Abstract
We present a system for random mutagenesis in Escherichia coli for the evolution of targeted genes. To increase error rates of DNA polymerase I (Pol I) replication, we introduced point mutations in three structural domains that govern Pol I fidelity. Expression of error-prone Pol I in vivo results in strong mutagenesis of a target sequence encoded in a Pol I-dependent plasmid (8.1 × 10–4 mutations per bp, an 80,000-fold increase), with a preference for plasmid relative to chromosome sequence. Mutagenesis is maximal in cultures maintained at stationary phase. Mutations are evenly distributed and show a variety of base pair substitutions, predominantly transitions. Mutagenesis extends at least 3 kb beyond the 400–500 nt reportedly synthesized by Pol I. We demonstrate that our error-prone Pol I can be used to generate enzymes with distinct properties by generating TEM-1 β-lactamase mutants able to hydrolyze a third-generation lactam antibiotic, aztreonam. Three different mutations contribute to aztreonam resistance. Two are found in the extended-spectrum β-lactamases most frequently identified in clinical isolates, and the third (G276R) has not been previously described. Our system of targeted mutagenesis in E. coli should have an impact on enzyme-based applications in areas such as synthetic chemistry, gene therapy, and molecular biology. Given the structural conservation between polymerases, this work should also provide a reference for altering the fidelity of other polymerases.
DNA polymerases catalyze the template-directed incorporation of deoxynucleotides into a growing primer terminus. Errors in nucleotide incorporation by DNA polymerases are significant sources of mutation, and contribute to genetic diversity. The fidelity of replication by DNA polymerases is ensured by base pair complementarity, by dNTP-induced conformational changes, and, in some cases, by proofreading catalyzed by a 3′ → 5′ exonuclease domain (1).
DNA polymerase I (Pol I) is one of five different DNA polymerases that have been described so far in Escherichia coli. In vivo, Pol I plays a role in lagging-strand replication of chromosomal DNA, DNA repair, and ColE1 plasmid replication (2). Pol I has been intensively studied as a model polymerase, and extrapolation to a diversity of other DNA polymerases has been possible given the conservation in structure and catalytic mechanism across eukaryotic and prokaryotic DNA polymerases (3).
Here, we engineered a highly error-prone E. coli DNA Pol I, and used this mutant polymerase as the basis of an in vivo mutagenesis system for modification of targeted sequences. Generation of enzyme variants is important in both basic studies of enzyme structure and function, and in creation of improved derivatives for medicine and technology. Several approaches for enzyme modification have been described to date. One of these approaches is random nucleotide mutagenesis, which involves the generation of large libraries harboring random mutations in vitro (4) and the subsequent identification of mutants of interest. Given the inordinate numbers of possible amino acid substitutions, the generation of nonfunctional mutants, and the need to separately identify the mutants of interest, the number of mutants that can be effectively analyzed is typically only a small fraction of all possible mutations. Random mutant libraries may also be generated in vivo by using mutator strains. However, currently available strains have little specificity for targeting specific genes.
The mutagenesis system we describe here is based on the creation of a highly inaccurate E. coli Pol I that carries low-fidelity mutations in three distinct structural domains. Expression of this error-prone Pol I in vivo results in very strong mutagenesis of a ColE1 plasmid, whereas mutagenesis in the chromosome is significantly lower. Coupling of mutagenesis to positive genetic selection eliminates defective mutants from the mutant pool and allows accumulation of mutations with a positive effect on the phenotype of interest, which dramatically increases the number of mutations that can be analyzed. We tested our Pol I-based system for enzyme modification by targeting TEM-1 β-lactamase for mutagenesis and selecting for resistance to a third generation lactam antibiotic, aztreonam.
Materials and Methods
Strains. JS200 cells (SC-18 recA718 polA12 uvrA155 trpE65 lon-11 sulA1) were first described as SC18–12. SC-18 carries tetracycline resistance and is insensitive to λ phage. Cells were grown under appropriate antibiotic selection: tetracycline at 12.5 μg/ml, chloramphenicol (cm) at 30 μg/ml, and/or kanamycin (kan) at 50 μg/ml. For growth conditions, competent cell preparation, and transformation, see ref. 5.
Reporter Plasmid Construction. The reporter plasmids used to measure the reversion frequency of the β-lactamase gene were pLA230 and pLA2800, located 230 bp and 2,800 bp away from the pUC19 plasmid origin of replication (ori), respectively (6). The other reporters derived from plasmids pLA2800 and pGPSΔ (6) are as follows.
pLA3700. The entire npt (kanr) gene was amplified by using oligonucleotides 5′-CATCGGTACCTTAACCAATTCTGATTAGAAAAAC-3′ and 5′-GATGGGTACCCTAGATTTAAATGATATCGGATCC-3′ containing KpnI restriction sites. The 980-bp amplified fragment was cloned, restricted with KpnI, and inserted into the KpnI site of plasmid pLA2800. This adds one extra copy of npt and moves the stop codon to 3,691 bp of the ori.
