Abstract
The multidomain proapoptotic protein Bax of the Bcl-2 family is a central regulator for controlling the release of apoptogenic factors from mitochondria. Recent evidence suggests that the Bax-associating protein MOAP-1 may act as an effector for promoting Bax function in mitochondria. Here, we report that MOAP-1 protein is rapidly up-regulated by multiple apoptotic stimuli in mammalian cells. MOAP-1 is a short-lived protein (t1/2 ≈ 25 min) that is constitutively degraded by the ubiquitin-proteasome system. Induction of MOAP-1 by apoptotic stimuli ensues through inhibition of its polyubiquitination process. Elevation of MOAP-1 levels sensitizes cells to apoptotic stimuli and promotes recombinant Bax-mediated cytochrome c release from isolated mitochondria. Mitochondria depleted of short-lived proteins by cycloheximide (CHX) become resistant to Bax-mediated cytochrome c release. Remarkably, incubation of these mitochondria with in vitro-translated MOAP-1 effectively restores the cytochrome c releasing effect of recombinant Bax. We propose that apoptotic stimuli can facilitate the proapoptotic function of Bax in mitochondria through stabilization of MOAP-1.
Keywords: apoptosis, Bcl-2 family, proteasome, DNA damage
Mitochondria are the major organelles involved in the signal transduction and biochemical execution of apoptosis (1). Proteins of the Bcl-2 family are central transducers of survival and apoptotic signals (2). They act at mitochondria by regulating the permeability and integrity of the mitochondrial outer membranes, thereby controlling the release of apoptogenic factors. The Bcl-2 family consists of three major subfamilies of prosurvival or proapoptotic molecules. The BH3-only proteins (Bim, Bad, Bid, Bik, Noxa, Puma, and Hrk) serve as sentinels for the initiation of apoptosis by modulating the function of the multidomain prosurvival (Bcl-2, Bcl-w, Mcl-1, Bcl-XL, and A1/Bfl-1) or proapoptotic Bcl-2 members (Bax and Bak) (3–5). The prosurvival family members prevent cell death mainly by interrupting oligomerization of Bax/Bak, largely through binding and sequestering activator BH3 domains and thereby preventing their interaction with Bax/Bak (6, 7). Although the molecular details about how the Bcl-2 family of proteins regulates mitochondrial apoptotic signaling remain to be resolved, the multidomain proapoptotic molecules Bax and Bak have been shown to be the essential effectors responsible for the execution of apoptosis mediated through multiple signals (6, 8, 9).
Modulator of apoptosis (MOAP-1), initially named MAP-1, was identified as a binding partner of Bax in a yeast two hybrid screen (10). MOAP-1 contains a BH3-like motif and is capable of triggering apoptosis in mammalian cells when overexpressed (10, 11). Knocking down MOAP-1 by RNAi confers inhibition of apoptotic signaling triggered by multiple apoptotic stimuli and promotes anchorage-independent growth of tumor cells (11). Remarkably, isolated mitochondria from MOAP-1 knockdown cells were highly resistant to the cytochrome c (Cyto c)-releasing effect of recombinant Bax, suggesting that MOAP-1 may act as an effector for facilitating Bax function in mitochondria (11). Interestingly, it has recently been demonstrated that the tumor suppressor RASSF1A specifically links cell death receptor- (12) and activated K-Ras-mediated (13) apoptotic signaling to Bax activation through binding to MOAP-1.
The ubiquitin-proteasome system (UPS) plays important roles in regulating many cellular and physiological processes, including apoptosis. Many key regulators in the apoptosis and survival pathway, such as p53, NF-κB, the IAP family, are known to be regulated by UPS (14, 15). Accumulating evidence also suggests that UPS has a role in directly regulating the levels of certain pro- and antiapoptotic members of the Bcl-2 family, including Bik, Bim, A1/Bfl-1, and Mcl-1 (16–19). Here, we present evidence to show that rapid inhibition of UPS-dependent degradation of MOAP-1 by multiple apoptotic stimuli may play an important role in facilitating Bax function in mitochondria.
Results
MOAP-1 Protein Is Rapidly Up-Regulated by Multiple Apoptotic Stimuli.
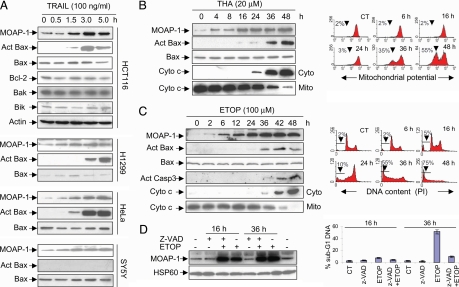
MOAP-1 is proapoptotic in mammalian cells when overexpressed (10, 11). Reduction of MOAP-1 levels by RNAi knockdown suppresses apoptosis triggered by multiple apoptotic stimuli (11). Because MOAP-1 appears to be a low-abundance protein in mammalian cells (ref. 11 and data not shown) and the level of MOAP-1 could be an important determinant for influencing the sensitivity of mammalian cells to apoptotic signals, we decided to evaluate the potential effect of apoptotic stimuli on MOAP-1 protein levels in mammalian cells. Using a combined immunoprecipitation (IP)/Western blot analysis as described in ref. 20 for detection of other low-abundance proteins, MOAP-1 protein levels were first found to be rapidly up-regulated in HCT116, H1299, SY5Y, and HeLa cells upon tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) treatment (Fig. 1A). The induction of MOAP-1 by TRAIL displayed a fast kinetics similar to Bax activation in the HCT116, H1299 and HeLa cells (Fig. 1A). Interestingly, although TRAIL failed to induce Bax activation (Fig. 1A) and apoptosis (data not shown) in SY5Y cells, it was effective in triggering up-regulation of MOAP-1 protein in these cells (Fig. 1A).
