Abstract
Myotubularin-related protein 7 (MTMR7) is a member of the myotubularin (MTM) family. The cDNA encoding the mouse MTMR7 contains 1,983 bp, and the predicted protein has a deduced molecular mass of 75.6 kDa. Northern and Western blot analyses showed that MTMR7 is expressed mainly in brain and mouse neuroblastoma N1E-115 cells. Recombinant MTMR7 dephosphorylated the D-3 position of phosphatidylinositol 3-phosphate and inositol 1,3-bisphosphate [Ins(1,3)P2]. The substrate specificity of MTMR7 is different than other MTM proteins in that this enzyme prefers the water-soluble substrate. Immunofluorescence showed that MTMR7 is localized in Golgi-like granules and cytosol, and subcellular fractionation showed both cytoplasmic and membrane localization of MTMR7 in N1E-115 cells. An MTMR7-binding protein was found in an anti-MTMR7 immunoprecipitate from N1E-115 cells and identified as MTM-related protein 9 (MTMR9) by tandem mass spectrometry. The coiled-coil domain of MTMR9 was sufficient for binding to MTMR7. The binding of MTMR9 increased the Ins(1,3)P2 phosphatase activity of MTMR7. Our results show that MTMR7 forms a complex with MTMR9 and dephosphorylates phosphatidylinositol 3-phosphate and Ins(1,3)P2 in neuronal cells.
Myotubular myopathy is an X-linked severe congenital disorder characterized by hypotonia and respiratory insufficiency. Mutations in myotubularin 1 (MTM1) cause the disorder as identified by positional cloning (1). At least 13 MTM1-like proteins have been found in the human genome, and they are termed MTM-related proteins (MTMRs) (2–4). MTMR2 is mutated in a recessive form of Charcot-Marie-Tooth disease type 4B, a demyelinating neurological disorder (5). Because mutations in different MTM proteins cause distinct disorders, it seems that MTM proteins are not redundant (6).
MTM proteins contain a dual-specificity protein tyrosine phosphatase (PTP) motif, and MTM1, MTMR1, MTMR2, MTMR3, MTMR4, and MTMR6 have been shown to dephosphorylate the D-3 position of phosphatidylinositol (PtdIns) 3-phosphate [PtdIns(3)P] as the preferred substrate (7–11). Some MTM proteins are thought to be enzymatically inactive because they lack a conserved cysteine residue in the PTP motif that is required for activity. These include MTMR5 [SET-binding factor 1 (Sbf-1)], MTMR9 (LIP-STYX), MTMR10, MTMR11, MTMR12 [3-phosphatase adaptor protein (3-PAP)], and MTMR13 (Sbf-2).
PtdIns(3)P phosphatases were isolated before the discovery of MTM proteins. Types I and II PtdIns(3)P phosphatase were shown to be composed of a homo- and a heterodimer, respectively (12). Recently, MTM1 and MTMR12 were shown to be the subunits of type II 3-phosphatase (13, 14). MTMR2 has also been found to bind MTMR5 and MTMR12 (14, 15). Therefore, inactive MTM proteins may bind to and regulate the localization and/or activity of the enzymatic molecules as adapter proteins. MTMR5-deficient mice have impaired spermatogenesis and germ-cell differentiation (16), indicating that the adapter proteins are functionally important.
Other conserved structures of MTM proteins are a SET interaction domain (SID) and predicted coiled-coil domain. The SET domain is named from the Drosophila proteins Su(var)3-9, Enhancer-of-zeste, and Trithorax, which contain the motif of unknown function (17). MTMR5 was isolated as a SET domain-binding protein of the protooncogene product Hrx by using yeast two-hybrid screening (18). The region that was essential for the binding of MTMR5 to the SET domain was identified and termed the SID (18). A splicing isoform of MTMR12 that lacks the SID failed to interact with MTM1 (14). A deletion of the coiled-coil domain abolished the interaction between MTMR2 and MTMR5 (15).
The major substrate of MTM proteins, PtdIns(3)P, is formed by the phosphorylation of PtdIns by PtdIns 3-kinase, and this enzyme is found on the surface of early endosomes (19). In Saccharomyces cerevisiae, disruption of yeast PtdIns 3-kinase (Vps34p) results in decreased cellular PtdIns(3)P levels and defects in vacuolar protein sorting (20–24). Vps34p is the sole PtdIns 3-kinase in yeast, and it phosphorylates only PtdIns (23). A temperature-sensitive yeast vps34 mutant decreased PtdIns(3)P levels within 1 min after Vps34p was inactivated (22). A defect of yeast MTM (Ymr1P) increased PtdIns(3)P content in yeast (7). The PtdIns 3-kinase inhibitor wortmannin inhibits several membrane-trafficking events in mammalian cells (25), suggesting that PtdIns 3-kinase and PtdIns(3)P also play a role in membrane trafficking in higher organisms. The data suggest that PtdIns(3)P levels are regulated in cells and that MTM proteins are involved in the process.
We analyzed MTMR7 to define the function of MTM proteins in cells further. It is specifically expressed in brain and neuronal cells and dephosphorylates PtdIns(3)P and Ins(1,3)P2. We further found an MTMR7-binding protein and identified it as MTMR9. MTMR6 also binds to MTMR9. The relationship among MTMR6, -7, and -9 and their biological function are discussed in this article.
