Abstract
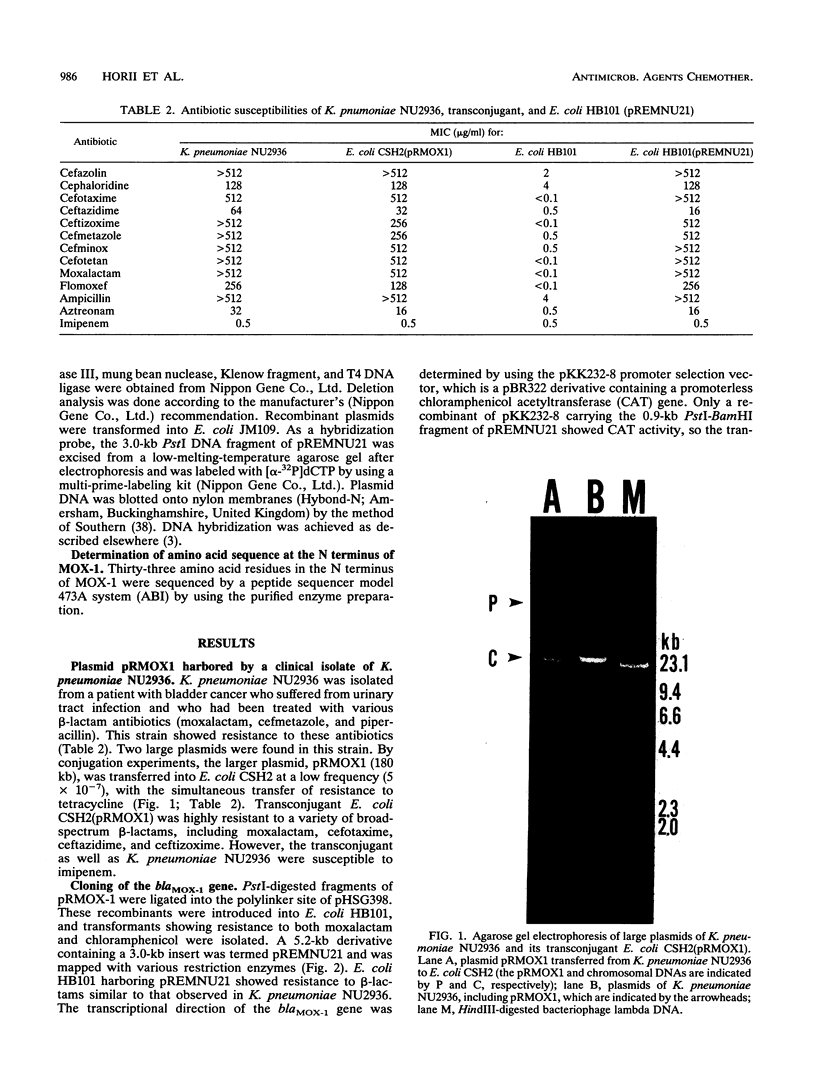
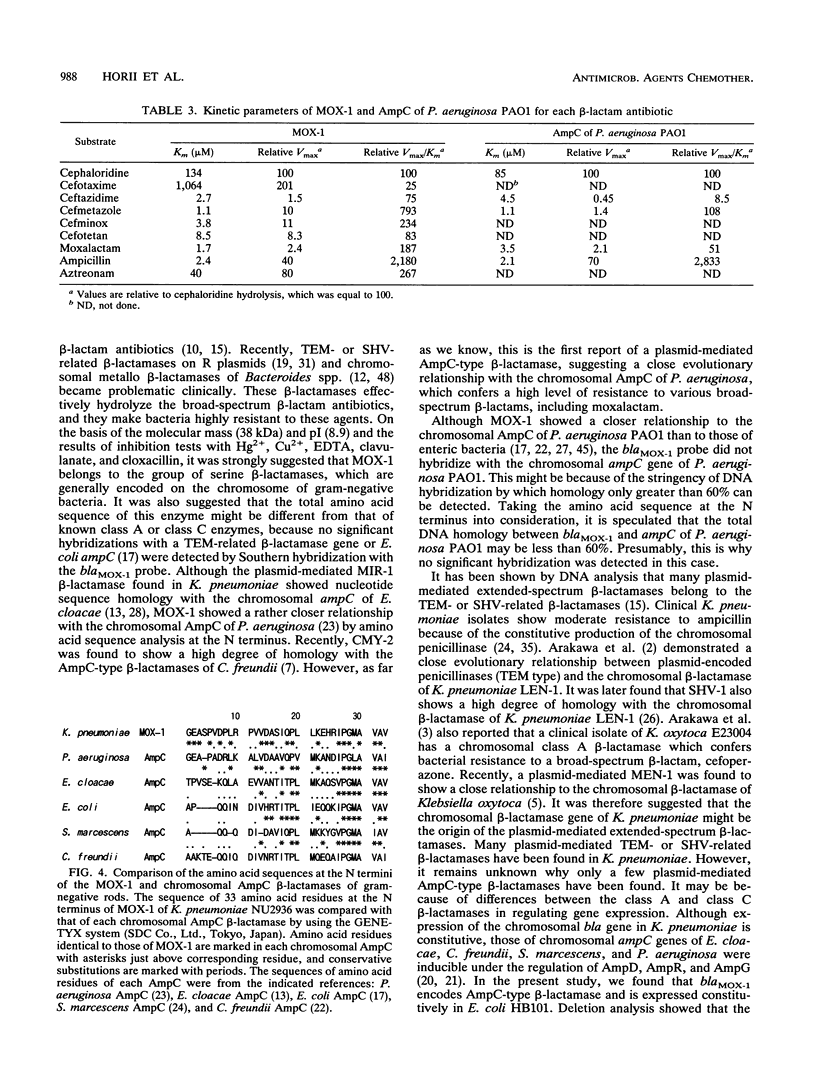
Klebsiella pneumoniae NU2936 was isolated from a patient and was found to produce a plasmid-encoded beta-lactamase (MOX-1) which conferred resistance to broad spectrum beta-lactams, including moxalactam, flomoxef, ceftizoxime, cefotaxime, and ceftazidime. Resistance could be transferred from K. pneumoniae NU2936 to Escherichia coli CSH2 by conjugation with a transfer frequency of 5 x 10(-7). The structural gene of MOX-1 (blaMOX-1) was cloned and expressed in E. coli HB101. The MIC of moxalactam for E. coli HB101 producing MOX-1 was > 512 micrograms/ml. The apparent molecular mass and pI of this enzyme were calculated to be 38 kDa and 8.9, respectively. Hg2+ and Cu2+ failed to block enzyme activity, and the presence of EDTA in the reaction buffer did not reduce the enzyme activity. However, clavulanate and cloxacillin, serine beta-lactamase inhibitors, inhibited the enzyme activity competitively (Kis = 5.60 and 0.35 microM, respectively). The kinetic study of MOX-1 suggested that it effectively hydrolyzed broad-spectrum beta-lactams. A hybridization study confirmed that blaMOX-1 is encoded on a large resident plasmid (pRMOX1; 180 kb) of strain NU2936. By deletion analysis, the functional region was localized within a 1.2-kb region of the plasmid. By amino acid sequencing, 18 of 33 amino acid residues at the N terminus of MOX-1 were found to be identical to those of Pseudomonas aeruginosa AmpC. These findings suggest that MOX-1 is a plasmid-mediated AmpC-type beta-lactamase that provides enteric bacteria resistance to broad-spectrum beta-lactams, including moxalactam.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ambler R. P. The structure of beta-lactamases. Philos Trans R Soc Lond B Biol Sci. 1980 May 16;289(1036):321–331. doi: 10.1098/rstb.1980.0049. [DOI] [PubMed] [Google Scholar]
- Arakawa Y., Ohta M., Kido N., Fujii Y., Komatsu T., Kato N. Close evolutionary relationship between the chromosomally encoded beta-lactamase gene of Klebsiella pneumoniae and the TEM beta-lactamase gene mediated by R plasmids. FEBS Lett. 1986 Oct 20;207(1):69–74. doi: 10.1016/0014-5793(86)80014-x. [DOI] [PubMed] [Google Scholar]
- Arakawa Y., Ohta M., Kido N., Mori M., Ito H., Komatsu T., Fujii Y., Kato N. Chromosomal beta-lactamase of Klebsiella oxytoca, a new class A enzyme that hydrolyzes broad-spectrum beta-lactam antibiotics. Antimicrob Agents Chemother. 1989 Jan;33(1):63–70. doi: 10.1128/aac.33.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann B. J. Pedigrees of some mutant strains of Escherichia coli K-12. Bacteriol Rev. 1972 Dec;36(4):525–557. doi: 10.1128/br.36.4.525-557.