Fig. 1.
Apoptotic stimuli up-regulate MOAP-1 protein during the early phase of apoptotic signaling. (A) Levels of endogenous MOAP-1 protein were rapidly up-regulated by TRAIL. The indicated cells were treated with TRAIL for various periods of time. RIPA lysates were subjected to IP with the rabbit anti-MOAP-1 antibody (R5), followed by IB with the mouse anti-MOAP-1 antibody (M6). Actin was used as an internal control to demonstrate that equal amount of total proteins was used for IP. The levels of Bcl-2, Bak, and Bik in the total lysates of HCT116 cells were also measured with their respective antibodies. Bax activation (Act Bax) was monitored by using a conformation-specific Bax antibody (N-20). (B and C) Induction of MOAP-1 by THA and ETOP occurred during the early phase of apoptotic signaling. HCT116 cells (B) or 293T cells (C) were treated with 20 μM THA or 100 μM ETOP, respectively, for the indicated periods of time. MOAP-1 levels, Bax activation (Act Bax), Capase 3 activation (Act Casp3), and Cyto c release were monitored (B and C Left). For detection of Cyto c release from mitochondria, the cells were fractionated into heavy membrane fractions enriched with mitochondria (Mito) and cytosolic (Cyto) fractions. THA-induced mitochondrial depolarization (B Right) or ETOP-induced DNA fragmentation (C Right) were analyzed by flow cytometry as described in Materials and Methods. Data shown are representative of at least three independent experiments. (D) Caspase inhibition fails to suppress the elevation of MOAP-1 protein induced by ETOP. 293 T cells were pretreated with 10 μM z-VAD for 1 h before being subjected to 100 μM ETOP treatment for 16 h or 36 h. (Left) MOAP-1 levels were analyzed as in A. (Right) DNA fragmentation was analyzed as in C. Results are presented as percentage of apoptotic cells (mean ± SD, n = 3).
To assess the effect of other apoptotic stimuli on MOAP-1 levels, a number of cell lines were subjected to treatment with a series of apoptotic stimuli, including the endoplasmic reticulum stress inducer thapsigargin (THA), DNA-damaging agents, serum withdrawal, or the PKC inhibitor staurosporine. Except staurosporine, all apoptotic stimuli tested were able to rapidly enhance MOAP-1 levels in mammalian cell lines, including SY5Y, HCT116, HepG2, 293T, H1299, and HeLa cells [Fig. 1 B–D Left and supporting information (SI) Fig. 6 and data not shown]. Although the up-regulation of MOAP-1 levels by DNA-damaging stimuli displayed a similar kinetic as that of p53 induction in SY5Y and A2780 cells, which are known to harbor wild-type p53 (21) (SI Fig. 6 B and C), similar effect was also observed in the p53 mutant cell 293T (SI Fig. 6A) and p53 null cell H1299 (SI Fig. 6D), suggesting that the up-regulation of MOAP-1 by DNA-damaging stimuli involves a p53-independent mechanism. MOAP-1 has been shown to be a mitochondria-enriched protein (11). The accumulation of MOAP-1 protein upon etoposide (ETOP) (SI Fig. 6E) or TRAIL treatment (data not shown) was mainly seen in the heavy membrane fraction containing mitochondria. MCF-7 cells appear to have higher basal levels of MOAP-1 than other mammalian cell types (data not shown). Consistent with our previous observations (11), most apoptotic stimuli tested, with the exception of ETOP and camptothecin, were relatively ineffective for inducing up-regulation of MOAP-1 protein in these cells (data not shown). In contrast to rapid induction of apoptosis by TRAIL, apoptosis induced by THA, DNA-damaging agents and serum withdrawal appears much slower (Fig. 1 B and C and data not shown). Nevertheless, the up-regulation of MOAP-1 by multiple apoptotic stimuli was readily detected before those commitment events of apoptotic signaling, such as Bax activation, Cyto c release from mitochondria, mitochondrial potential changes and the appearance of subG1 DNA content (Fig. 1 B and C; data not shown). The broad spectrum caspase inhibitor z-VAD failed to inhibit MOAP-1 up-regulation by ETOP (Fig. 1D Left) and TRAIL (data not shown), whereas it effectively blocked DNA fragmentation (Fig. 1D Right), indicating that the effect of apoptotic stimuli on MOAP-1 up-regulation is not dependent on a positive feedback mechanism driven by sustained caspase activation.
Apoptotic Stimuli Stabilize MOAP-1 Protein.
To study the mechanism by which apoptotic stimuli promote an increase in MOAP-1 protein levels, we first measured MOAP-1 mRNA levels by real-time PCR. No significant difference was noted in MOAP-1 mRNA levels between control and TRAIL- or ETOP-treated cells (SI Fig. 7A). Translational regulation by the 5′ or 3′ UTR of mRNAs has been found to play an important role in the regulation of levels of certain proteins by apoptotic stimuli (22). To exclude this possibility, cells were transiently transfected with a HA-tagged MOAP-1-expressing plasmid, in which only the coding region of MOAP-1 is transcribed under the control of the CMV promoter. Similar to the endogenous MOAP-1 protein, the levels of HA-tagged MOAP-1 protein were rapidly increased by treatment with TRAIL or ETOP (SI Fig. 7B).