Materials and Methods
cDNA Constructs. We used human and mouse brain Marathon-Ready cDNAs (CLONTECH) as templates to amplify human MTMR6, mouse MTMR7, and mouse MTMR9 cDNAs by PCR. Full-length human MTMR2 and human MTMR12 constructs were from Gregory Taylor (University of Michigan Medical School, Ann Arbor) and Harshal Nandurkar (St. Vincent's Hospital, Melbourne), respectively. PCR products were subcloned by using the TOPO-TA cloning kit (Invitrogen). The cDNAs were sequenced by the Protein and Nucleic Acid Chemistry Laboratory (Washington University, St. Louis), and sequence data were obtained from both strands. The cDNA sequences of MTMR2/MTMR6/MTMR7/MTMR9/MTMR12 corresponded to GenBank accession numbers NM_004685/ NM_004685/AK081973/BC046275/AY028703. Full-length MTM cDNAs were subcloned into pCMV5-Myc, pCMV5-HA, or pCNF-FLAG, expression vectors for expression in mammalian cells. GATEWAY cloning systems vectors (Invitrogen) were used to create N-terminal GST full-length MTMR7 and N-terminal (His)6-tagged MTMR9 according to manufacturer instructions. The Bac-to-Bac baculovirus expression system (Invitrogen) was used for expression of MTMR7 and MTMR9 according to manufacturer instructions. Domains of these proteins were also expressed in bacteria by using pGEX-6P-1 (Amersham Pharmacia Biosciences) as follows: MTMR9-SID (amino acids 350–482), MTMR9-coil (coiled-coil domain, amino acids 475–545), MTMR7-SID-coil (amino acids 355–554), and MTMR9-SID-coil (amino acids 350–545). All of these domain constructs harbored a GST tag at the N terminus.
Northern Blot Analysis. Multitissue Northern blot membranes were purchased from CLONTECH. A PstI–PstI fragment of MTMR7 cDNA (from –14 to 610 bp) was used as an [α-32P]dCTP-labeled 624-bp probe. Hybridization was performed following manufacturer protocol (CLONTECH).
Cell Culture and Expression of Recombinant Proteins in Tissue-Culture Cells. COS-7 and N1E-115 cells were cultured in DMEM (Mediatech, Herndon, VA) containing 10% FCS and 60 μg/ml kanamycin at 37°C in a humidified atmosphere of 95% air/5% CO2. Mammalian expression constructs were transfected into COS-7 cells by using Lipofectamine 2000 (Invitrogen) following manufacturer protocol, and cells were harvested 48 h after transfection. N1E-115 cells were differentiated into neuronal form by serum starvation for 3 days by using dishes and glass coverslips that were treated with sterilized 3% polyethyleneimine (Sigma) in 0.1 M borate buffer (pH 8.4) for at least 6 h. They then were washed twice with sterilized deionized water and medium, respectively, just before plating.
Western Blot Analysis. Cells were harvested in a lysis buffer (50 mM Tris·HCl, pH 7.5/150 mM NaCl/1 mM EDTA) supplemented with a protease inhibitor mixture (Roche Molecular Biochemicals) and sonicated briefly. Mouse tissues were homogenized in the same buffer with a Potter–Elvehjem homogenizer and sonicated for 30 sec on ice. All cell homogenates were centrifuged at 10,000 × g for 20 min at 4°C. The protein concentration of cell lysates was measured (Bio-Rad) and adjusted to 2.5 μg/μl with SDS sample buffer. Growing N1E-115 cells were fractionated into cytosol and membrane fractions as described by Poussu et al. (26). Equal proportions of extracts were run on SDS/PAGE.
Protein Expression and Purification. GST-MTMR7 and (His)6-MTMR9 were prepared by using the Bac-to-Bac baculovirus expression system (Invitrogen). Bovine p85α and GST-bovine p110α PtdIns 3-kinase baculovirus were from Bart Vanhaesebroeck (Ludwig Institute for Cancer Research, London) and coinfected into Sf9 cells. GST-tagged SID or/and coiled-coil proteins were expressed in the Escherichia coli strain BL21-CodonPlus (DE3)-RIL (Stratagene). The cells containing GST proteins were collected with lysis buffer, supplemented with 10 μg/ml leupeptin and 10 μg/ml aprotinin, and sonicated for 30 sec on ice. All cell homogenates were centrifuged at 30,000 × g for 30 min at 4°C. The cell lysates were mixed with glutathione-Sepharose 4B beads (Amersham Pharmacia Biosciences) and rotated for 3 h at 4°C. The beads were washed three times with the same buffer, and GST proteins were eluted with 50 mM reduced glutathione at 4°C. Sf9 cells expressing (His)6-MTMR9 were collected with buffer (50 mM Tris·HCl, pH 7.5/300 mM NaCl/10 mM imidazole/10 μg/ml leupeptin/10 μg/ml aprotinin). Baculovirus (His)6-MTMR9 was purified by using nickel-agarose beads (Qiagen, Valencia, CA) according to manufacturer protocol. The beads were washed three times with the same buffer and eluted with buffer containing 250 mM imidazole. For enzyme assays of MTMR7 and MTMR9 complex, Sf9 cell lysate containing GST-MTMR7 was divided into two tubes and rotated with glutathione-Sepharose 4B beads for 2 h at 4°C. Purified (His)6-MTMR9 protein then was added to one tube, and both were rotated for 1 h. Proteins were eluted by using glutathione as described above. After SDS/PAGE, proteins were stained with Coomassie blue, and protein concentration was determined by densitometry.