pLA2200. A 1,517-bp SalI fragment was excised from plasmid pLA3700, and the plasmid backbone was religated. This brought the stop codon to 2,174 bp of the ori.
pLA1400. The npt gene (amplified by using oligonucleotides containing SacI and AflIII adapters, 5′CATCGAGCTCTTAACCAATTCTGATTAGAAAAAC-3 and 5′-GATGACATGTCTAGATTTAAATGATATCGGATCC-3′) was cloned into the SacI and Aflll sites of pLA2800. This brings the β-lactamase ochre codon to 1,403 bp from the ori.
pLA700. The β-lactamase reporter was amplified by using oligonucleotides 5′-CATCGATATCTTACCAATGCTTAATCAGTG-3′ and 5′-GATGACTAGTCCCTATTTGTTTATTTTTCT-3′ and cloned into the SwaI and SpeI sites of pGPSΔ, placing the stop codon in the β-lactamase gene only 709 nt away from the ori.
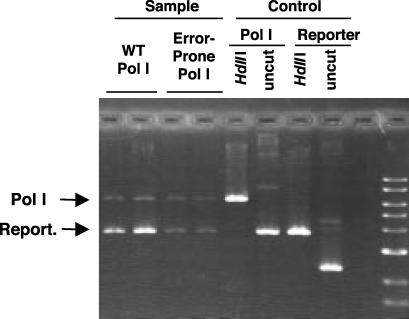
Quantitation of Target Gene Mutagenesis. To accurately measure mutation frequency at the target sequence in the plasmid, we first quantitated the amount of reporter plasmid. We used the Pol I plasmid, which is Pol I-independent (7), as an internal control. JS200 cells transformed with the Pol I plasmid (encoding either wild-type or A424D I709N A759R Pol I) and with the reporter plasmid pLA230, were grown overnight at 30°C; duplicate 10–5 dilutions were grown, in parallel, in 2XYT at 37°C for 17 h. Plasmids from each of these duplicate cultures were extracted by using the Perfectprep miniprep kit (Eppendorf) and linearized with HindIII, which has a unique site on both plasmids. These restriction digests containing linearized Pol I and reporter plasmids were run on a 1% agarose gel; the ratio of reporter plasmid to Pol I plasmid was reduced to 2.0 in cells expressing the error-prone Pol I, compared with a ratio of 18.5 in cells expressing wild-type Pol I. Similar results were obtained upon transformation with retrieved plasmids, comparing the number of kan-resistant colonies (reporter) to the number of cm-resistant ones (Pol I plasmid). The estimated copy number of pUC19, a plasmid that carries the same origin of replication as our reporter plasmid, is 100 copies per cell (2). pSC101 plasmids, which provide an origin of replication to our Pol I plasmid, are present at four to six copies per cell (2). This results in an estimate of only 10 copies per cell for our reporter plasmid. Under optimized conditions for mutagenesis, we observe an average frequency of reversions at the ochre stop codon of 1.9 × 10–2 per cell in different experiments. With an estimated 10 target plasmids per cell, this results in a reversion frequency of 1.9 × 10–3 per codon. Considering that two of the nine possible base pair substitutions at the ochre codon are not permissible, this is equivalent to 2.44 × 10–3 mutations per codon. Assuming an even distribution of mutations, this translates to 8.1 × 10–4 mutations per bp.
β-Lactamase Reversion and Rifampin Resistance Assays. E. coli JS200 cells carrying the Pol I plasmid (cmr) were transformed with the reporter plasmid (kanr), pLA230 unless otherwise indicated (6). Single cm-, kan-resistant colonies were grown in 5 ml LB tetracycline with kan and cm at 30°C to OD600 ≈ 0.5. For “initial” mutagenesis, a 1:50 dilution was grown to OD600 ≈ 0.7 in LB and plated. For “optimized” mutagenesis, the cultures were diluted 1:105 in 2XYT media prewarmed at 37°C. These new cultures were left in a water bath at 37°C for 15 min, grown for 15–17 h in a 37°C shaker, and plated. Reversion of the β-lactamase ochre codon was detected by plating the appropriate dilution of the saturated cultures onto kan, cm plates containing 50 μg/ml carbenicillin. For rifampin resistance, cells were plated in 25 μg/ml rifampin. The results are expressed as frequency relative to viable colonies (grown on kan cm alone) and represent the average of at least two clones that are carried independently.
Sequencing of Aztreonam-Resistant Clones. Individual colonies were picked from aztreonam plates and grown in the presence of 10 μg/ml aztreonam, under permissive conditions (30°C, OD600 ≈ 0.5) to avoid further mutagenesis. Plasmids were extracted from 3 ml of each of these cultures and resuspended in 50 μl of water. They were sequenced by using the following primers: Blac-5, 5′-TTACGGTTCCTGGCCTTTTGC-3′; Blac-6, 5′-GGTTGAGTACTCACCAGTCAC-3′; Blac-7, 5′-TCCGATCGTTGTCAGAAGTAA-3′; and Blac-8, 5′-CCATTTCCACCCCTCCCAGTT-3′. The sequence was analyzed by using sequencher software.