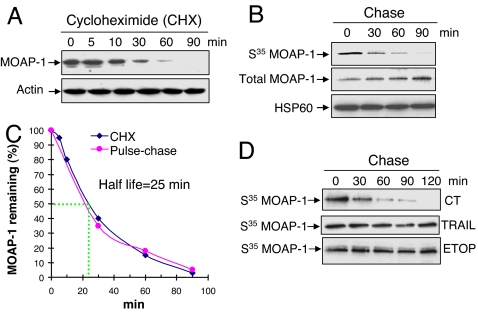
Because neither transcriptional nor translational mechanism appears to have a significant role in mediating MOAP-1 up-regulation by apoptotic stimuli, we next explored possible posttranslational mechanism underlying MOAP-1 regulation. We first determined the half-life of MOAP-1 in mammalian cells. The protein synthesis inhibitor CHX was added to the 293T cells to block the de novo protein synthesis. The remaining levels of MOAP-1 in the cells at different time points after CHX treatment were monitored (Fig. 2A). In addition, 293T cells were also subjected to pulse–chase analysis (Fig. 2B). Both approaches yielded similar results and the half-life of MOAP-1 is estimated to be ≈25 min in 293T cells (Fig. 2C). To ensure that the fast turnover of MOAP-1 is not limited to 293T cells, the time-dependent decay of MOAP-1 by CHX was measured in COS-1, SY5Y, HepG2 and HCT116 cells. The half-life of MOAP-1 was estimated to be between 20–30 min among these four cell lines (SI Fig. 8). In contrast, Bax is a stable protein, because its level remained unchanged upon CHX treatment even up to 16 h (SI Fig. 8 and data not shown). Interestingly, TRAIL or ETOP treatment led to a pronounced extension of MOAP-1 half-life compared with the untreated control (Fig. 2D). These data together suggest that protein stabilization, possibly by inhibition of its degradation, is likely to be a predominant mechanism underlying the up-regulation of MOAP-1 induced by apoptotic stimuli.
Fig. 2.
MOAP-1 is a short-lived protein that can be stabilized by apoptotic stimuli. (A) Inhibition of de novo protein synthesis caused rapid elimination of endogenous MOAP-1 protein. 293T cells were incubated with 50 μg/ml CHX for the indicated periods of time. MOAP-1 protein levels were monitored as in Fig. 1. Actin was used as an internal control. (B) Pulse–chase analysis for estimating the half-life of MOAP-1. 293T cells labeled with S35-methionine/cysteine were chased for the indicated periods of time. RIPA lysates were subjected to IP with anti-MOAP-1 antibody (R5). Immunoprecipitates were analyzed by autoradiography (for S35 MOAP-1) or by IB with anti-MOAP-1 antibody (for total MOAP-1). HSP60 was used as an internal control. (C) The half-life of MOAP-1 is estimated to be ≈25 min. The relative amount of total MOAP-1 in A or S35-labeled MOAP-1 in B was quantified by densitometry and plotted with respect to time. MOAP-1 level at time 0 was defined as 100%. (D) TRAIL or ETOP treatment extended the half-life of MOAP-1. H1299 cells were pretreated with 50 ng/ml TRAIL for 1 h or 100 μM ETOP for 3 h. Pulse–chase assay was then performed as in B. The same concentration of TRAIL or ETOP was maintained during the entire period of pulse–chase analysis.
Apoptotic Stimuli Inhibit Polyubiquitination of MOAP-1, Which Is Required for Its Degradation by Proteasome.
Many proteases are known to be involved in regulating protein stability in mammalian cells (23). To determine which protease(s) might be participating in MOAP-1 regulation, 293T cells were subjected to treatment with diverse protease inhibitors. Among all of them, only the proteasome inhibitors (i.e., MG132, LLnL, epoxomycin and lactacystin) were found to dramatically elevate endogenous MOAP-1 protein levels through extension of its half-life in all of the cell lines tested, including the human primary foreskin cell FS-4 (SI Fig. 9 and data not shown), suggesting that the proteasome system plays a major role in the fast turnover of MOAP-1. Interestingly, levels of MOAP-1 protein being up-regulated by MG132 were not increased further upon ETOP (SI Fig. 9F) or TRAIL treatment (data not shown), providing additional evidence that apoptotic stimuli up-regulate MOAP-1 primarily through affecting its stability, instead of increasing MOAP-1 mRNA levels.
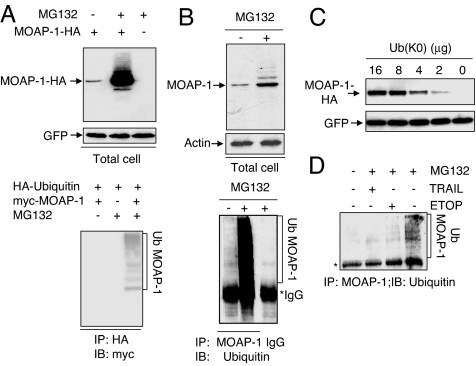
The stabilizing effect of proteasome inhibitors on MOAP-1 led us to explore further whether MOAP-1 is a direct substrate for ubiquitination. Transiently expressed HA-tagged MOAP-1 was significantly up-regulated by MG132. Furthermore, in addition to the band corresponding to unmodified MOAP-1, a series of additional, slower migrating forms of the protein were observed in the cells treated with MG132 (Fig. 3A Upper). It is possible that the higher molecular weight bands represent the polyubiquitinated forms of MOAP-1. To test this hypothesis, 293T cells were cotransfected with myc-tagged MOAP-1 and HA-tagged ubiquitin (Ub). Polyubiquitinated forms of myc-tagged MOAP-1 were readily observed in the transfected cells in the presence of MG132 (Fig. 3A Lower). A similar experiment was performed to detect the polyubiquitinated forms of endogenous MOAP-1, but in this instance MOAP-1 was immunoprecipitated with anti-MOAP-1 antibody and polyubiquitinated forms of MOAP-1 were detected by anti-Ub antibody. In the presence of MG132, polyubiquitinated forms of endogenous MOAP-1 protein were readily seen in SY5Y cells (Fig. 3B), 293T, HCT116, COS-1 and H1299 cells (data not shown). To assess the requirement of polyubiquitination of MOAP-1 for its degradation by the proteasome, HA-tagged MOAP-1 was cotransfected with the plasmid expressing the mutant Ub [Ub(K0)], with all of its lysines mutated to arginines (24). Levels of MOAP-1 were elevated in a dose-dependent manner directly proportional to the amount of Ub(K0) plasmid transfected, suggesting that polyubiquitin chain formation is necessary for efficient degradation of MOAP-1 by the proteasome (Fig. 3C).