Enzyme Assays. The D-3 position-radiolabeled phosphoinositide substrates were made by using 100 μM PtdIns, PtdIns 4-phosphate, PtdIns 5-phosphate, or PtdIns 4,5-bisphosphate, bovine p85α, and GST-bovine p110α PtdIns 3-kinase/1 μCi/ml [α-32P]ATP (1 Ci = 37 GBq)/1 μM ATP/0.5 mM phosphatidyl serine in 50 μl of 50 mM Tris·HCl, pH 7.5, for 20 min at 37°C. The reaction was stopped by adding 150 μl of 1 M HCl. Lipids were extracted in chloroform/methanol (2:1, vol/vol), and the products were separated by TLC with chloroform/methanol/ammonia/water (90:90:7:22, vol/vol). Radiolabeled phosphoinositides were scraped from TLC plates and extracted into chloroform. For the lipid phosphatase assay, 3.5 μg of GSTMTMR7 was incubated with 100 μM substrate containing 5,000 cpm, 0.5 mM phosphatidyl serine/10 mM MgCl2 in 50 μl of 50 mM Tris·HCl, pH 7.5, for 15 min at 37°C. Reactions were stopped by the addition of 150 μl of 1 M HCl, and lipids were extracted in chloroform/methanol (2:1, vol/vol). The products were analyzed by TLC and autoradiography. Assays of water-soluble substrates contained 100 μM inositol polyphosphates, 89 ng of GST-MTMR7, and 10 mM MgCl2 in 50 μl of 50 mM Tris·HCl, pH 7.5, incubated for 10 min at 37°C. The Malachite-green assay was carried out as described by Maehama et al. (27). The activity of the MTMR7 and MTMR9 complex was determined in buffer containing 100 mM sodium acetate, 50 mM Bis-Tris, and 50 mM Tris, pH 6.0 (27).
Anti-MTMR7 Antibody. A rabbit polyclonal antibody was raised by using a peptide from mouse MTMR7 corresponding to amino acids 552–660. The antibody was purified on GST-MTMR7 bound to cyanogen bromide-activated Sepharose 4B beads (Sigma). Antiserum was mixed with the beads, rotated for3hat4°C, and after extensive washing, eluted with 0.1 M glycine (pH 2.5) and neutralized with 1 M Tris·HCl (pH 9.0).
Immunofluorescence. N1E-115 cells cultured on glass coverslips were fixed with 10% formaldehyde in PBS for 10 min at room temperature, washed three times with PBS, incubated for 5 min in 0.2% Triton X-100 in PBS, washed five times with PBS, incubated with 3% BSA and 10% calf serum in PBS for 30 min, and washed five times with PBS. N1E-115 cells then were incubated with anti-MTMR7 and anti-γ-adaptin antibodies (BD Biosciences) for 1 h. After five washes with PBS, the cells were incubated with Alexa Fluor 488 goat anti-rabbit IgG and Alexa Fluor 594 goat anti-mouse IgG (Molecular Probes) for 30 min. The cells were washed five times with PBS, mounted in GEL/MOUNT (Biomedia, Foster City, CA), and observed with a confocal microscope.
Isolation of Binding Proteins. N1E-115 cells grown to 90% confluency in 100-mm dishes were incubated in l-methionine- and l-cysteine-free DMEM (Invitrogen) that contained 10% dialyzed FCS and 100 μCi/ml of Tran35S-label (ICN) for3hat37°C. The cells were washed three times with PBS and harvested with 500 μl of lysis buffer, supplemented with 1% Triton X-100 and protease inhibitor mixture (Roche Molecular Biochemicals). The cells were sonicated and centrifuged at 10,000 × g for 20 min at 4°C. The cell lysate was mixed with 10 μg of anti-MTMR7 antibody or rabbit IgG (Sigma) and rotated for 1 h at 4°C, followed by the addition of 50 μl of a 50% slurry of protein A-Sepharose beads. After being rotated for 1 h at 4°C, the beads were washed five times with the same buffer containing 600 mM NaCl and then mixed with SDS sample buffer. To isolate a large amount of the MTMR7-binding protein, N1E-115 cells were grown to 90% confluency on 40 150-mm dishes, and cell lysate was mixed with 700 μg of anti-MTMR7 and 1 ml of a 50% slurry of protein A-Sepharose beads. The immunoprecipitate was run on SDS/PAGE. The gel was stained with Bio-Safe Coomassie stain (Bio-Rad), and the protein band was cut out, trypsinized, and analyzed by tandem mass spectrometry at the Protein and Nucleic Acid Chemistry Laboratory.
Immunoprecipitation and GST Pull-Down Assay. COS-7 cells that overexpressed epitope-tagged proteins were harvested with lysis buffer supplemented with 1% Triton X-100 and protease inhibitor mixture (Roche Molecular Biochemicals) and sonicated for 30 sec on ice. After centrifugation, anti-hemagglutinin (HA) (Sigma) or anti-FLAG (Sigma) antibody-conjugated beads was added to the lysate and rotated for 2 h at 4°C. GST pull-down assays used the GST-tagged domains bound to glutathione-Sepharose 4B beads mixed with COS-7 cell lysates containing Myc-tagged MTM proteins. Samples were rotated overnight at 4°C, and beads were washed five times with the same buffer containing 600 mM NaCl and then mixed with SDS sample buffer and analyzed by Western blotting.
Results
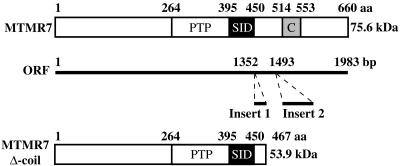
cDNA Sequence of Mouse MTMR7. In the GenBank database, two types of mouse MTMR7 cDNA sequences are found. One type (two entries) contains inserts 1 and 2 in its ORF, whereas the other four did not (Fig. 1). When the sequence of the two inserts was translated into amino acid sequence, insert 1 was found to contain several stop codons in all three frames. To evaluate the significance of the inserts, the genome sequences of mouse and human MTMR7 were compared, and their intron–exon boundaries were determined (Tables 1 and 2, which are published as supporting information on the PNAS web site, www.pnas.org). In the mouse genome, insert 1 was found between exons 11 and 12, and insert 2 was found between exons 12 and 13. The human MTMR7 genome sequence contained a part of insert 2 but did not contain insert 1. We conclude that the cDNA that contains the inserts is a splicing isoform unique to mouse MTMR7 and termed it MTMR7 Δ-coil, because the stop codons in insert 1 result in deletion of the coiled-coil domain. We used the mouse MTMR7 cDNA that did not contain the inserts for further studies. MTMR2 also has splicing isoforms (28), and we have also found several splicing isoforms of MTMR1 and MTMR6 (unpublished data). The significance of those splicing isoforms is uncertain.
