments to confirm the copy number of rat Mrg genes. We used probes that covered most of the coding regions of rMrgA, rMrgB2, rMrgB4, rMrgB8, rMrgC, and rMrgD. Because the coding region of each Mrg is contained within a single exon (11), the number of bands of equivalent intensity on a Southern blot approximates the number of genes. Control experiments using mouse genomic DNA indicated that the rat Mrg probes were capable of hybridizing to at least 10 genes in the mMrgA and mMrgC subfamilies [Fig. 1B, lanes rMrgA (M) and rMrgC (M)]. Analysis of rat genomic DNA [Fig. 1B (R)] revealed one band each for rMrgA, rMrgC, rMrgB8 (B8 subdivision), and rMrgD; at least four strong bands for rMrgB2 (B2 subdivision), and two strong bands for rMrgB4 (B4 subdivision). The weaker bands detected by the rMrgB4 probe are likely due to cross-hybridization with other rMrgB2-like genes (note size similarities between MrgB2 and MrgB4 lanes). The number of bands detected by Southern blot analysis was therefore well correlated with the number of genes identified by our database searches.
Identification of Gerbil Mrg Family Members. The contrasting results in rat and mouse raised the question of whether the diversity of murine Mrgs represents the exception, or rather the rule, among rodents. To address this question, we characterized Mrgs from a third murid rodent, the Mongolian gerbil (Meriones unguiculatus). Because genomic sequence data are currently not available for this species, our approach was restricted to experimental analysis.
First, using degenerate PCR primers and gerbil liver genomic DNA as the template, we identified gerbil orthologs of MrgB1, MrgB4, and MrgD. On the basis of a phylogenetic analysis, gerbil (g)MrgB1 and gMrgB4 are located in the B2 and B4 subdivisions, respectively (Fig. 6). Despite numerous attempts, we were unable to amplify gerbil MrgA or MrgC sequences with degenerate primers.
We therefore conducted a Southern blot analysis of gerbil genomic DNA using rat MrgA and MrgC probes. That these rat probes strongly cross-hybridized to their murine orthologs under our hybridization conditions (Fig. 1B) suggested they would likely cross-hybridize to their gerbil orthologs as well. Consistent with this expectation, the rat MrgA and MrgC as well as the MrgD probes cross-hybridized to gerbil DNA, revealing single intense bands of 7.5, 1.5, and 8 kb, respectively (Fig. 1B, lanes G). The two additional weak bands revealed by the rMrgC probe likely represent cross-hybridization to gMrgA and/or a restriction fragment of gMrgC. These data suggest that, like the rat, the gerbil has a single copy of MrgA and MrgC and a single copy of MrgD, like all mammals thus far examined.
Gerbil DNA was also probed with rMrgB probes. We detected a total of three bands that cross-hybridized to both the rMrgB2 and rMrgB4 probes, albeit with different intensities to each (Fig. 1B). The 3.5- and 11-kb bands correspond to gMrgB1 and gMrgB4, respectively, as determined by probing duplicate blots with gerbil MrgB1 and MrgB4 DNA probes (data not shown). The 1.1-kb band did not hybridize to the rMrgB8 probe, and its identity is unknown. It could represent an additional gerbil MrgB gene or a restriction fragment of gMrgB1 or gMrgB4. Taken together, these data suggest that gerbil has at least two, and possibly three, MrgB genes, a number significantly less than mouse or rat (Fig. 1C).
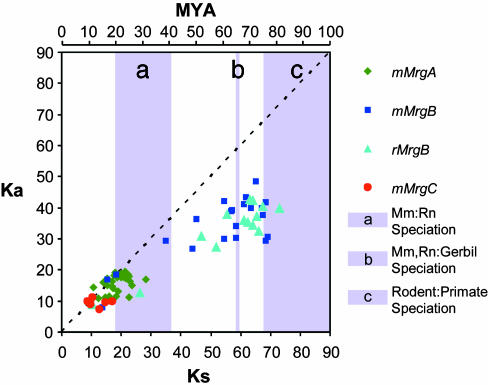
Expansion of the MrgA, MrgB, and MrgC Subfamilies Occurred at Different Times During Rodent Evolution. The foregoing data suggested that the murine genome contains a far greater number of Mrgs than either the rat or the gerbil. This difference could reflect an evolutionary contraction of the family that occurred in the latter two species or a selective expansion in the mouse. To distinguish between these alternatives, we determined the evolutionary times at which expansions of the different Mrg subfamilies occurred in mice, in relation to the times of speciation of rat, mouse, and gerbil (see Methods). These calculations suggested that the mouse MrgA and MrgC subfamilies each diverged from their respective common ancestors 10–25 MYA (Fig. 2), corresponding to a time shortly after, or coincident with, the speciation of rats and mice, and ≈25–45 million years after the speciation of gerbils from the rat–mouse lineage (21–23). Thus the larger sizes of the MrgA and MrgC subfamilies in mouse are consistent with an evolutionarily late selective expansion in that species.