Fig. 3.
Apoptotic stimuli suppress polyubiquitination of MOAP-1. (A) Proteasome inhibition caused accumulation of polyubiquitinated forms of transiently expressed MOAP-1 (Ub MOAP-1). (Upper) HA-MOAP-1 or control vector was cotransfected with pEGFP into 293T cells. Sixteen hours after transfection, the cells were treated with 10 μM MG132 for another sixteen hours. RIPA lysates were analyzed by IB, using anti-HA antibody. The levels of GFP were used to monitor transfection efficiency. (Lower) 293T cells were transfected with indicated plasmids. Sixteen hours after transfection, the cells were left untreated or treated with MG132 for another 16 h. RIPA lysates were subjected to IP with anti-HA-conjugated beads, followed by IB with anti-myc antibody. (B) Proteasome inhibition caused accumulation of ubiquitinated forms of endogenous MOAP-1 (Ub MOAP-1). SY5Y cells were either left untreated or treated with 10 μM MG132 for 16 h. (Upper) RIPA lysates were analyzed by IB with anti-MOAP-1 or anti-actin (loading control) antibodies. (Lower) RIPA lysates were subjected to IP with MOAP-1 antibody or control IgG, followed by IB with anti-Ub antibody. (C) MOAP-1 protein levels were elevated by inhibition of polyubiquitin chain formation. 293T cells were cotransfected with MOAP-1-HA and pEGFP in combination with indicated amounts of plasmid expressing the lysine-less Ub mutant [Ub(K0)]. Sixteen hours after transfection, the cells were harvested, and RIPA lysates were analyzed by IB with anti-HA antibody. The levels of GFP were used to monitor transfection efficiency. (D) TRAIL and ETOP inhibit polyubiquitination of MOAP-1. H1299 cells were treated with 50 ng/ml TRAIL for 1 h or 100 μM ETOP for 3 h before MG132 was added, and the cells were incubated for another 12 h. The cells were harvested and analyzed as in B.
To identify the domain in MOAP-1 that might be essential for coupling MOAP-1 to UPS, a series of MOAP-1 deletion mutants were generated (SI Fig. 10A). All of the deletion mutants that contain the center portion of MOAP-1 (amino acids 141–190) have a short half-life (data not shown) and they were all dramatically up-regulated by MG132 (SI Fig. 10B). Moreover, the polyubiquitination of the M5 mutant (MOAP-1 amino acids 115–190) was readily detected in the presence of MG132 (SI Fig. 10D). Levels of this mutant was effectively elevated by either TRAIL or ETOP treatment (SI Fig. 10C), suggesting that the center region of MOAP-1 contains a functional domain sufficient for mediating the stabilization effect on MOAP-1 by apoptotic stimuli. Interestingly, the degradable property contained within the M5 mutant is transferable to a heterologous protein GST that is not normally regulated by UPS (SI Fig. 10 E–G).
To investigate whether apoptotic stimuli have any effect on the ubiquitination process of MOAP-1, H1299 cells were pretreated with TRAIL or ETOP before incubating with MG132 to promote accumulation of ubiquitinated forms of endogenous MOAP-1. Both stimuli significantly reduced accumulation of ubiquitinated forms of MOAP-1 (Fig. 3D), suggesting that the effect of apoptotic stimuli on stabilizing MOAP-1 protein is likely to be mediated through inhibition of its polyubiquitination process. The inhibition of polyubiquitination of MOAP-1 by apoptotic stimuli is unlikely to be a general phenomenon during the early phase of apoptotic signaling, because the global accumulation of total polyubiquitinated proteins in H1299 cells induced by MG132 is not inhibited by ETOP or TRAIL treatment (SI Fig. 11A). Furthermore, although the levels of BH3-only protein Bik can be dramatically elevated in the presence of proteasome inhibitors (ref. 16 and SI Fig. 11B), neither ETOP nor TRAIL could up-regulate levels of Bik protein (SI Fig. 11B) and inhibit accumulation of polyubiquinated forms of Bik stimulated by MG132 (SI Fig. 11C).
Elevating MOAP-1 Protein Levels Sensitizes Mammalian Cells to Apoptotic Stimuli.