Fig. 1.
MTMR7 and its splicing isoform. The diagrams show predicted MTMR7 (Top) and its splicing isoform protein (MTMR7 Δ-coil) (Bottom). The translation of the splicing isoform mRNA is terminated within an insert 1 and yields MTMR7 Δ-coil. The amino acid numbers are indicated on the top of each diagram. (Middle) The ORF of mouse MTMR7 and its splicing isoform. Nucleotide numbers are depicted on the top of the diagram. PTP, PTP domain; C, coiled-coil domain. PTP and coiled-coil domains are defined by smrt (http://smart.embl-heidelberg.de).
The coding sequence of mouse MTMR7 is 1,983 bp and predicts a protein of 75.6 kDa. The amino acid sequence of mouse MTMR7 is similar to that of mouse MTMR6 with 56.8% identical amino acids. The amino acid sequence of MTMR7 contains a potentially functional PTP motif, an SID, and a coiled-coil domain as observed in other MTM proteins (Fig. 1).
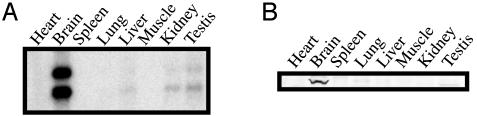
Tissue Distribution of MTMR7. Northern blot analysis showed that mouse MTMR7 is expressed in brain, liver, kidney, and testis (Fig. 2A). Its expression was especially high in brain, and Western blot analysis of mouse tissues also showed that MTMR7 is expressed mainly in brain (Fig. 2B). The antibody also detected MTMR7 in N1E-115 mouse neuroblastoma cells (data not shown). When cells were differentiated into a neuronal form, the expression level of MTMR7 did not change (data not shown).
Fig. 2.
Tissue distribution of mouse MTMR7. (A) Northern blot analysis of mouse MTMR7. An MTMR7 cDNA probe detected ≈2.7- and 3.5-kb bands in brain, liver, kidney, and testis. (B) Western blot analysis of mouse MTMR7 protein. The anti-MTMR7 antibody detected a single 85-kDa band in mouse brain lysate. In each lane, 50 μg of protein was loaded.
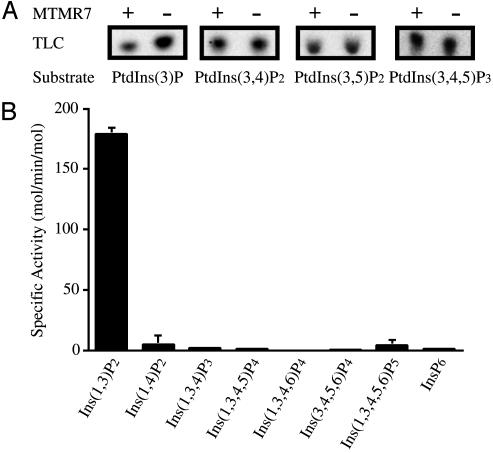
Enzymatic Activity of MTMR7. Previously characterized MTM proteins specifically dephosphorylate the D-3 position of PtdIns(3)P (7–11). MTMR7 contains a CX5R motif that is essential for the phosphatase activity of MTM proteins. We prepared recombinant MTMR7 and determined its substrate specificity as shown in Fig. 3A. MTMR7 reduced the level of PtdIns(3)P substrate, indicating the dephosphorylation of the D-3 position of PtdIns(3)P, whereas the other substrates remained unchanged. We conclude that MTMR7 is a PtdIns(3)P phosphatase. Using a Malachite-green assay method, we further determined its specific activity using PtdIns(3)P. The specific activity of MTMR7 was 14.7 mol/min per mol, which is much lower than that of MTM1, MTMR1, and MTMR2 but similar to that of MTMR6 (10). We also examined the inositol polyphosphate 3-phosphatase activity of MTMR7. As shown in Fig. 3B, MTMR7 specifically dephosphorylated Ins(1,3)P2 and did not show any activity with other inositol polyphosphates. The specific activity with Ins(1,3)P2 substrate is >10-fold higher than that with the lipid PtdIns(3)P under the conditions used.
Fig. 3.
Enzymatic activity of MTMR7. (A) Radiolabeled PtdIns substrates (100μM) were incubated with or without GST-MTMR7 (3.5 μg) at 37°C for 15 min, and the remaining substrate was analyzed by TLC. (B) The substrate specificity of MTMR7 with water-soluble inositol polyphosphates. GST-MTMR7 (89 ng) was incubated with 100 μM of each substrate at 37°C for 10 min. Released inorganic phosphate was determined by a dye-assay method (27). PtdIns(3,4)P2, PtdIns 3,4-bisphosphate; PtdIns(3,5)P2, PtdIns 3,5-bisphosphate; PtdIns(3,4,5)P3, PtdIns 3,4,5-trisphosphate; Ins(1,4)P2, inositol 1,4-bisphosphate; Ins(1,3,4)P3, inositol 1,3,4-trisphosphate; Ins(1,3,4,5)P4, inositol 1,3,4,5-tetrakisphosphate; Ins(1,3,4,6)P4, inositol 1,3,4,6-tetrakisphosphate; Ins(3,4,5,6)P4, inositol 3,4,5,6-tetrakisphosphate; Ins(1,3,4,5,6)P5, inositol 1,3,4,5,6-pentakisphosphate; InsP6, inositol 1,2,3,4,5,6-hexakisphosphate.