Fig. 2.
Pairwise synonymous (Ks) and nonsynonymous (Ka) nucleotide substitutions per 100 sites between mouse and rat Mrg subfamily members. Each point represents a single pairwise comparison between Mrgs of the mMrgA (green diamonds), mMrgB (dark blue squares), rMrgB (light blue triangles), or mMrgC (red circles) subfamily. The dashed line marks a Ka/Ks ratio of 1.0 or neutral selection. Points below the line are considered to be under negative selection (Ka/Ks ratio <1.0) and points above, under positive selection (Ka/Ks ratio >1.0). The scale at the top of the graph relates Ks values to evolutionary divergence time in MYA. a, The shaded bar marks the approximate time when rats last shared a common ancestor with mice 20–41 MYA (21–23). b, The shaded bar indicates the approximate time when gerbils last shared a common ancestor with rats and mice 66 MYA (21, 22). c, The shaded bar indicates the approximate time when rodents and primates last shared a common ancestor 75–115 MYA (21–23).
In contrast to the results for the MrgA and MrgC subfamilies, the divergence times for MrgB gene pairs generally occurred before rat–mouse speciation but fell over a much broader window of evolutionary time spanning 10–80 MYA (Fig. 2). Pairwise (rat–rat and mouse–mouse) comparisons between members of the three different MrgB subdivisions (B2, B4, and B8) yielded average divergence times of 71 ± 6, 66 ± 4, and 70 ± 3 MYA (±SD) for B2–B4, B2–B8, and B4–B8 comparisons, respectively. The small variability in these numbers, combined with their similar absolute values, suggests that the B2, B4, and B8 subdivisions originated from a single ancestral MrgB gene ≈65–70 MYA. That this divergence occurred shortly before or during the time that rats and mice were predicted to have speciated from gerbils is consistent with our identification of one gerbil MrgB gene in each of the B2 and B4 subdivisions. The average divergence time calculated for rat or mouse MrgB members within each subdivision was 34 ± 17 MYA, consistent with the idea that these subdivisions expanded after the divergence of rats and mice from the gerbil lineage. Thus, it appears that at least two subdivisions of the MrgB family are likely present in all rodents but differ in size due to additional expansion events (Fig. 1C).
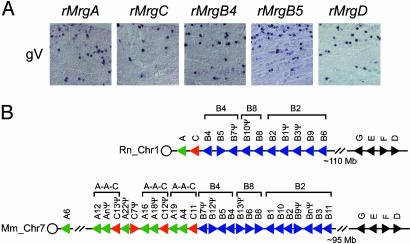
Mrgs Expressed in Sensory Neurons Are Positioned Adjacent to One Another in the Genome. To determine whether rat Mrgs are expressed in sensory neurons like their murine counterparts, we performed in situ hybridization experiments with tissue from newborn and adult rats. These experiments indicated that rMrgA, rMrgB4, rMrgB5, rMrgC, and rMrgD were all expressed strongly in newborn dorsal root ganglia (DRGs), adult DRGs, and trigeminal ganglia (gV; Figs. 3 and 4; data not shown). Others have similarly reported expression of rMrgA and rMrgC exclusively in adult rat DRG neurons (12, 20). In contrast, we did not detect expression of rMrgB1, rMrgB2, rMrgB3, rMrgB6, or rMrgB8 in sensory neurons. Although we have not looked exhaustively, none of our rMrg probes clearly hybridized to any other tissues or organs in postnatal day 0 animals. As in the rat, mMrgB4 and mMrgB5 were likewise expressed in adult mouse DRG neurons (see below), although not in newborn DRG neurons (11). Taken together, these data indicate that genes within the MrgB B4 subdivision, but not the B2 or B8 subdivisions, are expressed in sensory neurons like MrgA, MrgC, and MrgD.
Fig. 3.