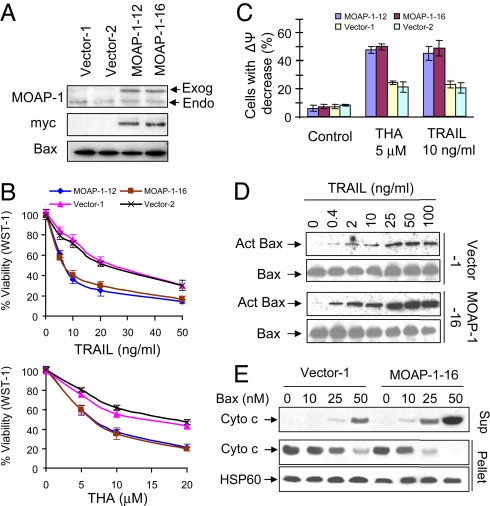
Reduction of MOAP-1 levels by RNAi knockdown approach was shown to inhibit Bax-dependent apoptotic signaling (11). Although transiently overexpressed MOAP-1 is able to trigger apoptosis on its own (10, 11), it is unclear whether higher basal levels of MOAP-1 in mammalian cells would sensitize cells to apoptotic signals. To evaluate this, HCT116 clonal lines stably expressing exogenous myc-tagged MOAP-1 (HCT116 myc-MOAP-1 cells) were generated. The expression levels of myc-MOAP-1 in stable clonal lines are much lower compared with the expression upon transient overexpression conditions (data not shown), but their levels were significantly higher than the levels of endogenous MOAP-1 (Fig. 4A). Moreover, both forms of MOAP-1 were tightly controlled by UPS and effectively up-regulated by apoptotic stimuli in the stable lines (data not shown). HCT116 myc-MOAP-1 cells were healthy and displayed normal growth and morphology (data not shown). These cells, however, were more sensitive than vector cells to TRAIL- and THA-induced apoptosis as determined by WST-1 assay (Fig. 4B) and mitochondrial potential changes (Fig. 4C). Interestingly, although staurosporine failed to up-regulate MOAP-1 in HCT116 cells (data not shown), higher levels of MOAP-1 had similar effect on heightening the sensitivity of HCT116 cells to the apoptotic effect of staurosporine as to TRAIL and THA (data not shown). These data suggest that elevation of MOAP-1 protein levels has a general effect on sensitizing cells to apoptotic stimuli. In comparison with vector cells, higher levels of activated Bax were detected upon treatment with TRAIL in the cells stably expressing exogenous myc-MOAP-1 (Fig. 4D). Mitochondria in heavy membrane fractions isolated from HCT116 myc-MOAP-1 cells were clearly more sensitive than those from vector cells to the Cyto c releasing effect of recombinant Bax (Fig. 4E), further supporting the idea that the levels of MOAP-1 correlate positively to sensitivity of mitochondria to the Cyto c releasing effect of Bax. To extend our analysis on the effect of higher levels of MOAP-1 expression on apoptosis to other cell lines, MCF-7 clonal lines stably expressing myc-MOAP-1 were generated. As in HCT116 cells, higher basal levels of MOAP-1 also sensitized MCF-7 cells to apoptotic stimuli (SI Fig. 12 and data not shown).
Fig. 4.
Higher levels of MOAP-1 sensitize the HCT116 cells to multiple apoptotic stimuli. (A) MOAP-1 expression in the HCT116 vector control and myc-MOAP-1 stable clonal lines. RIPA lysates were immunoprecipitated with anti-MOAP-1 or anti-Bax antibodies, followed by IB with anti-MOAP-1, anti-Bax or anti-myc antibodies. The arrows indicate the endogenous (Endo) or exogenous myc-MOAP-1(Exog). (B and C) Cell death analysis of HCT116 vector control or HCT116 myc-MOAP-1 cells subjected to treatment with TRAIL or THA. Cells grown in 96-well plate were treated with indicated concentrations of THA for 36 h or TRAIL for 16 h. Cell viability after treatment was determined by the WST-1 assay (B). Results are presented as percentage of control (mean ± SD, n = 3). Cells grown in 6-well plate were treated with 5 μM THA for 36 h or 10 ng/ml TRAIL for 16 h, harvested and stained with Mito-tracker Red for analysis of mitochondrial membrane potential changes by flow cytometry (C). (D) Higher levels of MOAP-1 promote TRAIL-induced Bax activation. Vector 1# and MOAP-1 16# cells were treated with indicated concentrations of TRAIL for 5 h. Bax activation (Act Bax) was analyzed by using a conformation-specific Bax antibody (N-20). (E) Higher levels of MOAP-1 increase the sensitivity of isolated mitochondria to recombinant Bax-induced Cyto c release. Heavy membrane fractions containing mitochondria isolated from Vector-1 or MOAP-1–16 cells were treated with recombinant Bax, followed by centrifugation. The supernatants (sup) and pellets were immunoblotted with anti-Cyto c or HSP60 antibodies.
MOAP-1 Is a Key Short-Lived Protein to Promote Bax Function in Mitochondria.
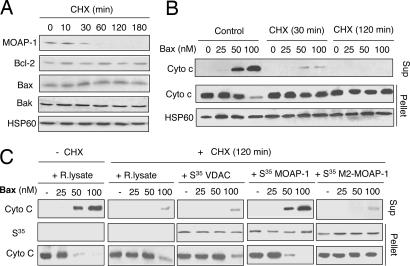
CHX treatment is known to have very diverse effects on apoptosis in ex vivo studies. It can significantly promote or block apoptosis when combined with different apoptotic stimuli in distinct cellular contexts (25, 26). The contribution of mitochondrial short-lived proteins as a whole in regulating the function of recombinant Bax in isolated mitochondria has not been explored. MOAP-1 knockdown by RNAi has been shown to attenuate recombinant Bax- and tBid-mediated Cyto c release (11). To test the possibility that depletion of short-lived proteins in mitochondria, including MOAP-1, by CHX would result in a similar phenotype as the MOAP-1 knockdown by RNAi on Bax- or tBid-mediated Cyto c release in isolated mitochondria, heavy membrane fractions containing mitochondria were isolated from the cells pretreated with CHX for various durations. Even 1 h of CHX treatment was sufficient to deplete MOAP-1 in the heavy membrane fractions to undetectable levels, whereas the levels of Bax, Bak Bcl-2 and HSP60 remained unchanged (Fig. 5A). Similar to mitochondria isolated from MOAP-1 knockdown cells (11), mitochondria in heavy membrane fractions isolated from cells that were pretreated with CHX for relatively short time (<2 h) were found to be resistant to the Cyto c releasing effect of recombinant Bax (Fig. 5B) and tBid (data not shown). Because the molecular mechanism of Bax function in mitochondria has not been fully elucidated, it is unclear whether any other short-lived mitochondrial protein(s), in addition to MOAP-1, may also be required to facilitate the apoptotic function of Bax. To address this, heavy membrane fractions containing mitochondria isolated from CHX-pretreated cells were preincubated with reticulate lysates or in vitro-translated MOAP-1, VDAC or an inactive mutant of MOAP-1 (M2 mutant, amino acids 1–115), before being exposed to recombinant Bax protein. Remarkably, exogenously added MOAP-1, but not the mitochondrial protein VDAC or the M2 mutant, was effective in restoring the Cyto c releasing effect of recombinant Bax (Fig. 5C).