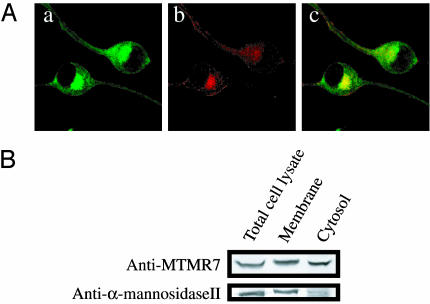
Cellular Localization of MTMR7 in N1E-115 Cells. We stained neuroblastoma N1E-115 cells with anti-MTMR7 antibody to determine the cellular localization of MTMR7. As shown in Fig. 4Aa, anti-MTMR7 antibody stained a cluster of granules close to the nucleus and the cytoplasm in N1E-115 cells. When NIH 3T3 cells were stained with anti-MTMR7 antibody, no signal was observed and no MTMR7 was seen by Western blotting of these cells (data not shown). Therefore, we conclude that the anti-MTMR7 antibody specifically detected endogenous MTMR7 in N1E-115 cells. When N1E-115 cells were costained with anti-MTMR7 and anti-γ-adaptin antibody, MTMR7-positive granules partially overlapped with γ-adaptin-positive granules (Fig. 4A). Therefore, MTMR7 may be enriched in Golgi or endosomes. N1E-115 cells were fractionated into cytosol and membrane fractions, and each was analyzed by Western blotting (Fig. 4B). MTMR7 was found in both cytosol and membrane fractions.
Fig. 4.
Cellular localization of MTMR7 in N1E-115 cells. (A) Immunostaining with anti-MTMR7 antibody. N1E-115 cells were stained with anti-MTMR7 antibody (a) and anti-γ-adaptin antibody (b), and then the two figures were merged (c). (B) N1E-115 cells were fractionated into cytosol and membrane fractions. In each fraction, endogenous MTMR7 was detected by anti-MTMR7 antibody. α-Mannosidase II was used as a marker of a membrane protein.
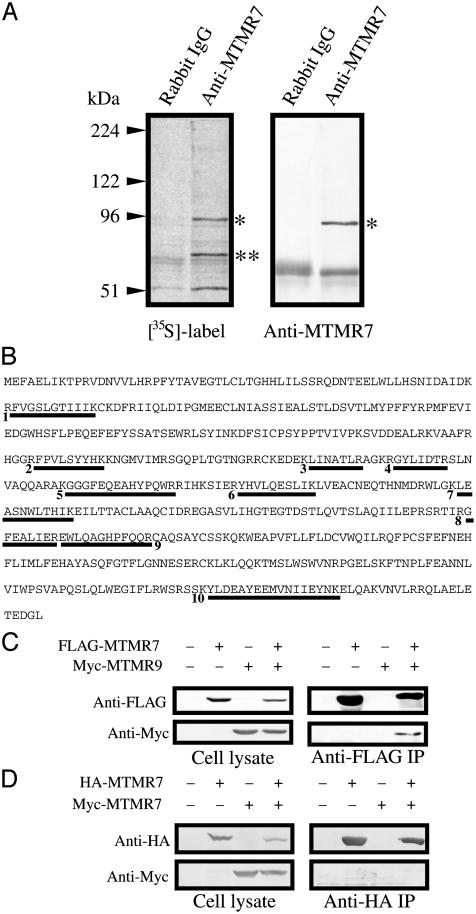
Identification of an MTMR7-Binding Protein. We next determined whether MTMR7 forms a complex with another protein. N1E-115 cells were labeled with [35S]methionine and [35S]cysteine, and MTMR7 was immunoprecipitated as shown in Fig. 5A. Anti-MTMR7 antibody immunoprecipitated two proteins (Fig. 5A Left). The result of Western blotting indicated that the upper band was MTMR7 (Fig. 5A Right). Therefore, the lower band was thought to be an MTMR7-binding protein. The density of the two bands is similar, suggesting that the complex consists of a 1:1 complex with almost all MTMR7 in a complex in vivo. To identify the MTMR7-binding protein, N1E-115 cells were grown on 40 150-mm dishes, and the binding protein was purified by using anti-MTMR7 antibody. The binding protein was analyzed by tandem mass spectrometry and identified as MTMR9 (Fig. 5B). We identified 10 peptides from MTMR9 that spanned 115 of 546 amino acids comprising 21% of MTMR9. FLAG-tagged MTMR7 and Myc-tagged MTMR9 were coexpressed in COS-7 cells and immunoprecipitated by anti-FLAG antibody (Fig. 5C). Anti-Myc antibody detected the MTMR9 in the immunoprecipitate. This result confirmed the data of tandem mass spectrometry. Because type I PtdIns(3)P phosphatase is a homodimer (12), we investigated whether MTMR7 formed a homodimer. HA-tagged MTMR7 and Myc-tagged MTMR7 were also coexpressed in COS-7 cells, and their binding was examined (shown in Fig. 5D). There was no homodimer formed.
Fig. 5.