Phenotypic Characterization of Individual TEM β-Lactamase Mutants. The sites HaeIII, ScaI, and FspI (unique within the target plasmid) were used to subclone each individual mutation identified from our aztreonam selections within the β-lactamase ORF into the vector encoding β-lactamase and to generate different possible mutant combinations. These constructs were transformed into JS200 cells carrying the wild-type Pol I plasmid to maintain the same genetic background without inducing further mutagenesis, and grown in the absence of any β-lactam. Cells were diluted to the standard inoculum of 105 colony-forming units (cfu)/ml and grown for 16 h in the presence of increasing concentrations of aztreonam. We established IC50 values by plotting OD600 against drug concentration and finding the concentration that reduces survival by 50%. To account for variations in individual clone expression, two independent experiments were performed and averaged out. The standard deviation between the two experiments was in the order of 30%. To characterize the effect of these mutations on an optimal substrate, the standard inoculum of 105 cells per ml was grown in 1.5 mg/ml carbenicillin, a concentration that is very close to the minimal inhibitory concentration, and OD600 values were taken. These experiments were done twice independently, and we present the average and standard error of the mean.
Mutation Spectrum. JS200 cells expressing plasmid-encoded D424A I709N A759R Pol I and carrying the reporter plasmid pLA230 were grown under optimized conditions for mutagenesis and plated in 50 μg/ml carbenicillin. Plasmids were isolated from single colonies, and the sequence at the ochre codon was determined by using oligonucleotide Blac-5. Secondary mutations were confirmed by using oligonucleotides Blac-6, Blac-7, and Blac-8
Results
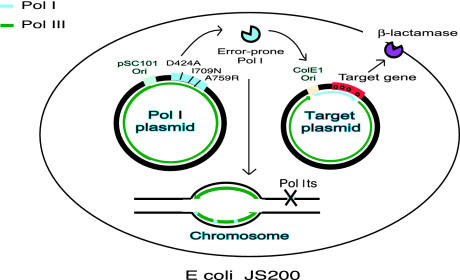
Two-Plasmid System for in Vivo Mutagenesis. Fig. 1 illustrates the two-plasmid system for random mutagenesis in vivo. The host is JS200, an E. coli strain that carries a chromosomally encoded, temperature-sensitive Pol I allele. JS200 cells are transformed with two plasmids. One plasmid is a low-copy plasmid with a pSC101 (Pol I-independent) origin of replication encoding error-prone Pol I. The second one is a ColE1 (Pol I-dependent) multicopy plasmid encoding a target sequence for mutagenesis. In this work we used pLA230 as a reporter for target plasmid mutagenesis. This plasmid encodes a TEM-1 β-lactamase with an ochre (TAA) codon located 230 bp downstream from the origin of replication (6). Carbenicillin-resistant colonies detect mutations within the ochre codon that cause reversion to an active β-lactamase. Thus, the frequency of carbenicillin resistance reflects the frequency of ochre codon mutagenesis in the target plasmid.
Fig. 1.
Two-plasmid system for in vivo mutagenesis. JS200 (polAts) cells were sequentially transformed with two plasmids. Plasmid I carries the polA gene under control of the tac promoter; it is a low-copy plasmid with a pSC101 origin of replication and carries a cm-resistance marker (25). Plasmid II carries the target gene placed in close proximity downstream of a pUC19 (ColE1-type) origin of replication; it is high-copy and carries a kan-resistance marker. Error-prone Pol I is expressed from plasmid I and initiates replication of plasmid II encoding the targeted gene downstream from ori; it has been reported that Pol I synthesizes the first 400–500 nt before a switch to the more accurate and processive Pol III (12). Pol III is the main replicative polymerase and is responsible for replicating the majority of chromosomal DNA. Pol I catalyzes Okazaki fragment joining and plays a role in DNA repair (2). Sequences synthesized by Pol I and Pol III are indicated in light blue and green, respectively.
Engineering of a Highly Error-Prone DNA Pol I. To achieve very high error rates, we generated a Pol I carrying amino acid replacements in three distinct structural domains that govern fidelity. We have previously shown that expression of E. coli Pol I containing specific point mutations at I709 results in detectable mutagenesis of our pLA230 reporter in vivo (6). I709 is located in motif A, a conserved sequence in the palm domain of the polymerase active site. Low-fidelity mutations at this position are postulated to enhance misincorporation by enlarging the substrate-binding pocket (8). Mutagenesis by I709 low-fidelity mutants was dramatically enhanced by concomitant inactivation of proofreading activity, conferred by the D424A mutation in the exonuclease domain (6). To increase Pol I error rates further, we analyzed the effect of concurrent mutation in the O-helix, a conserved sequence (motif B) that closes into the polymerase active site on dNTP binding. Low-fidelity mutations in the O-helix may stabilize the enzyme in the closed conformation, favoring misincorporation (3). Amino acids A759 and S756 were chosen for replacement, based on homology to Thermus aquaticus (Taq) Pol I T664 and A661, where specific substitutions increase the error rate of the polymerase. Both A759R and S756E significantly increased mutagenesis of the reporter in our in vivo assay, and this effect was greatly enhanced by inactivation of proofreading (Table 2 and data not shown).