Fig. 5.
MOAP-1 is a key short-lived protein to facilitate Bax-induced Cyto c release from isolated mitochondria. (A) Effects of CHX on the levels of Bcl-2 family proteins in heavy membrane fractions containing mitochondria. HCT116 cells were treated with 50 μg/ml CHX for the indicated periods of time. The levels of MOAP-1, Bcl-2, Bak, Bax, and HSP60 were analyzed by IB. (B) Mitochondria isolated from CHX-pretreated cells were resistant to Bax-mediated Cyto c release. Heavy membrane fractions containing mitochondria isolated from the control or CHX-pretreated HCT116 cells were incubated with recombinant Bax followed by centrifugation. The supernatants (sup) and pellets were immunoblotted with anti-Cyto c or HSP60 antibodies. (C) In vitro-translated MOAP-1 protein effectively restored the sensitivity of mitochondria from CHX-pretreated cells to the Cyto c releasing effect of Bax. Heavy membrane fractions containing mitochondria from the cells pretreated with CHX for 2 h were preincubated with PBS, rabbit reticulate lysate (R. lysate), S35-labeled (1 × 104 cpm/methionine) in vitro-translated MOAP-1, VDAC, or the M2 mutant of MOAP-1 (amino acids 1–115) for 20 min. The mitochondria was washed twice, treated with recombinant Bax, and analyzed as in B.
Discussion
It has been reported that proteasome activity is compromised in cells exposed to apoptotic stimuli, because some of the key subunits in the 19S regulatory complex are cleaved by activated caspases (27). As a consequence, levels of proteins that are constitutively degraded by UPS would increase and large amount of ubiquitinated proteins would be accumulated in the apoptotic cells (27). Interestingly, it has also been demonstrated that proteasome is required for initiation of apoptosis triggered by certain stimuli (16, 19, 28). Our data clearly showed that up-regulation of MOAP-1 is not a consequence of apoptosis, but it is rather an early event in the apoptotic signaling process engaged by multiple stimuli.
Ubiquitination is a pivotal step to mark proteins for degradation by the proteasome. Distinct E3 ligases associate with their specific substrate proteins and transfer the activated Ub to the target proteins through the isopeptide bond formed between the carboxyl-terminal Gly (G76) of Ub and the ε-NH2 group of lysine residue in substrate proteins. Although MOAP-1 has 17 internal lysine residues, the three lysine residues at the center domain of MOAP-1 (K161, K163, and K164) are the obvious candidate lysines that might be critical for the assembly of polyubiquitin chains, because polyubiquitination of the MOAP-1 M5 mutant (amino acids 115–190) can be readily detected. However, mutation of a single or all three of these lysines to arginine did not significantly suppress the ubiquitination of MOAP-1 (data not shown). Further systematic mutation analyses of lysine residues in full length MOAP-1 protein yielded similar results (data not shown) and it appears that, similar to certain Ub substrates (29, 30), no specific essential lysine residue for the polyubiquitination can be identified, suggesting that a unconventional mechanism may be involved in MOAP-1 polyubiquitination (29, 30).
A variety of mechanisms, including posttranslational modifications, such as phosphorylation, sumolyation, regulation by specific deubiquitination enzymes, and association with a third negative regulatory partner, have all been shown to be involved in affecting protein ubiquitination and its subsequent degradation by the proteasome (31). Multiple apoptotic stimuli stabilize MOAP-1 protein probably through inhibition of its ubiquitination-dependent degradation process. The underlying molecular steps and regulators involved, however, could be distinct for individual stimulus. The precise mechanism by which individual apoptotic stimulus inhibits the ubiquitination and degradation of MOAP-1 remains to be investigated.
Several mitochondrial proteins, such as ARTS (32) and Mcl-1 (16, 19), which are involved in the regulation of mitochondria-dependent apoptotic signaling, have been shown to be substrates of UPS. It is possible that a significant number of mitochondrial proteins that play important roles in mediating apoptotic or survival signaling are short-lived proteins. Interestingly, heavy membrane fractions containing mitochondria depleted of short-lived proteins by CHX are resistant to recombinant Bax- and tBid-mediated Cyto c release, demonstrating that the net effect of depleting short-lived proteins in mitochondria is to reduce rather than heighten the sensitivity of mitochondria to recombinant Bax-mediated Cyto c release. Surprisingly, incubation of the heavy membrane fractions containing mitochondria from CHX-pretreated cells with in vitro translated MOAP-1 protein alone was found to be sufficient to restore the ability of recombinant Bax to release Cyto c. These data lend further support to the idea that MOAP-1 plays an effector role for Bax function in mitochondria as suggested from our earlier data (11) and raises an intriguing possibility that MOAP-1 could be a key short-lived mitochondrial protein required for facilitating Bax function in mitochondria.
The first clinically tested proteasome inhibitor, Bortezomib, has recently been successfully launched for cancer the treatment of multiple myeloma (33, 34). It is noteworthy that proteasome inhibitors generally exhibit high toxicity to cells, presumably because of their activities in inhibiting a broad spectrum of proteins regulated by UPS. Interestingly, an experimental approach based on the crystal structure of MDM2 bound to a peptide from its substrate p53, has been successfully used to identify the small molecular weight compound Nutlin-3 that specifically blocks the interaction between MDM2 and p53. Indeed, degradation of p53 by MDM2 was abolished, and the p53 pathway became active in cells upon treatment with Nutlin-3 (35). Because the center domain of MOAP-1 (amino acids 116–190) is as sensitive as the wild-type protein to the regulatory effect of UPS, this region of MOAP-1 might contain the docking site for its specific E3 ligase. It is anticipated that chemical compounds with activity in interfering interaction between MOAP-1 and its E3 ligase should have a specific effect in stabilizing MOAP-1 protein. Because higher levels of MOAP-1 sensitize cells to Bax-mediated apoptotic signaling, these compounds might be potential antitumor agents. Mitochondria are increasingly being recognized as promising targets for cancer therapy (36, 37), identification of UPS as a mechanism for regulating stability of MOAP-1 would thus afford the opportunity for conceptualizing therapeutic strategies aimed at altering the functional activity of Bax in mitochondria.