Isolation and identification of MTMR7-binding protein. (A) N1E-115 cells were labeled with [35S]methionine and [35S]cysteine and immunoprecipitated with anti-MTMR7 antibody. (Left) Autoradiograph. (Right) Western blot. Single and double asterisks indicate MTMR7 and the MTMR7-binding protein, respectively. The numbers depicted on the left are molecular mass markers. (B) The amino acid sequence of mouse MTMR9. The binding protein was digested by trypsin and analyzed by tandem mass spectrometry. The data obtained were searched against the NCBInr database by using the mascot search program (Matrix Science, London). Ten tryptic peptides that matched MTMR9 are underlined. (C) Interaction of MTMR7 and MTMR9. FLAG-tagged MTMR7 and Myc-tagged MTMR9 were coexpressed in COS-7 cells. The FLAG-tagged protein was purified with anti-FLAG antibody beads, and Myc-tagged MTMR9 was detected with anti-Myc antibody. (D) Analysis of homodimer formation of MTMR7. Myc- and HA-tagged MTMR7 were coexpressed in COS-7 cells. HA-tagged protein was immunoprecipitated (IP) with anti-HA antibody-conjugated beads, and immunoprecipitates were analyzed by Western blotting.
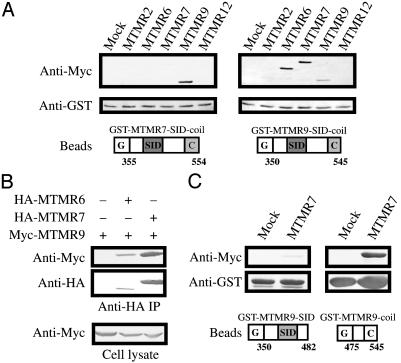
MTMR7 and MTMR9 Interaction Domains. MTMR5 binds to the protooncogene product Hrx via an SID, and that domain is conserved in all MTM proteins (2). Therefore, the SID was a candidate for the module that connects MTMR7 and MTMR9. However, another recent study indicated that the putative coiled-coil domain is also important for the dimerization of MTM proteins (15). Therefore, GST-MTMR7-SID-coil and GST-MTMR9-SID-coil beads were prepared, and their binding to MTMR2, MTMR6, MTMR7, MTMR9, and MTMR12 was examined (Fig. 6A). The GST-MTMR7-SID-coil construct specifically bound to MTMR9, and GST-MTMR9-SID-coil construct bound to MTMR7. In addition, GST-MTMR9-SID-coil beads bound to MTMR6. GST-MTMR9-SID-coil also bound to MTMR9 weakly, suggesting that MTMR9 may form homodimers. Because the GST pull-down assay showed that MTMR6 also bound to MTMR9, these two proteins were coexpressed in COS-7 cells, and their binding was examined by immunoprecipitation as shown in Fig. 6B. Although the expression level of MTMR6 was much less than that of MTMR7, MTMR6 bound to full-length MTMR9 in amounts comparable to its binding to MTMR7 in vivo.
Fig. 6.
Interaction of MTMR6, MTMR7, and MTMR9. (A) Myc-tagged MTM proteins were expressed in COS-7 cells. Cell lysates were mixed with GSTMTMR7-SID-coil or GST-MTMR9-SID-coil protein beads, and a GST pull-down assay was performed. Mock, empty pCMV5-Myc vector is transfected; G, GST; C, coiled-coil domain. The numbers depicted under the diagrams of GST proteins indicate the amino acid numbers of each construct. (B) MTMR6 associates with full-length MTMR9. HA-tagged MTMR6 and MTMR7 were transfected into COS-7 cells with Myc-tagged MTMR9. HA-tagged proteins were immunoprecipitated (IP). Immunoprecipitates were analyzed by Western blotting with anti-Myc antibody. (C) Identification of MTMR7-binding domain of MTMR9. Myc-tagged MTMR7 was expressed in COS-7 cells. Cell lysates were mixed with GST-MTMR9-SID or GST-MTMR9-coil protein bound to glutathione-Sepharose 4B beads, and a GST pull-down assay was performed.
We prepared GST-MTMR9-SID and GST-MTMR9-coiled-coil beads to define the binding site of MTMR9. The interaction with Myc-tagged MTMR7 was examined by a GST pull-down assay. GST-MTMR9-coil beads interacted with Myc-tagged MTMR7 (Fig. 6C Right). Myc-tagged MTMR7 faintly bound to GST-MTMR9-SID, but it was negligible compared with GSTMTMR9-coil (Fig. 6C Left). These data show that the coiled-coil domain is sufficient for MTMR9 to interact with MTMR7 in vitro.
Effect of MTMR9 Binding on MTMR7. MTMR5 and MTMR12 are reported to determine the cellular localization of their binding partners (14, 15). Therefore, we coexpressed FLAG-tagged MTMR7 and Myc-tagged MTMR9 in COS-7 cells and observed their cellular localization. When cells were fractionated, FLAG-tagged MTMR7 was found in both cytosol and membrane fractions and coexpression of Myc-tagged MTMR9 did not alter this (data not shown). In COS-7 cells, ectopically expressed MTMR7 and MTMR9 were observed in cytosol by immunof luorescence, and coexpression of those proteins showed no difference in localization from single expressions (data not shown).
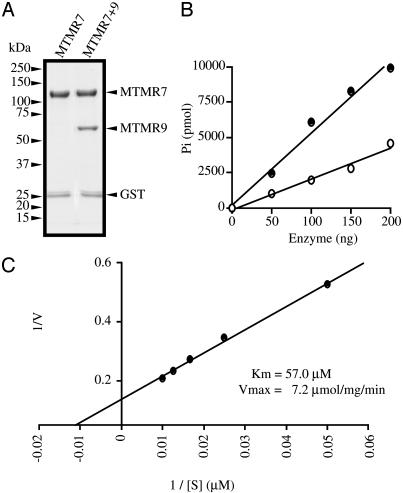
We further prepared a complex of GST-MTMR7 and (His)6-MTMR9 to define a function for MTMR9. The amount of protein was estimated by densitometry, and ≈80% of MTMR7 formed a complex with MTMR9 in our enzyme preparation based on densitometry of the bands in Fig. 7A. We assayed MTMR7 and the complex as shown in Fig. 7B. The complex had 2.5 times the activity of MTMR7 under these conditions. When assays were carried out at pH 5.5, the difference was even greater (5-fold), mainly because of the decreased activity of free MTMR7 at the lower pH (data not shown). The kinetics of hydrolysis of Ins(1,3)P2 by the complex is shown in Fig. 7C. Immunoprecipitation showed that almost all MTMR7 is complexed with MTMR9 in N1E-115 cells (Fig. 5A), suggesting that the activity of MTMR7 is regulated by MTMR9.