Table 2. Mutation spectrum of secondary mutations in the β-lactamase gene.
|
100-500*
|
500-1,050*
|
100-1,050*
|
||||
|---|---|---|---|---|---|---|
| Mutations | n | % | n | % | n | % |
| Transitions | ||||||
| AT → GC | 10 | 33.3 | 7 | 36.8 | 17 | 34.7 |
| GC → AT | 13 | 43.3 | 9 | 47.4 | 22 | 44.9 |
| Transversions | ||||||
| AT → TA | 5 | 16.7 | 2 | 10.5 | 7 | 14.3 |
| AT → CG | 1 | 3.3 | 0 | 0 | 1 | 2.0 |
| GC → TA | 1 | 3.3 | 1 | 5.3 | 2 | 4.1 |
| GC → CG | 0 | 0 | 0 | 0 | 0 | 0 |
| Total | 30 | 100 | 19 | 100 | 49 | 100 |
These data exclude frameshifts, which were not scored in this assay.
Location of sequence is relative to the origin of replication.
Analysis of in Vivo Mutagenesis by Error-Prone Pol I. An optimal system for random in vivo mutagenesis would yield frequent mutations throughout a targeted gene and few mutations outside the target. We varied culture conditions to enhance mutagenic performance according to these criteria. We identified a number of factors with a positive effect, including the choice of rich medium, prewarming the medium before inoculation, and reducing the initial inoculum. The critical variable, although, was growing the culture to saturation rather than maintaining it in exponential phase.
Fig. 2 shows the results of mutagenesis assays performed under our optimized conditions (“Optimized”); for comparison, we also used the conditions previously described (“Initial”) (6). The frequency of Pol I-mediated mutagenesis was determined, both in the target plasmid (filled columns) and in the chromosome (open columns). Rifampin resistance, which detects point mutations in the rpoB gene (9), was our indicator of chromosomal mutagenesis. Table 1 summarizes the results. The A759R mutation in the O-helix yields similar levels of target plasmid mutagenesis as I709N, confirming that A759 is a critical determinant of fidelity in E. coli Pol I. Strikingly, expression of the I709N A759R D424A triple mutant results in mutagenesis frequencies that are more than additive, i.e., 6–26 times above those of each double mutant. Mutagenesis of the target plasmid in this experiment was 10,000 times above that yielded by wild-type Pol I, but in other experiments the increase in mutagenesis was as great as 200,000-fold (see Fig. 4B, for example). Five independent experiments performed under optimized conditions gave on average an 80,000-fold increase in mutagenesis. Note also that this apparent synergism is not significantly affected by culture conditions, as we found an increase in plasmid mutagenesis of 8- to 12-fold over each double mutant under initial conditions as well.
Fig. 2.
D424A I709N A759R Pol I achieves efficient and preferential mutagenesis, and mutagenesis is sensitive to culture conditions. JS200 cells transformed with a plasmid expressing Pol I and a plasmid encoding the β-lactamase reporter were grown under “initial” or “optimized” conditions as described in Materials and Methods. The plot represents carbenicillin (as an indicator of plasmid mutagenesis, solid columns) or rifampin resistance (as a readout for chromosomal mutagenesis, white columns) as a function of different Pol I low-fidelity mutants. Each point was plated in triplicate, and error bars represent standard error of the mean (P < 0.05).
Table 1. Error-prone Pol I achieves high levels of mutagenesis, with a preference for the targeted plasmid.
|
Mutagenesis*
|
Targeting†
|
|||
|---|---|---|---|---|
| Pol I | Initial | Optimized | Initial | Optimized |
| Wild type | 1.9 × 10-7 | 1.4 × 10-7 | 1 | 1 |
| D424A 1709N | 8.6 × 10-5 | 3.7 × 10-4 | 12.5 | 413 |
| A759R | 3.0 × 10-6 | ND | 14.2 | ND |
| D424A A759R | 5.9 × 10-5 | 7.9 × 10-5 | 4.9 | 68 |
| 1709N A759R | 3.6 × 10-4 | ND | 0.54 | ND |
| D424A1709N A759R | 7.0 × 10-4 | 2.1 × 10-3 | 1.0 | 390 |
ND, not determined.
By β-lactamase reversion assay.
The relative rate of plasmid versus chromosomal mutagenesis for each mutant polymerase was obtained after normalizing to that found for the wild-type polymerase. This factors in the different nature of the assays [forward assay (Rifr) versus reversion assay (β-lactamase)], and the difference in target numbers [single copy (Rifr) versus a multicopy (β-lactamase)].
Fig. 4.