Correlated expression and chromosomal localization of rodent Mrgs. (A) Expression analysis of rat Mrgs in adult trigeminal ganglia (gV). In situ hybridization was performed with antisense digoxigenin-labeled riboprobes. (B) Chromosomal arrangement of rat and mouse Mrgs. Analyses of the January 21, 2003, assembly of the rat genome and the February 24, 2003 (National Center for Biotechnology Information mouse build 30), assembly of the mouse genome revealed that most of the Mrg family members were located within two discrete regions of rat chromosome 1 and mouse chromosome 7. These two regions encompass the MrgABC cluster (760 kb in size from rat assembly NW_043369; 1.2 Mb in size from mouse assemblies NT_039420-NT_039423) and the MrgDEFG cluster (1.9 Mb in size from rat assemblies NW_043404-NW_043405; 1.6 Mb in size from mouse assembly NT_039437). The circle marks the relative position of the centromere. Triangles denote the direction of transcription and indicate the relative position of each gene on the chromosome. This figure is not drawn to scale. Brackets indicate the location of the three MrgB subdivisions. Several of the mouse A-A-C repeats are also highlighted. The mouse A-C cluster begins with MrgA6 and ends with a misassembled fragment of MrgC11. We did not plot all of the mMrgA and mMrgC genes because of obvious inaccuracies in the mouse assembly.
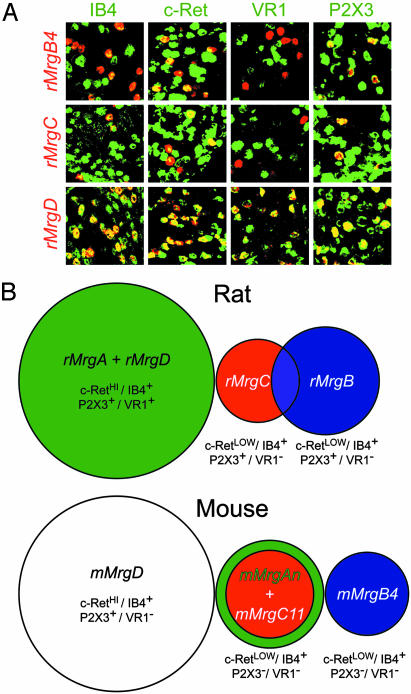
Fig. 4.
Analysis of Mrg expression in adult rat and mouse DRG neurons. (A) Coexpression of rat Mrgs with various sensory neuron markers. With the exception of IB4, all gene combinations were detected by double-label in situ hybridization (ISH) with the indicated antisense cRNA probe. Fluorescein-conjugated G. simplicifolia IB4-lectin was applied to sections after the ISH procedure to detect IB4-binding cells. (B) Summary of the rat and mouse Mrg expression domains in adult DRG sensory neurons. The sizes of the circles in the Venn diagrams are proportional to the sizes of the cell populations. Our results of double-label ISH among mMrgAs, mMrgB4, mMrgC11, mMrgD, and several nociceptive sensory neuron markers are also indicated (11, 13).
The sensory neuron-specific expression of rat Mrgs raised the possibility that they might be clustered together within the genome, like olfactory and vomeronasal GPCRs (6, 24, 25). Using the draft rat genome assembly as a guide, we found that most of the rMrg family members map to rat chromosome 1 (Fig. 3B, Rn). The rMrgA, rMrgB, and rMrgC genes are found together within a 760-kb cluster (the MrgABC cluster), and the rMrgD, rMrgE, rMrgF, and rMrgG genes are found together within a 1.9-Mb cluster (the MrgDEFG cluster). All of the Mrgs from the MrgABC cluster expressed in sensory neurons are adjacent to one another in the rat genome. Within this cluster, the MrgBs are arranged along the chromosome in a centromeric to telomeric orientation, within phylogenetically defined B4, B8, and B2 subdivisions.
Analysis of the assembled mouse genome revealed a similar arrangement of mMrgs into phylogenetically segregated mMrgABC and mMrgDEFG clusters on syntenic regions of mouse chromosome 7 (Fig. 3B, Mm). Furthermore, the murine mMrgA and mMrgC genes were generally found as a repeat of (A-A-C)n. This arrangement may explain why there are roughly twice as many mMrgAs (n = 22) as mMrgCs (n = 14) (11). The clustering of Mrgs with sensory neuron-specific expression suggests the presence of a locus control region and/or that the gene duplication events expanding these subfamilies included local cell type-specific transcriptional regulatory elements.