Fig. 7.
Effect of the binding of MTMR9 to MTMR7. (A) Purification of recombinant MTMR7 and MTMR9 complex. The complex of MTMR7 and MTMR9 was prepared as described in Materials and Methods. Proteins were run on SDS/PAGE and stained with Coomassie blue. (B) Comparison of the Ins(1,3)P2 phosphatase activity of MTMR7 and its complex with MTMR9. Open circles indicate MTMR7, and filled circles represent the MTMR7 and MTMR9 complex. Enzymes were incubated with 100 μM substrate for 10 min at 37°C. The amount of enzyme was normalized to GST-MTMR7. (C) Lineweaver–Burk plot of GST-MTMR7 and (His)6-MTMR9 complex for hydrolysis of Ins(1,3)P2. The enzymatic activity of the MTMR7 and MTMR9 complex was determined at pH 6.0.
Discussion
We find that MTMR7 binds MTMR9 both in vivo in N1E-115 cells and in vitro with recombinant proteins. MTMR7 dephosphorylated PtdIns(3)P in vitro similar to other known active MTM proteins. However, its lipid phosphatase activity is much less than that of other MTM proteins, and it prefers the water-soluble substrate Ins(1,3)P2. The substrate preference was not altered by binding to MTMR9 (data not shown). No function of Ins(1,3)P2 is yet known, and thus the function of MTMR7 remains unknown. Cellular localization of MTMR7 implies that the complex of MTMR7 and MTMR9 dephosphorylates PtdIns(3)P at late endosomes. Almost all endogenous MTMR7 was bound to MTMR9, and some MTMR7 was found in a membrane fraction in N1E-115 cells (Fig. 4B and 5A Left). Therefore, some complexes of MTMR7 and MTMR9 are bound to membranes. PtdIns 5-phosphate has been shown to increase the PtdIns(3)P phosphatase activity of MTM1 and MTMR6 (29). Therefore, it is possible that some modification of the complex itself or other intracellular factors increase the lipid phosphatase activity of MTMR7 in vivo.
The SID is conserved in all MTM family members, suggesting that this domain is important for function. We showed that MTMR7 bound to MTMR9 and that the SID-coil region of each was involved in the association. We had supposed that the two MTMR proteins would bind via an SID, because that domain mediates the binding of MTMR5 and Hrx (18). However, GST-MTMR9-SID did not interact with MTMR7, and GSTMTMR9-coil did (Fig. 6C). We conclude that the coiled-coil domain is sufficient for binding. The MTMR7 coiled-coil domain may also participate in complex formation. Deletion and point mutation of either coiled-coil domain of MTMR2 and MTMR5 abolished the binding (15). However, there are no data that show that MTM proteins interact exclusively via each coiled-coil domain (e.g., the two domains alone bind to each other). We found that MTM1 binds to MTMR12 and that a splicing isoform of MTMR12 that lacked the SID did not bind to MTM1 (14). The crystal structures of the heterodimeric proteins may help to elucidate the interactions of MTM family proteins.
We found that enzymatically inactive MTMR9 complexes with both MTMR6 and MTMR7. MTMR5 and MTMR12 also associate with active MTM proteins (14, 15). The concept of adaptor proteins was originally suggested when types I and II PtdIns(3)P phosphatases were isolated from rat brain (12). In that case, the type I enzyme was a homodimer of 65 kDa, whereas the type II was comprised of a 65- and 78-kDa adaptor subunit. A cDNA encoding the 78-kDa adaptor protein was cloned recently and designated 3-phosphatase adaptor protein (3-PAP) and later as MTMR12 (2, 13). We recently found that a binding partner of MTMR12 is MTM1 and that the complex affects the cellular localization of MTM1 (14). The nature of the type I homodimer isolated from rat brain remains uncertain. Schaletzky et al. (29) showed that MTM1 oligomerizes in the presence of substrate, thus type I 3-phosphatase may be an MTM1 homodimer.
The amino acid sequences of MTMR6 and MTMR7 are 53.4% identical. Both proteins bind to MTMR9. According to the results of Northern and Western blotting, the expression of MTMR7 is restricted to brain. The expression of MTMR6 is ubiquitous and is also expressed in brain (6). The effect of MTMR9 on MTMR6 activity remains to be determined.
Supplementary Material
Acknowledgments
We thank Monita Wilson and Shao-Chun Chang for careful reading of the manuscript. This work was supported by National Institutes of Health Grants HL 55772 and HL 16634.
Abbreviations: MTM, myotubularin; MTMR, MTM-related protein; PTP, protein tyrosine phosphatase; PtdIns, phosphatidylinositol; PtdIns(3)P, PtdIns 3-phosphate; SET, Su(var)3-9, Enhancer-of-zeste, and Trithorax; Sbf, SET-binding factor; Ins(1,3)P2, inositol 1,3-bisphosphate; SID, SET interaction domain; HA, hemagglutinin.