Distribution of mutagenesis in the target plasmid. (A) Location of secondary mutations within 650 bp of target sequence. JS200 cells expressing the D424A I709N A759R polymerase and carrying the reporter plasmid pLA230 were grown under optimized conditions for mutagenesis and plated in 50 μg/ml carbenicillin. Single carbenicillin-resistant colonies were grown as described in Materials and Methods.(B) Mutagenesis is distance-sensitive but remains significant for at least 3.7 kb. JS200 cells were transformed with a plasmid containing D424A I709N A759R Pol I and with plasmids pLA230, pLA700, pLA1600, pLA2200, pLA2800, or pLA3700 as reporters. Cultures were grown under optimized conditions for mutagenesis and plated in the presence or absence of carbenicillin. Each point represents the average of two independent cultures, each plated in duplicate.
Mutagenesis is elevated in the reporter plasmid relative to the chromosome (Fig. 2, “Initial”). This bias probably reflects the limited extent of Pol I replication in the chromosome relative to ColE1 plasmid replication and is in line with previous reports (6, 10). Optimized cell culture conditions significantly enhanced this bias for ColE1 plasmid mutations (Fig. 2 and Table 1). In the case of the I709N A759R D424A triple mutant, optimized conditions increased plasmid targeting by 400-fold. We conclude that under our optimized conditions, mutagenesis preferentially occurs in the ColE1 plasmid.
To estimate the mutation load, we established the number of target plasmids present in cells expressing I709N A759R D424A Pol I (see Materials and Methods). In these cultures, we found only ≈10 copies per cell (Fig. 3). This is in contrast with ≈100 copies per cell in cultures expressing the wild-type Pol I. Assuming an even distribution of mutations throughout the targeted gene, this translates to a mutation frequency of 8.1 × 10–4 per bp (see Materials and Methods).
Fig. 3.
Reporter plasmid is less abundant in cells expressing D424A I709N A759R Pol I. JS200 cells transformed with the Pol I and reporter plasmids were grown under “optimized” conditions as described in Materials and Methods. These plasmids were restricted with HindIII (with a unique site on both plasmids) and run on a 1% agarose gel. In cells expressing wild-type Pol I, pLA230 plasmid signal is on average 18.6-fold greater than that of the Pol I plasmid. In cells expressing D424A I709N A759R Pol I, the signal of the reporter plasmid is only 2.0-fold that of the Pol I plasmid.
Mutation Spectrum. To characterize the nucleotide changes associated with I709N A759R D424A Pol I expression, we sequenced 158 carbenicillin-resistant clones. Of the 158 clones, 155 showed mutations in the TAA ochre codon, which gives an overall specificity of 98% for our reversion assay. Two mutations replaced A for G at the second position, resulting in an opal stop codon (TGA), and expression is likely caused by partial suppression by tRNAtrp (11). All permissible substitutions were observed, with a predominance of TTA (38%) and CAA (23%) substitutions. To establish a complete spectrum of base pair substitutions, we sequenced at least 650 bp for all 158 clones, from positions 100 to 750 downstream of ori. Forty-six secondary mutations were detected within this 650-bp interval. Of these, 40 were located in the ORF, of which 22 were silent. Only two mutations came up twice in this analysis, indicating a high degree of diversity.
Because Pol I is believed to copy the first 400–500 nt downstream of ori before a switch to the more accurate Pol III (12), we expected a higher frequency of mutations in this sequence. In contrast with this expectation, we observed a relatively even distribution of mutations along the length of the 650 bp sequenced (Fig. 4A). Comparing mutations between 100 and 500 bp with mutations between 500 and 750 bp, we detected a similar frequency of hits and a comparable spectrum (Table 2), suggesting the two sets of mutations had been generated by a common mechanism. Table 2 shows the mutation spectrum of the 46 secondary mutations and of three additional ones found further downstream. We detected a bias toward transitions (80%) with a predominance of GC → AT mutations (56%). This argues against a major involvement of SOS polymerases, which exhibit a bias toward AT → GC transitions and AT → CG transversions (Pol IV; ref. 13) or toward different transversions (Pol V; ref. 14). These results also present a profile that is distinct from simple mismatch repair overload; cells defective in mismatch repair show a stronger bias for transitions (96%), with a majority of AT → GC mutations (66%) (15). Thus, the mutation spectrum indicates that the majority of mutagenesis occurring within the first ≈700 bp downstream from the origin of replication is likely caused by Pol I errors during plasmid replication.
When the reporter gene was placed further downstream from the plasmid origin of replication, we did detect a significant decrease in the frequency of β-lactamase reversion (Fig. 4B). However, as shown, mutagenesis remained significantly elevated for at least 3 kb following an initial decrease. This finding indicates that large genes may be efficiently mutagenized in cells expressing I709N A759R D424A Pol I.