To obtain more clues about how the MrgABC cluster could have evolved, we searched assembled genomic sequences surrounding the rat and mouse MrgABC cluster for repetitive elements with the repeatmasker program (http://ftp.genome.washington.edu). For comparison, we searched a similar stretch of assembled genomic DNA surrounding the rat and mouse MrgD and MrgF cluster. This search revealed that the mouse and rat MrgABC clusters are intercalated with very large amounts of LINE1/L1 retrotransposon sequences (mouse MrgABC cluster = 43.2% L1; rat MrgABC cluster = 48.3% L1) (Figs. 7 and 8, which are published as supporting information on the PNAS web site). In contrast, the rat and mouse MrgD and MrgF cluster contains very few L1 elements or other repeats (mouse MrgDF cluster = 0.76% L1; rat MrgDF cluster = 0.58% L1). We also noticed that L1 retrotransposon sequences were found only at the 5′ end of the rMrgA coding exon but were found at the 5′ and 3′ ends of the coding exon of most mMrgAs (Figs. 7 and 8). These repeat elements may have played a role in the expansion of the MrgABC cluster (see Discussion).
Similar Subpopulations of Nociceptive Sensory Neurons Are Defined by Mrg Receptor Expression in Rat and Mouse. Nociceptive primary sensory neurons fall into many different subclasses. These subclasses can be distinguished on the basis of their function, neurotrophin dependence, and expression of molecular markers (26, 27). One such subclass expresses the glial cell line-derived neurotrophic factor (GDNF) receptor c-Ret and binds Griffonia simplicifolia isolectin IB4 (28). This GDNF-dependent subset has been implicated in neuropathic and inflammatory pain, conditions for which current analgesics are inadequate (29–34). In the mouse, Mrgs are expressed exclusively within this subset of nociceptors (11). We therefore wished to determine whether the restriction of Mrg expression to this neuronal subclass was conserved in the rat.
As in the mouse, we found that all rMrgB4+, rMrgC+, and rMrgD+ (all of which are rMrgA+; see below) neurons were also IB4 binding+ and c-Ret+ (Fig. 4A; Table 1, which is published as supporting information on the PNAS web site). Within this population, rMrgD+ cells were approximately two and three to four times more numerous than rMrgB4+ or MrgC+ cells, respectively, similar to the situation in mouse (Table 1). As in the mouse, the MrgD+ subset of c-Ret+ cells coexpressed the purinergic receptor P2X3 (11). The main difference between rat and mouse was that in the rat, all rMrgD+ cells [as well as a minority (7%) of rMrgC+ cells] express the capsaicin receptor VR1 (Fig. 4A and Table 1), whereas most or all Mrg-expressing cells are VR1– in the mouse (11). In addition, in the mouse, only mMrgD+ neurons coexpress the purinergic receptor P2X3, whereas in the rat, all Mrg+ neurons are P2X3+ (Fig. 4A). Taken together, these studies indicate that in rats, as in mice, Mrgs are restricted to the GDNF-dependent subset of nociceptive sensory neurons but display subtle differences in the other signaling molecules that they coexpress (Fig. 4B).
In the mouse, Mrg expression defines multiple subsets of neurons within the IB4+/Ret+ population (11). Because rats have a much smaller number of Mrgs, we next performed a series of double-label in situ hybridization experiments to determine how many adult DRG cell types they distinguish (Fig. 9 and Table 2, which are published as supporting information on the PNAS web site, for quantification). Briefly, we found that rMrgD and rMrgA are 100% coexpressed and define a single cell type that does not express rMrgB4 or rMrgC. A second nonoverlapping cell type was defined by coexpression of rMrgB4 and rMrgB5 (Table 2) and a third by unique expression of rMrgC. A fourth cell type was defined by coexpression of rMrgC and rMrgB4 in 27% of the MrgC+ cells (Fig. 4B and Table 2). Thus, the number of distinct neuronal subtypes defined by the combinatorial expression of rMrgA, rMrgB, rMrgC, and rMrgD is on the same order as the number of genes (Fig. 4B).
Because we had not previously detected expression of mMrgB4 or mMrgB5 in newborn mouse DRG (11), these rat data prompted us to reexamine the coexpression of Mrgs in adult mouse DRG. These experiments revealed subtle differences between mouse and rat in the relative distribution of these receptors. For example, in the rat, rMrgB4 and rMrgC are partially coexpressed; however, the mouse orthologs mark two nonoverlapping populations of murine DRG neurons (Fig. 9). Furthermore, unlike the rat, where rMrgD is coexpressed with rMrgA, mMrgD is not coexpressed with any mMrgA genes thus far examined in the adult mouse (Fig. 4B). Conversely, whereas rMrgC never overlaps with rMrgA in the rat, all mMrgC11+ cells coexpress mMrgA3 in mouse (although some mMrgA3+ cells are mMrgC11–). We used the mMrgA3 probe as representative of the murine MrgA family, because it gave the strongest signal in adult mouse DRG tissue. However, these observations were confirmed by using mixed probes containing mMrgA1, mMrgA2, mMrgA3, and mMrgA4 (data not shown). These data therefore suggest that MrgA genes are always coexpressed with another Mrg, but that the companion gene differs in rat and mouse (Fig. 4B).