Materials and Methods
Cell Culture, Transfection, and Generation of Stable Lines.
Cells were cultured in 5% CO2 in RPMI 1640, DMEM, or Mycoy's 5A medium supplemented with 10% FBS. Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA) was used for transfection. For generation of stable cell lines expressing exogenous myc-tagged MOAP-1, HCT116 or MCF-7 cells were transfected with pIRESneo vector or pIRESneo myc-MOAP-1 construct and selected in 1.25 mg/ml G418 (Gibco, Carlsbad, CA). Individual clonal lines were evaluated for the expression of myc-MOAP-1.
Determination of Protein Half-Life in Vivo.
A pulse–chase assay was performed as described in ref. 16. Cells were harvested, and cell lysates were subjected to IP analysis as described below. CHX, a protein synthesis inhibitor, was also used to estimate protein half-life as described in ref. 24.
Immunoblotting (IP) and Immunoprecipitation (IP).
Cells (≈1 × 107) were harvested and resuspended in 500 μl of RIPA buffer (50 mM Hepes, pH 7.4/150 mM NaCl/1% Nonidet P-40/0.1% SDS/0.25% Na-deoxycholate/1 mM EDTA/1 mM Na3VO4) supplemented with “complete” protease inhibitors (Roche, Indianapolis, IN), followed by sonication for 30 min at 4°C. After centrifugation to remove insolubles (350,000 × g for 20 min), protein content in cell lysates was determined (Bio-Rad, Hercules, CA). Equal amount of proteins for each cell lysate was subjected to IB or IP as described in ref. 10. Bax activation was analyzed as described in ref. 11.
Cell-Death Assay and Flow Cytometry Analysis.
WST-1 assay (Roche) was used to measure cell viability. For detection of subG1 DNA, cells were fixed in cold 70% ethanol, stained with 50 μg/ml propidium iodide, and analyzed with a FACScan flow cytometer (BD Biosciences, San Jose, CA). Mitochondrial potential change, measured by MitoTracker Red staining, was performed in accordance with the manufacturer's instructions (Molecular Probes, Eugene OR).
In Vitro Cyto c Release.
Equal amounts of heavy membrane fractions containing mitochondria were incubated with recombinant Bax or tBid at 30°C for 30 min, followed by centrifugation at 7,500 × g for 10 min at 4°C. Supernatants and pellets were subjected to Western blot analysis as described in ref. 11.
Supporting Information.
Real-time PCR, subcellular fractionation, expression and purification of recombinant Bax and t-Bid proteins, and further details on antibodies, chemicals, and plasmid construction are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. M. Pagano (New York University School of Medicine, New York, NY) for the Ub(K0) expression plasmid; Drs. B. L. Tang, S. L. Chan, and S. S. Lee for valuable comments about the manuscript. V.C.Y. is an adjunct staff member of the Department of Pharmacology, National University of Singapore. This work was supported by grants from the Singapore Agency for Science, Technology, and Research.
Abbreviations
- Cyto c
cytochrome c
- ETOP
etoposide
- CHX
cycloheximide
- IB
immunoblotting
- IP
immunoprecipitation
- MOAP-1
modulator of apoptosis
- THA
thapsigargin
- TRAIL
tumor necrosis factor-related apoptosis-inducing ligand
- UPS
the ubiquitin/proteasome system
- Ub
ubiquitin.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0700007104/DC1.
References
- 1.Wang X. Genes Dev. 2001;15:2922–2933. [PubMed] [Google Scholar]
- 2.Chan SL, Yu VC. Clin Exp Pharmacol Physiol. 2004;31:119–128. doi: 10.1111/j.1440-1681.2004.03975.x. [DOI] [PubMed] [Google Scholar]
- 3.Danial NN, Korsmeyer SJ. Cell. 2004;116:205–219. doi: 10.1016/s0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- 4.Strasser A. Nat Rev Immunol. 2005;5:189–200. doi: 10.1038/nri1568. [DOI] [PubMed] [Google Scholar]
- 5.Willis SN, Adams JM. Curr Opin Cell Biol. 2005;17:617–625. doi: 10.1016/j.ceb.2005.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng EH, Wei MC, Weiler S, Flavell RA, Mak TW, Lindsten T, Korsmeyer SJ. Mol Cell. 2001;8:705–711. doi: 10.1016/s1097-2765(01)00320-3. [DOI] [PubMed] [Google Scholar]
- 7.Certo M, Del Gaizo Moore V, Nishino M, Wei G, Korsmeyer S, Armstrong SA, Letai A. Cancer Cell. 2006;9:351–365. doi: 10.1016/j.ccr.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 8.Lindsten T, Ross AJ, King A, Zong WX, Rathmell JC, Shiels HA, Ulrich E, Waymire KG, Mahar P, Frauwirth K, et al. Mol Cell. 2000;6:1389–1399. doi: 10.1016/s1097-2765(00)00136-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wei MC, Zong WX, Cheng EH, Lindsten T, Panoutsakopoulou V, Ross AJ, Roth KA, MacGregor GR, Thompson CB, Korsmeyer SJ. Science. 2001;292:727–730. doi: 10.1126/science.1059108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tan KO, Tan KM, Chan SL, Yee KS, Bevort M, Ang KC, Yu VC. J Biol Chem. 2001;276:2802–2807. doi: 10.1074/jbc.M008955200. [DOI] [PubMed] [Google Scholar]