Evolution of β-Lactamase in Continuous Culture. To test the capacity of our system to evolve proteins with distinct properties, we chose TEM-1 β-lactamase as a target and used aztreonam as a selective agent. Aztreonam is a monobactam antibiotic distantly related to penicillin, the natural substrate of TEM-1. TEM-1 β-lactamase exhibits very low activity against aztreonam, but mutant variants that confer resistance have emerged in clinics (16). In this experiment, controls included cells expressing (i) wild-type Pol I and wild-type β-lactamase; (ii) D424A I709N A759R Pol I and a target plasmid carrying a large β-lactamase deletion; and (iii) wild-type Pol I and the deleted β-lactamase. Two independent selections were carried out under optimized mutagenic conditions. After one round of mutagenesis without selection, a 1:10 dilution of the culture was incubated with 0.5 μg/ml aztreonam (IC99 = 0.2 μg/ml). Viable cells were expanded by growing a 1:10 dilution at the same concentration of drug, after which another 1:10 dilution was grown in 32 μg/ml aztreonam. None of the controls showed detectable growth at this concentration of aztreonam, indicating that the presence of both error-prone Pol I and β-lactamase was essential for development of resistance. Surviving cells were plated at a higher concentration of aztreonam (64 μg/ml). Plasmids were obtained from single colonies, and the TEM-1 β-lactamase gene was directly sequenced by using specific primers.
Twenty-three sequences were obtained from each selection. All 46 surviving clones contain mutations at the same two positions most frequently observed in extended-spectrum β-lactamases identified in clinical isolates (24), namely E104 (changed to K) and R164 (changed to H in selection 1 and to S in selection 2). All 23 sequences obtained in selection 1 carried a silent mutation in the T180 codon. One sequence carried an additional, nonsynonymous mutation, G267R. This mutant also carried the silent mutation at position T180, which strongly suggests that the G267R mutation occurred in plasmids already carrying the E104K R164H amino acid substitutions. In selection 2, the E104K R164S double mutation carried no detectable silent mutations, and the E104 R164 wild-type sequence was also detected among the plasmid population before subcloning.
Phenotypic Analysis of Resulting Mutations. To rule out mutations elsewhere in the vector and to generate single, double, and triple amino acid substitutions, we subcloned each of the mutations found in our aztreonam selections into the wild-type target plasmid. For phenotypic analysis, the mutants were transformed in JS200 cells expressing wild-type Pol I (to avoid further mutagenesis). IC50 values for all mutants (representing an average of two individual clones) are shown in Table 3. The E104K and R164H/S single mutations appear to confer some resistance, but G267R alone does not. This argues that the G267R mutation could only have been obtained by using an iterative approach. In combination, each of these mutations makes a significant contribution to aztreonam resistance.
Table 3. β-Lactam resistance phenotypes of TEM-1 mutants.
| β-Lactamase | % Carbenicillin*† survival‡ | Fold aztreonam resistance* |
|---|---|---|
| Deletion | 1.3 ± 0.6 × 10-1§ | 0.57 |
| Wild type | 100.0 ± 9.0 | 1.00 |
| E104K | 86.4 ± 7.3§ | 2.70 |
| R164H | 87.9 ± 21 | 2.18 |
| R164S | 67.6 ± 12§ | 2.53 |
| G267R | 63.6 ± 0.7§ | 0.84 |
| E104K R164H | 76.2 ± 9.6§ | 43.7 |
| E104K R164S | 74.3 ± 2.9§ | 74.7 |
| E104K G267R | 66.9 ± 5.8§ | 2.87 |
| R164H G267R | 54.8 ± 1.8§ | 2.70 |
| E104K R164H G267R | 62.0 ± 4.9§ | 67.8 |
| E104K R164S G267R | 64.5 ± 5.8§ | 160.0 |
IC50, average of two experiments, normalized to wild type.
At a concentration of 1.5 mg/ml, which caused minimal growth inhibition in cells transfected with wild-type (2.1%, with a standard deviation of 6.3).
OD600, the error represents standard error of the mean (P < 0.05).
Statistically significant difference from wild type (P < 0.05).
Phenotypic analysis of our aztreonam resistance mutations also provides some insight into the evolutionary pathways seen in the two independent selections. The R164S amino acid substitution confers slightly enhanced aztreonam protection relative to R164H. In selection 1, stronger selective pressure may have contributed to the loss of the wild-type plasmid in bacteria that harbored the R164H mutation, increasing the number of double mutants susceptible to additional mutagenesis. The introduction of a third mutation, G267R, further enhanced resistance, to a level comparable to that of E104K R164S.
Discussion
We describe the rapid evolution of β-lactamase in vivo as an experimental application of Pol I random mutagenesis to evolution of enzymes. This approach has been outlined by us (6) and others (10), but, to our knowledge, it has not been successfully implemented before. The generation of a highly error-prone Pol I was critical for success. Our error-prone Pol I combines mutations in the proofreading domain and in motifs A and B in the polymerase domain. We showed a synergistic effect in vivo of motif A and motif B (O-helix) low-fidelity mutations. This observation supports models of replication in which motif A and the O-helix make distinct contributions to fidelity.