Discussion
Expression of the Mrg family of GPCRs revealed a previously unanticipated degree of molecular diversity among murine nociceptive sensory neurons (11). Humans, in contrast, express a smaller number of related genes (12). On the one hand, Mrg diversity in mice could reflect aspects of sensory physiology and/or neuronal connectivity that are generic to rodents but different from that in humans. On the other hand, it could reflect genomic expansion events that are an atypical feature of mice. In an effort to distinguish these possibilities, we characterized the complement of Mrgs expressed in two related rodent species. Surprisingly, our data suggest that rats and gerbils each have a single MrgA and MrgC gene, and that these subfamilies underwent a relatively recent expansion in mice, primarily via local gene duplication events. However, like mice, rats and gerbils contain several MrgB genes and one MrgD gene. These and other findings suggest that Mrg diversity and function in rodents can be reduced to a core set of approximately four different genes, defining a comparable number of nociceptive neuron subtypes. These observations reduce the complexity of Mrg diversity in rodents to a level more closely approximating the limited Mrg diversity in humans.
The Localized Expansion of Murine MrgA and MrgC Genes May Be Driven by Interspersed Retrotransposons and Nonhomologous Recombination Events. What mechanism(s) underlie the selective and localized expansion of the mMrgA and mMrgC subfamilies in mice? The high frequency of interspersed L1 retrotransposons in the mouse MrgA-C cluster (>40% L1 sequence) suggests that such repeats could have facilitated unequal crossover events that expanded these subfamilies (Fig. 5). Consistent with this idea, Mrgs are generally arranged in a head-to-tail (5′ to 3′) fashion. This gene arrangement, coupled with the fact that phylogenetically related genes are adjacent, strongly supports an unequal crossover mechanism for expansion (24, 25, 35). However, such a mechanism by itself would not explain why a similar MrgA-C expansion did not occur in rat, because the rat also contains a similarly high frequency of L1 retrotransposons surrounding the rMrgA and rMrgC genes (≈48%). One possibility is that the expansion of the murine genes could be due to local L1 retrotransposition of the mMrg sequence, which seems unlikely, however, because the average mMrgA transcriptional unit (first and second exon ≈14 kb; Fig. 8) is significantly larger than the average length of extraneous DNA transposed by L1 elements (36).
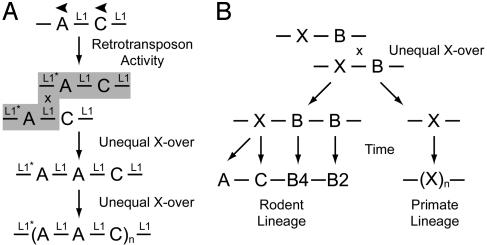
Fig. 5.
Possible mechanisms for Mrg expansion. (A) Idealized mechanism for the expansion of the mouse MrgA and MrgC (-A-C-) gene cluster. First, an L1 retrotransposon inserts into the 3′ end of the ancestral murine MrgA gene (L1*). At a later date, an unequal crossover event occurs between this new L1* and preexisting intergenic L1 sequences, creating the initial (A-A-C) repeat. Last, additional rounds of unequal crossover take place due to the large amount of homologous L1 sequence in the local genomic environment. (B)An unequal crossover event could explain why rodent and primate Mrg families are related but not orthologous. Assume that the common ancestor of primates and rodents contained single MrgX and MrgB genes. Unequal crossover could resolve into -X-B-B- and -X-containing chromosomes. In the rodent lineage, the -X-gene may have evolved into MrgA and MrgC genes, because they appear to be more closely related to human MrgXs than to rodent MrgBs (Fig. 6 and ref. 11). In humans, the -X-gene likely underwent additional rounds of unequal crossover to create the clustered MrgX/SNSR subfamily.
A more likely explanation is that expansion of the murine MrgA-C cluster was initiated by de novo L1 retrotransposition into the ancestral MrgA-C cluster during murine evolution, followed by unequal crossover with preexisting L1 sequences to duplicate the ancestral mMrgA gene and create an (A-A-C) repeat (Fig. 5A) (37). Additional rounds of localized unequal crossover could have created the present day (A-A-C)n repeats, explaining why there are roughly twice as many mMrgAs (n = 22) as mMrgCs (n = 14) (11). Our observation that mouse MrgAs have L1 sequences at the 5′ and 3′ ends of their coding exons, but that rat MrgA has an L1 sequence only at the 5′ end of its coding exon, supports the idea that a unique retrotransposition event took place during murine evolution. Furthermore, the broader clustering of divergence times calculated for mMrgAs versus the more recent and compact clustering of divergence times for mMrgCs (Fig. 2) is consistent with the idea that the ancestral mMrgA duplicated first, followed by duplication of the (A-A-C) repeat. This model therefore takes into account the high level of ongoing retrotransposition as well as the large number of local gene family expansions observed in the mouse genome (10).