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthélémy M., Péduzzi J., Bernard H., Tancrède C., Labia R. Close amino acid sequence relationship between the new plasmid-mediated extended-spectrum beta-lactamase MEN-1 and chromosomally encoded enzymes of Klebsiella oxytoca. Biochim Biophys Acta. 1992 Jul 13;1122(1):15–22. doi: 10.1016/0167-4838(92)90121-s. [DOI] [PubMed] [Google Scholar]
- Bauernfeind A., Grimm H., Schweighart S. A new plasmidic cefotaximase in a clinical isolate of Escherichia coli. Infection. 1990 Sep-Oct;18(5):294–298. doi: 10.1007/BF01647010. [DOI] [PubMed] [Google Scholar]
- Brosius J. Plasmid vectors for the selection of promoters. Gene. 1984 Feb;27(2):151–160. doi: 10.1016/0378-1119(84)90136-7. [DOI] [PubMed] [Google Scholar]
- Bush K. Characterization of beta-lactamases. Antimicrob Agents Chemother. 1989 Mar;33(3):259–263. doi: 10.1128/aac.33.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush K., Sykes R. B. Methodology for the study of beta-lactamases. Antimicrob Agents Chemother. 1986 Jul;30(1):6–10. doi: 10.1128/aac.30.1.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuchural G. J., Jr, Malamy M. H., Tally F. P. Beta-lactamase-mediated imipenem resistance in Bacteroides fragilis. Antimicrob Agents Chemother. 1986 Nov;30(5):645–648. doi: 10.1128/aac.30.5.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galleni M., Lindberg F., Normark S., Cole S., Honore N., Joris B., Frere J. M. Sequence and comparative analysis of three Enterobacter cloacae ampC beta-lactamase genes and their products. Biochem J. 1988 Mar 15;250(3):753–760. doi: 10.1042/bj2500753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honoré N., Nicolas M. H., Cole S. T. Inducible cephalosporinase production in clinical isolates of Enterobacter cloacae is controlled by a regulatory gene that has been deleted from Escherichia coli. EMBO J. 1986 Dec 20;5(13):3709–3714. doi: 10.1002/j.1460-2075.1986.tb04704.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacoby G. A., Medeiros A. A. More extended-spectrum beta-lactamases. Antimicrob Agents Chemother. 1991 Sep;35(9):1697–1704. doi: 10.1128/aac.35.9.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarlier V., Nicolas M. H., Fournier G., Philippon A. Extended broad-spectrum beta-lactamases conferring transferable resistance to newer beta-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns. Rev Infect Dis. 1988 Jul-Aug;10(4):867–878. doi: 10.1093/clinids/10.4.867. [DOI] [PubMed] [Google Scholar]