Optimizing culture conditions was another critical factor. Growing cultures to saturation was the single most important variable in optimization contributing to enhanced mutagenesis targeted to a ColE1 plasmid sequence (Fig. 2). Selective enhancement in plasmid mutagenesis did not correlate with an increase in target plasmid copy number, which is also ≈10 plasmids per cell in exponential cultures (not shown). The observed selective enhancement of plasmid mutagenesis may be caused by continued plasmid replication after chromosomal replication arrest. This would further restrict error-prone Pol I replication to ColE1 plasmid sequences and would also preferentially fix mutations introduced by nontargeted mechanisms such as mismatch repair and SOS mutagenesis to the plasmid. These two mechanisms may contribute to the enhancement in the frequency mutagenesis observed in saturated cultures. Under saturation conditions, mismatch repair is likely limiting (17) and SOS polymerases are expressed in the absence of exogenous DNA damage (18). The weighting toward transitions, however, argues against a predominance of SOS mutations.
Cells expressing error-prone Pol I harbor only an estimated 10 ColE1 plasmids per cell, compared with 100 plasmids in cells expressing wild-type Pol I. This decreases the size of our mutant libraries by 10-fold. On the other hand, a lower copy number of target plasmids may improve selection efficiency, as each new mutation potentially represents a larger fraction of expressed protein, and thus may have a larger impact on the phenotype. The decrease in plasmid copy number suggests that the three point mutations that we introduced to reduce polymerase fidelity may also decrease catalytic efficiency. In addition, our preliminary evidence suggests that the D424A I709N A759R mutations in Pol I may compromise RNAII primer recognition for initiation of ColE1 DNA synthesis (19) (data not shown).
We estimate a mutation frequency in our target sequence of 8.1 × 10–4 per bp, although with considerable variation between different experiments. This frequency is comparable to the highest mutation frequency reported in vivo, 5.0 × 10–4 in the XL1-Red strain (20). Unlike our host, this and other efficient mutator strains are deficient in major DNA repair pathways, which slows their growth and severely reduces transformation efficiency (20). Because mutagenesis in these strains is not restricted in any way to the sequence of interest, growth in culture invariably results in widespread genomic mutagenesis that may obscure phenotypic expression from mutations in the target gene (21, 22) and decrease the efficiency of mutagenesis with prolonged passage in culture (20).
Mutations were well distributed across the 650 bp sequenced (Fig. 4A). No qualitative or quantitative difference in mutagenesis was detected between the first 500 bp, which comprises sequence currently believed to be synthesized by Pol I (12), and the next 250 bp downstream, which is presumably synthesized by the more accurate Pol III. This suggests that Pol I leader synthesis may extend to ≈700 nt in the pLA230 reporter plasmid. This is longer than generally accepted, but the lack of the primosome assembly sequence (pas) site in our reporter plasmid may have altered Pol III replisome assembly. Mutagenesis in cells expressing D424A I709N A759R Pol I decreases in sequences located further from the plasmid origin of replication as expected if mutations were introduced through errors in Pol I replication of the plasmid (Fig. 4B). Intriguingly, the frequency of mutagenesis in sequences located far from the plasmid origin of replication fails to decrease to background levels. A direct interpretation would be that these mutations are also introduced by error-prone Pol I, which would imply a more extensive role for Pol I in plasmid replication than was previously supposed. A contribution from other mutagenic pathways in the cell is conceivable, especially in saturated cultures that are known to be promutagenic (23).
Pol I mutagenesis and aztreonam selection resulted in the rapid identification of TEM-1 β-lactamase mutants with altered substrate specificity in two independent selections. Aztreonam resistance increased by 150-fold. We identified resistance mutations at three positions in the β-lactamase protein, which has a probability of 10–10 (21), and two alternative pathways of accumulation. The E104K and R164S/H substitutions are the most prevalent among clinical isolates (24), whereas G267R, which further increases resistance by the other two, has not been previously reported (www.lahey.org/studies/temtable.htm). The absence of neutral mutations is consistent with the proposed mechanism of accumulation of beneficial mutations by sequential selection (22).
The G267R mutation is located in the loop connecting the B5 β-sheet to the N-terminal H11 helix. This loop undergoes a shift in the presence of the third most frequent mutation in clinical isolates, G238S. G267R may lead to increased aztreonam resistance in the presence of the E104K and R164H/S mutations by inducing a structural rearrangement in this loop resembling that of G238S mutants. The observed increase in sensitivity to carbenicillin associated with G267R (Table 3) is consistent with this proposition. Beyond helping to understand lactam resistance, the G267R mutation may arise in the field with continued pressure from extended-spectrum antibiotics such as aztreonam.
In summary, we present an example of random mutagenesis by Pol I applied to enzyme modification. Our system offers critical advantages over existing mutator strains for the construction and selection of mutant libraries in vivo, notably a healthy host, a diverse spectrum and good distribution of mutations, significant restriction to the target sequences, and iterative selection in culture. Thus, our system of rapid enzyme modification should facilitate progress in the improvement of complex biosynthetic or degradative pathways, and in overcoming rate-limiting steps for industrial and chemical applications. Given the structural conservation between polymerases, this work should also provide a reference for altering the fidelity of other polymerases.