References
- 1.Laporte, J., Hu, L. J., Kretz, C., Mandel, J.-L., Kioschis, P., Coy, J. F., Klauck, S. M., Poustka, A. & Dahl, N. (1996) Nat. Genet. 13, 175–182. [DOI] [PubMed] [Google Scholar]
- 2.Wishart, M. J. & Dixon, J. E. (2002) Trends Cell Biol. 12, 579–585. [DOI] [PubMed] [Google Scholar]
- 3.Senderek, J., Bergmann, C., Weber, S., Ketelsen, U.-P., Schorle, H., Rudnik-Schöneborn, S., Büttner, R., Buchheim, E. & Zerres, K. (2003) Hum. Mol. Genet. 12, 349–356. [DOI] [PubMed] [Google Scholar]
- 4.Azzedine, H., Bolino, A., Taïeb, T., Birouk, N., Di Duca, M., Bouhouche, A., Benamou, S., Mrabet, A., Hammadouche, T., Chkili, T., et al. (2003) Am. J. Hum. Genet. 72, 1141–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bolino, A., Muglia, M., Conforti, F. L., LeGuern, E., Salih, M. A. M., Georgiou, D.-M., Christodoulou, K., Hausmanowa-Petrusewicz, I., Mandich, P., Schenone, A., et al. (2000) Nat. Genet. 25, 17–19. [DOI] [PubMed] [Google Scholar]
- 6.Laporte, J., Blondeau, F., Buj-Bello, A., Tentler, D., Kretz, C., Dahl, N. & Mandel, J.-L. (1998) Hum. Mol. Genet. 7, 1703–1712. [DOI] [PubMed] [Google Scholar]
- 7.Taylor, G. S., Maehama, T. & Dixon, J. E. (2000) Proc. Natl. Acad. Sci. USA 97, 8910–8915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walker, D. M., Urbé, S., Dove, S. K., Tenza, D., Raposo, G. & Clague, M. J. (2001) Curr. Biol. 11, 1600–1605. [DOI] [PubMed] [Google Scholar]
- 9.Zhao, R., Qi, Y., Chen, J. & Zhao, Z. J. (2001) Exp. Cell Res. 265, 329–338. [DOI] [PubMed] [Google Scholar]
- 10.Kim, S.-A., Taylor, G. S., Torgersen, K. M. & Dixon, J. E. (2001) J. Biol. Chem. 277, 4526–4531. [DOI] [PubMed] [Google Scholar]
- 11.Laporte, J., Liaubet, L., Blondeau, F., Tronchère, H., Mandel, J.-L. & Payrastre, B. (2002) Biochem. Biophys. Res. Commum. 291, 305–312. [DOI] [PubMed] [Google Scholar]
- 12.Caldwell, K. K., Lips, D. L., Bansal, V. S. & Majerus, P. W. (1991) J. Biol. Chem. 266, 18378–18386. [PubMed] [Google Scholar]
- 13.Nandurkar, H. H., Caldwell, J. C., Whisstock, J. C., Layton, M. J., Gaudet, E. A., Norris, F. A., Majerus, P. W. & Mitchell, C. A. (2001) Proc. Natl. Acad. Sci. USA 98, 9499–9504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nandurkar, H. H., Layton, M., Laporte, J., Selan, C., Corcoran, L., Caldwell, K. K., Mochizuki, Y., Majerus, P. W. & Mitchell, C. A. (2003) Proc. Natl. Acad. Sci. USA 100, 8660–8665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim, S.-A., Vacratsis, P. O., Firestein, R., Cleary, M. L. & Dixon, J. E. (2003) Proc. Natl. Acad. Sci. USA 100, 4492–4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Firestein, R., Nagy, P. L., Daly, M., Huie, P., Conti, M. & Cleary, M. L. (2002) J. Clin. Invest. 109, 1165–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alvarez-Venegas, R. & Avramova, Z. (2002) Gene 285, 25–37. [DOI] [PubMed] [Google Scholar]
- 18.Gui, X., De Vio, I., Slany, R., Miyamoto, A., Firestein, R. & Cleary, M. L. (1998) Nat. Genet. 18, 331–337. [DOI] [PubMed] [Google Scholar]
- 19.Gillooly, D. J., Morrow, I. C., Lindsay, M., Gould, R., Bryant, N. J., Gaullier, J.-M., Parton, R. G. & Stenmark, H. (2000) EMBO J. 19, 4577–4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herman, P. K. & Emr, S. D. (1990) Mol. Cell. Biol. 10, 6742–6754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stack, J. H., DeWald, D. B., Takegawa, K. & Emr, S. D. (1995) J. Cell Biol. 129, 321–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wurmser, A. E. & Emr, S. D. (1998) EMBO J. 17, 4930–4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stenmark, H. & Gillooly, D. J. (2001) Cell. Dev. Biol. 12, 193–199. [DOI] [PubMed] [Google Scholar]
- 24.Gillooly, D. J., Simonsen, A. & Stenmark, H. (2001) Biochem. J. 355, 249–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Futter, C. E., Collinson, L. M., Backer, J. M. & Hopkins, C. R. (2001) J. Cell Biol. 155, 1251–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poussu, A., Lohi, O. & Lehto, V.-P. (2000) J. Biol. Chem. 275, 7176–7183. [DOI] [PubMed] [Google Scholar]
- 27.Maehama, T., Taylor, G. S., Slama, J. T. & Dixon, J. E. (2000) Anal. Biochem. 279, 248–250. [DOI] [PubMed] [Google Scholar]
- 28.Bolino, A., Marigo, V., Ferrera, F., Loader, J., Romio, L., Leoni, A., Di Duca, M., Cinti, R., Cecchi, C., Feltri, M. L., et al. (2002) Gene 283, 17–26. [DOI] [PubMed] [Google Scholar]
- 29.Schaletzky, J., Dove, S. K., Short, B., Lorenzo, O., Clague, M. J. & Barr, F. A. (2003) Curr. Biol. 13, 504–509. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.