Such an expansion mechanism could also explain, in principle, why all of the murine MrgC genes except mMrgC11 are pseudogenes, whereas numerous mMrgA sequences are maintained as expressed ORFs (11, 13). Because the initial (A-A-C) cluster contained the ancestral mMrgA gene, each additional duplication of the (A-A-C) cluster should have included at least one expressed mMrgA. In contrast, the first exon of mMrgC11 is located at the boundary of the (A-A-C) cluster, thus transcriptional regulatory elements in the mMrgC11 gene could have been damaged or eliminated after additional duplications of the (A-A-C) cluster. This would prevent duplicated mMrgCs from being expressed and thereby eliminate selection pressure to maintain functional genes. Consistent with this idea, mMrgC11 is the only mouse MrgC that encodes a functional and expressed receptor, is more similar to rMrgC than to any other mouse MrgC (Fig. 6), and is located in the same “ancestral” chromosomal location as rMrgC (11, 13).
An unequal crossover mechanism could also account for why humans have MrgX genes rather than orthologs of rodent MrgAs, MrgBs, and MrgCs. After such nonhomologous meiotic recombination events, one homologous chromosome gains a gene (or genes), whereas the other loses a gene (or genes) (35). It is tempting to speculate that a nonreciprocal crossover event within a primordial MrgX-MrgB cluster yielded recombination products differentially preserved by unique selective pressures in the rodent and primate lineages (Fig. 5B). In a similar fashion, red and green cone opsins, which arose from an unequal crossover event, were selectively maintained on the X chromosome of Old World primates, because trichromacy provides a selective advantage (3). Despite this nonorthologous conservation of coding sequence, members of the human MrgX subfamily and rodent MrgA, MrgB, and MrgC subfamilies are all expressed in nociceptive sensory neurons, supporting the idea that common promoter and/or enhancer elements were preserved during the evolution of these families.
The Functional Significance of mMrgA Sequence Diversity in Mice. Does the fact that mice express more MrgAs than rats or gerbils imply that this diversity has a physiological significance unique to mice? Our analysis indicates that most intact MrgA coding sequences are under neutral or weak negative selection pressure (Ka/Ks ≤ 1; Fig. 2), arguing against the idea that expansion of the mMrgA family was driven by positive selection for diversification of receptor coding sequences. However, this expansion could reflect positive selection for differential transcription of duplicated MrgA genes. Evidence for such selection, however, is difficult to glean from inspection of noncoding sequences, because calculations based on third-position changes do not apply. The relative conservation of coding sequences among the expanded family of mMrgAs, taken together with the fact that other rodent species examined retain a single MrgA gene, suggests that the various murine MrgA receptors may have similar or equivalent functions in vivo. In support of this view, both mMrgA1 and mMrgA4 can be activated by related RF-amide neuropeptides (11).
If the murine MrgAs have similar functions, the problem of Mrg diversity in mice would reduce to a core group of approximately four receptors (MrgA, MrgB, MrgC, and MrgD). In rats as in mice, these four receptors define a similar number of distinct neuronal subtypes (Fig. 4B). Because these receptors all are restricted to the GDNF-dependent subset of small-diameter nociceptive neurons in both rodent species, they are likely to play a conserved functional role in rodent nociception. In humans, the related hMrgXs/SNSRs are also specifically expressed in subsets of small-diameter sensory neurons (12), although whether they are restricted to the GDNF-dependent subset is not yet clear. Furthermore, like mMrgAs and mMrgC11, these human receptors are activated by RF/Y-(G)/amide-containing neuropeptides (11–13). Thus, despite the evolutionary divergence of the rodent MrgABC and human MrgX/SNSR subfamilies, some aspects of Mrg function in nociceptive neurons are likely to be conserved between these mammalian species. A better understanding of these conserved functions may aid in the development of Mrg-specific agonists or antagonists as novel pain therapeutics.