- Jaurin B., Grundström T. ampC cephalosporinase of Escherichia coli K-12 has a different evolutionary origin from that of beta-lactamases of the penicillinase type. Proc Natl Acad Sci U S A. 1981 Aug;78(8):4897–4901. doi: 10.1073/pnas.78.8.4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kado C. I., Liu S. T. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol. 1981 Mar;145(3):1365–1373. doi: 10.1128/jb.145.3.1365-1373.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitzis M. D., Billot-Klein D., Goldstein F. W., Williamson R., Tran Van Nhieu G., Carlet J., Acar J. F., Gutmann L. Dissemination of the novel plasmid-mediated beta-lactamase CTX-1, which confers resistance to broad-spectrum cephalosporins, and its inhibition by beta-lactamase inhibitors. Antimicrob Agents Chemother. 1988 Jan;32(1):9–14. doi: 10.1128/aac.32.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korfmann G., Sanders C. C. ampG is essential for high-level expression of AmpC beta-lactamase in Enterobacter cloacae. Antimicrob Agents Chemother. 1989 Nov;33(11):1946–1951. doi: 10.1128/aac.33.11.1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindberg F., Lindquist S., Normark S. Inactivation of the ampD gene causes semiconstitutive overproduction of the inducible Citrobacter freundii beta-lactamase. J Bacteriol. 1987 May;169(5):1923–1928. doi: 10.1128/jb.169.5.1923-1928.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindberg F., Normark S. Sequence of the Citrobacter freundii OS60 chromosomal ampC beta-lactamase gene. Eur J Biochem. 1986 May 2;156(3):441–445. doi: 10.1111/j.1432-1033.1986.tb09601.x. [DOI] [PubMed] [Google Scholar]
- Lodge J. M., Minchin S. D., Piddock L. J., Busby S. J. Cloning, sequencing and analysis of the structural gene and regulatory region of the Pseudomonas aeruginosa chromosomal ampC beta-lactamase. Biochem J. 1990 Dec 15;272(3):627–631. doi: 10.1042/bj2720627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto H., Sawai T., Tazaki T., Yamagishi S., Mitsuhashi S. Characterization of the chromosomally mediated penicillinase in Klebsiella pneumoniae. Jpn J Microbiol. 1972 May;16(3):169–176. doi: 10.1111/j.1348-0421.1972.tb00645.x. [DOI] [PubMed] [Google Scholar]
- Matsumoto Y., Ikeda F., Kamimura T., Yokota Y., Mine Y. Novel plasmid-mediated beta-lactamase from Escherichia coli that inactivates oxyimino-cephalosporins. Antimicrob Agents Chemother. 1988 Aug;32(8):1243–1246. doi: 10.1128/aac.32.8.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier J., Levesque R. C. Cloning of SHV-2, OHIO-1, and OXA-6 beta-lactamases and cloning and sequencing of SHV-1 beta-lactamase. Antimicrob Agents Chemother. 1990 Aug;34(8):1577–1583. doi: 10.1128/aac.34.8.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura K., Yoshida T. Nucleotide sequence of the Serratia marcescens SR50 chromosomal ampC beta-lactamase gene. FEMS Microbiol Lett. 1990 Aug;58(3):295–299. doi: 10.1111/j.1574-6968.1990.tb13992.x. [DOI] [PubMed] [Google Scholar]