- 11.Tan KO, Fu NY, Sukumaran SK, Chan SL, Kang JH, Poon KL, Chen BS, Yu VC. Proc Natl Acad Sci USA. 2005;102:14623–14628. doi: 10.1073/pnas.0503524102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baksh S, Tommasi S, Fenton S, Yu VC, Martins LM, Pfeifer GP, Latif F, Downward J, Neel BG. Mol Cell. 2005;18:637–650. doi: 10.1016/j.molcel.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 13.Vos MD, Dallol A, Eckfeld K, Allen NP, Donninger H, Hesson LB, Calvisi D, Latif F, Clark GJ. J Biol Chem. 2006;281:4557–4563. doi: 10.1074/jbc.M512128200. [DOI] [PubMed] [Google Scholar]
- 14.Jesenberger V, Jentsch S. Nat Rev Mol Cell Biol. 2002;3:112–121. doi: 10.1038/nrm731. [DOI] [PubMed] [Google Scholar]
- 15.Vaux DL, Silke J. Nat Rev Mol Cell Biol. 2005;6:287–297. doi: 10.1038/nrm1621. [DOI] [PubMed] [Google Scholar]
- 16.Nijhawan D, Fang M, Traer E, Zhong Q, Gao W, Du F, Wang X. Genes Dev. 2003;17:1475–1486. doi: 10.1101/gad.1093903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nikrad M, Johnson T, Puthalalath H, Coultas L, Adams J, Kraft AS. Mol Cancer Ther. 2005;4:443–449. doi: 10.1158/1535-7163.MCT-04-0260. [DOI] [PubMed] [Google Scholar]
- 18.Kucharczak JF, Simmons MJ, Duckett CS, Gelinas C. Cell Death Differ. 2005;12:1225–1239. doi: 10.1038/sj.cdd.4401684. [DOI] [PubMed] [Google Scholar]
- 19.Willis SN, Chen L, Dewson G, Wei A, Naik E, Fletcher JI, Adams JM, Huang DC. Genes Dev. 2005;19:1294–1305. doi: 10.1101/gad.1304105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Linares LK, Hengstermann A, Ciechanover A, Muller S, Scheffner M. Proc Natl Acad Sci USA. 2003;100:12009–12014. doi: 10.1073/pnas.2030930100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ronca F, Chan SL, Yu VC. J Biol Chem. 1997;272:4252–4260. doi: 10.1074/jbc.272.7.4252. [DOI] [PubMed] [Google Scholar]
- 22.Holcik M, Sonenberg N. Nat Rev Mol Cell Biol. 2005;6:318–327. doi: 10.1038/nrm1618. [DOI] [PubMed] [Google Scholar]
- 23.Mayer RJ. Nat Rev Mol Cell Biol. 2000;1:145–148. doi: 10.1038/35040090. [DOI] [PubMed] [Google Scholar]
- 24.Bloom J, Amador V, Bartolini F, DeMartino G, Pagano M. Cell. 2003;115:71–82. doi: 10.1016/s0092-8674(03)00755-4. [DOI] [PubMed] [Google Scholar]
- 25.Hsu YT, Wolter KG, Youle RJ. Proc Natl Acad Sci USA. 1997;94:3668–3672. doi: 10.1073/pnas.94.8.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kreuz S, Siegmund D, Scheurich P, Wajant H. Mol Cell Biol. 2001;21:3964–3973. doi: 10.1128/MCB.21.12.3964-3973.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun XM, Butterworth M, MacFarlane M, Dubiel W, Ciechanover A, Cohen GM. Mol Cell. 2004;14:81–93. doi: 10.1016/s1097-2765(04)00156-x. [DOI] [PubMed] [Google Scholar]
- 28.Sohn D, Totzke G, Essmann F, Schulze-Osthoff K, Levkau B, Janicke RU. Mol Cell Biol. 2006;26:1967–1978. doi: 10.1128/MCB.26.5.1967-1978.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Demontis S, Rigo C, Piccinin S, Mizzau M, Sonego M, Fabris M, Brancolini C, Maestro R. Cell Death Differ. 2006;13:335–345. doi: 10.1038/sj.cdd.4401744. [DOI] [PubMed] [Google Scholar]
- 30.Herold MJ, Zeitz J, Pelzer C, Kraus C, Peters A, Wohlleben G, Berberich I. J Biol Chem. 2006;281:13663–13671. doi: 10.1074/jbc.M600266200. [DOI] [PubMed] [Google Scholar]
- 31.Pickart CM. Cell. 2004;116:181–190. doi: 10.1016/s0092-8674(03)01074-2. [DOI] [PubMed] [Google Scholar]
- 32.Lotan R, Rotem A, Gonen H, Finberg JP, Kemeny S, Steller H, Ciechanover A, Larisch S. J Biol Chem. 2005;280:25802–25810. doi: 10.1074/jbc.M501955200. [DOI] [PubMed] [Google Scholar]
- 33.Adams J. Nat Rev Cancer. 2004;4:349–360. doi: 10.1038/nrc1361. [DOI] [PubMed] [Google Scholar]
- 34.Nalepa G, Rolfe M, Harper JW. Nat Rev Drug Discov. 2006;5:596–613. doi: 10.1038/nrd2056. [DOI] [PubMed] [Google Scholar]
- 35.Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C, et al. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 36.Green DR, Kroemer G. Science. 2004;305:626–629. doi: 10.1126/science.1099320. [DOI] [PubMed] [Google Scholar]
- 37.Galluzzi L, Larochette N, Zamzami N, Kroemer G. Oncogene. 2006;25:4812–4830. doi: 10.1038/sj.onc.1209598. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.