Acknowledgments
We thank Dr. Akeo Shinkai for his generous assistance at the initial stages of this work. Thanks to Ann Blank, Eitan Glick, and Haiwei Guo for critical reading of the manuscript. We are grateful to Dr. Roel Schaaper for illuminating discussions and for the mutD strain. This work was supported by National Institutes of Health Grants CA78885 and ES 07032-25.
Abbreviations: Pol I, DNA polymerase I; Pol III, DNA polymerase III; ori, plasmid origin of replication; cm, chloramphenicol; kan, kanamycin.
References
- 1.Kunkel, T. A. & Bebenek, K. (2000) Annu. Rev. Biochem. 69, 497–529. [DOI] [PubMed] [Google Scholar]
- 2.Kornberg, A. & Baker, T. A. (1992) DNA Replication (Freeman, New York).
- 3.Patel, P. H., Suzuki, M., Adman, E., Shinkai, A. & Loeb, L. A. (2001) J. Mol. Biol. 308, 823–837. [DOI] [PubMed] [Google Scholar]
- 4.Cadwell, R. C. & Joyce, G. F. (1994) PCR Methods Appl. 3, S136–40. [DOI] [PubMed] [Google Scholar]
- 5.Camps, M. & Loeb, L. A. (2003) in Directed Enzyme Evolution Screening and Selection Methods, ed. Walker, J. M. (Humana, Totowa, NJ), pp. 11–18.
- 6.Shinkai, A. & Loeb, L. A. (2001) J. Biol. Chem. 276, 46759–46764. [DOI] [PubMed] [Google Scholar]
- 7.Cabello, F., Timmis, K. & Cohen, S. N. (1976) Nature 259, 285–290. [DOI] [PubMed] [Google Scholar]
- 8.Patel, P. H., Kawate, H., Adman, E., Ashbach, M. & Loeb, L. A. (2001) J. Biol. Chem. 276, 5044–5051. [DOI] [PubMed] [Google Scholar]
- 9.Jin, D. J. & Gross, C. A. (1988) J. Mol. Biol. 202, 45–58. [DOI] [PubMed] [Google Scholar]
- 10.Fabret, C., Poncet, S., Danielsen, S., Borchert, T. V., Ehrlich, S. D. & Janniere, L. (2000) Nucleic Acids Res. 28, E95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sandman, K. E. & Noren, C. J. (2000) Nucleic Acids Res. 28, 755–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Itoh, T. & Tomizawa, J. (1979) Cold Spring Harbor Symp. Quant. Biol. 43, 409–417. [DOI] [PubMed] [Google Scholar]
- 13.Kobayashi, S., Valentine, M. R., Pham, P., O'Donnell, M. & Goodman, M. F. (2002) J. Biol. Chem. 277, 34198–34207. [DOI] [PubMed] [Google Scholar]
- 14.Maor-Shoshani, A., Reuven, N. B., Tomer, G. & Livneh, Z. (2000) Proc. Natl. Acad. Sci. USA 97, 565–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schaaper, R. M. & Dunn, R. L. (1987) Proc. Natl. Acad. Sci. USA 84, 6220–6224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Petrosino, J., Cantu, C., III, & Palzkill, T. (1998) Trends Microbiol. 6, 323–327. [DOI] [PubMed] [Google Scholar]
- 17.Harris, R. S., Feng, G., Ross, K. J., Sidhu, R., Thulin, C., Longerich, S., Szigety, S. K., Hastings, P. J., Winkler, M. E. & Rosenberg, S. M. (1999) Mutat. Res. 437, 51–60. [PubMed] [Google Scholar]
- 18.Yeiser, B., Pepper, E. D., Goodman, M. F. & Finkel, S. E. (2002) Proc. Natl. Acad. Sci. USA 99, 8737–8741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cesareni, G., Helmer-Citterich, M. & Castagnoli, L. (1991) Trends Genet. 7, 230–235. [DOI] [PubMed] [Google Scholar]
- 20.Greener, A., Callahan, M. & Jerpseth, B. (1997) Mol. Biotechnol. 7, 189–195. [DOI] [PubMed] [Google Scholar]
- 21.Long-McGie, J., Liu, A. D. & Schellenberger, V. (2000) Biotechnol. Bioeng. 68, 121–125. [DOI] [PubMed] [Google Scholar]
- 22.Negri, M. C., Lipsitch, M., Blazquez, J., Levin, B. R. & Baquero, F. (2000) Antimicrob. Agents Chemother. 44, 2485–2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rosenberg, S. M. (2001) Nat. Rev. Genet. 2, 504–515. [DOI] [PubMed] [Google Scholar]
- 24.Orencia, M. C., Yoon, J. S., Ness, J. E., Stemmer, W. P. & Stevens, R. C. (2001) Nat. Struct. Biol. 8, 238–242. [DOI] [PubMed] [Google Scholar]
- 25.Takeshita, S., Sato, M., Toba, M., Masahashi, W. & Hashimoto-Gotoh, T. (1987) Gene 61, 63–74. [DOI] [PubMed] [Google Scholar]