Finally, although the mouse has been the mammalian genetic model of choice for humans, our results highlight the importance of comparing and analyzing additional rodent genomes before drawing evolutionary and functional inferences based on mouse–human differences in the size of particular gene families. This note of caution may be especially true for comparative studies of gene families, like the GPCRs, which have the potential to rapidly expand. Although such expansion may facilitate rapid functional adaptation and reproductive isolation (10, 24), it may also reflect genomic expansion events atypical of rodents, perhaps due to unique retrotransposition events occurring during evolution. Our data suggest that analysis of the completed rat genome may reveal additional instances of atypical expansions of murine gene families and argue for the sequencing of at least one additional rodent genome to serve as an outgroup for mouse–rat comparisons.
Supplementary Material
Acknowledgments
We thank Gaby Mosconi for laboratory management; Jung-Sook Chang for technical assistance; Mel Simon, Sang-Kyou Han, and Jong-Ik Hwang for helpful discussions throughout the course of this work; and Mel Simon and Cori Bargmann for comments on the manuscript. M.J.Z. was supported by the Cancer Research Fund of the Damon Runyon–Walter Winchell Foundation Fellowship (DRG-1581). X.D. is a postdoctoral fellow of the American Cancer Society, and D.J.A. is an Investigator of the Howard Hughes Medical Institute.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: Mrgs, Mas-related genes; SNSR, sensory neuron-specific receptors; GPCR, G protein-coupled receptor; MYA, million years ago; DRG, dorsal root ganglia; GDNF, glial cell line-derived neurotrophic factor.
Data deposition: The sequences described in this paper have been deposited in the GenBank database [accession nos. AF518238 –AF518249, AY266420 (rat Mrgs), and AY196926 –AY196928 (partial sequences for gerbil MrgB1, MrgB4, and MrgD)].
References
- 1.Young, J. M. & Trask, B. J. (2002) Hum. Mol. Genet. 11, 1153–1160. [DOI] [PubMed] [Google Scholar]
- 2.Lindemann, B. (2001) Nature 413, 219–225. [DOI] [PubMed] [Google Scholar]
- 3.Nathans, J. (1999) Neuron 24, 299–312. [DOI] [PubMed] [Google Scholar]
- 4.Wang, Y., Macke, J. P., Merbs, S. L., Zack, D. J., Klaunberg, B., Bennett, J., Gearhart, J. & Nathans, J. (1992) Neuron 9, 429–440. [DOI] [PubMed] [Google Scholar]
- 5.Vassalli, A., Rothman, A., Feinstein, P., Zapotocky, M. & Mombaerts, P. (2002) Neuron 35, 681–696. [DOI] [PubMed] [Google Scholar]
- 6.Lane, R. P., Cutforth, T., Axel, R., Hood, L. & Trask, B. J. (2002) Proc. Natl. Acad. Sci. USA 99, 291–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adams, M. D., Celniker, S. E., Holt, R. A., Evans, C. A., Gocayne, J. D., Amanatides, P. G., Scherer, S. E., Li, P. W., Hoskins, R. A., Galle, R. F., et al. (2000) Science 287, 2185–2195. [DOI] [PubMed] [Google Scholar]
- 8.Lander, E. S., Linton, L. M., Birren, B., Nusbaum, C., Zody, M. C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. (2001) Nature 409, 860–921. [DOI] [PubMed] [Google Scholar]
- 9.Venter, J. C., Adams, M. D., Myers, E. W., Li, P. W., Mural, R. J., Sutton, G. G., Smith, H. O., Yandell, M., Evans, C. A., Holt, R. A., et al. (2001) Science 291, 1304–1351. [DOI] [PubMed] [Google Scholar]
- 10.Waterston, R. H., Lindblad-Toh, K., Birney, E., Rogers, J., Abril, J. F., Agarwal, P., Agarwala, R., Ainscough, R., Alexandersson, M., An, P., et al. (2002) Nature 420, 520–562. [DOI] [PubMed] [Google Scholar]
- 11.Dong, X., Han, S., Zylka, M. J., Simon, M. I. & Anderson, D. J. (2001) Cell 106, 619–632. [DOI] [PubMed] [Google Scholar]
- 12.Lembo, P. M., Grazzini, E., Groblewski, T., O'Donnell, D., Roy, M. O., Zhang, J., Hoffert, C., Cao, J., Schmidt, R., Pelletier, M., et al. (2002) Nat. Neurosci. 5, 201–209. [DOI] [PubMed] [Google Scholar]
- 13.Han, S. K., Dong, X., Hwang, J. I., Zylka, M. J., Anderson, D. J. & Simon, M. I. (2002) Proc. Natl. Acad. Sci. USA 99, 14740–14745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Monnot, C., Weber, V., Stinnakre, J., Bihoreau, C., Teutsch, B., Corvol, P. & Clauser, E. (1991) Mol. Endocrinol. 5, 1477–1487. [DOI] [PubMed] [Google Scholar]
- 15.Young, D., Waitches, G., Birchmeier, C., Fasano, O. & Wigler, M. (1986) Cell 45, 711–719. [DOI] [PubMed] [Google Scholar]
- 16.Li, W. H., Wu, C. I. & Luo, C. C. (1985) Mol. Biol. Evol. 2, 150–174. [DOI] [PubMed] [Google Scholar]
- 17.Li, W. H. (1993) J. Mol. Evol. 36, 96–99. [DOI] [PubMed] [Google Scholar]
- 18.Pamilo, P. & Bianchi, N. O. (1993) Mol. Biol. Evol. 10, 271–281. [DOI] [PubMed] [Google Scholar]
- 19.Li, W. H., Luo, C. & Wu, C. (1985) in Molecular Evolutionary Genetics, ed. Macintyre, R. J. (Plenum, New York), pp. 1–94.
- 20.Bender, E., Buist, A., Jurzak, M., Langlois, X., Baggerman, G., Verhasselt, P., Ercken, M., Guo, H. Q., Wintmolders, C., Van den Wyngaert, I., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 8573–8578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar, S. & Hedges, S. B. (1998) Nature 392, 917–920. [DOI] [PubMed] [Google Scholar]
- 22.Kumar, S. & Subramanian, S. (2002) Proc. Natl. Acad. Sci. USA 99, 803–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O'hUigin, C. & Li, W. H. (1992) J. Mol. Evol. 35, 377–384. [DOI] [PubMed] [Google Scholar]
- 24.Young, J. M., Friedman, C., Williams, E. M., Ross, J. A., Tonnes-Priddy, L. & Trask, B. J. (2002) Hum. Mol. Genet. 11, 535–546. [DOI] [PubMed] [Google Scholar]
- 25.Zhang, X. & Firestein, S. (2002) Nat. Neurosci. 5, 124–133. [DOI] [PubMed] [Google Scholar]
- 26.Hunt, S. P. & Mantyh, P. W. (2001) Nat. Rev. Neurosci. 2, 83–91. [DOI] [PubMed] [Google Scholar]
- 27.Julius, D. & Basbaum, A. I. (2001) Nature 413, 203–210. [DOI] [PubMed] [Google Scholar]
- 28.Snider, W. D. & McMahon, S. B. (1998) Neuron 20, 629–632. [DOI] [PubMed] [Google Scholar]
- 29.Bennett, D. L., Michael, G. J., Ramachandran, N., Munson, J. B., Averill, S., Yan, Q., McMahon, S. B. & Priestley, J. V. (1998) J. Neurosci. 18, 3059–3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boucher, T. J., Okuse, K., Bennett, D. L., Munson, J. B., Wood, J. N. & McMahon, S. B. (2000) Science 290, 124–127. [DOI] [PubMed] [Google Scholar]
- 31.Bradbury, E. J., Burnstock, G. & McMahon, S. B. (1998) Mol. Cell Neurosci. 12, 256–268. [DOI] [PubMed] [Google Scholar]
- 32.Cockayne, D. A., Hamilton, S. G., Zhu, Q. M., Dunn, P. M., Zhong, Y., Novakovic, S., Malmberg, A. B., Cain, G., Berson, A., Kassotakis, L., et al. (2000) Nature 407, 1011–1015. [DOI] [PubMed] [Google Scholar]
- 33.Malmberg, A. B., Chen, C., Tonegawa, S. & Basbaum, A. I. (1997) Science 278, 279–283. [DOI] [PubMed] [Google Scholar]
- 34.Souslova, V., Cesare, P., Ding, Y., Akopian, A. N., Stanfa, L., Suzuki, R., Carpenter, K., Dickenson, A., Boyce, S., Hill, R., et al. (2000) Nature 407, 1015–1017. [DOI] [PubMed] [Google Scholar]
- 35.Li, W. H. (1997) Molecular Evolution (Sinauer, Sunderland, MA).
- 36.Goodier, J. L., Ostertag, E. M. & Kazazian, H. H., Jr. (2000) Hum. Mol. Genet. 9, 653–657. [DOI] [PubMed] [Google Scholar]
- 37.Fitch, D. H., Bailey, W. J., Tagle, D. A., Goodman, M., Sieu, L. & Slightom, J. L. (1991) Proc. Natl. Acad. Sci. USA 88, 7396–7400. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.