- Papanicolaou G. A., Medeiros A. A., Jacoby G. A. Novel plasmid-mediated beta-lactamase (MIR-1) conferring resistance to oxyimino- and alpha-methoxy beta-lactams in clinical isolates of Klebsiella pneumoniae. Antimicrob Agents Chemother. 1990 Nov;34(11):2200–2209. doi: 10.1128/aac.34.11.2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne D. J., Amyes S. G. Transferable resistance to extended-spectrum beta-lactams: a major threat or a minor inconvenience? J Antimicrob Chemother. 1991 Mar;27(3):255–261. doi: 10.1093/jac/27.3.255. [DOI] [PubMed] [Google Scholar]
- Petit A., Sirot D. L., Chanal C. M., Sirot J. L., Labia R., Gerbaud G., Cluzel R. A. Novel plasmid-mediated beta-lactamase in clinical isolates of Klebsiella pneumoniae more resistant to ceftazidime than to other broad-spectrum cephalosporins. Antimicrob Agents Chemother. 1988 May;32(5):626–630. doi: 10.1128/aac.32.5.626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippon A., Labia R., Jacoby G. Extended-spectrum beta-lactamases. Antimicrob Agents Chemother. 1989 Aug;33(8):1131–1136. doi: 10.1128/aac.33.8.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saino Y., Kobayashi F., Inoue M., Mitsuhashi S. Purification and properties of inducible penicillin beta-lactamase isolated from Pseudomonas maltophilia. Antimicrob Agents Chemother. 1982 Oct;22(4):564–570. doi: 10.1128/aac.22.4.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders C. C. Chromosomal cephalosporinases responsible for multiple resistance to newer beta-lactam antibiotics. Annu Rev Microbiol. 1987;41:573–593. doi: 10.1146/annurev.mi.41.100187.003041. [DOI] [PubMed] [Google Scholar]
- Sawai T., Yamagishi S., Mitsuhashi S. Penicillinases of Klebsiella pneumoniae and their phylogenetic relationship to penicillinases mediated by R factors. J Bacteriol. 1973 Sep;115(3):1045–1054. doi: 10.1128/jb.115.3.1045-1054.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirot D., Sirot J., Labia R., Morand A., Courvalin P., Darfeuille-Michaud A., Perroux R., Cluzel R. Transferable resistance to third-generation cephalosporins in clinical isolates of Klebsiella pneumoniae: identification of CTX-1, a novel beta-lactamase. J Antimicrob Chemother. 1987 Sep;20(3):323–334. doi: 10.1093/jac/20.3.323. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Spencer R. C., Wheat P. F., Winstanley T. G., Cox D. M., Plested S. J. Novel beta-lactamase in a clinical isolate of Klebsiella pneumoniae conferring unusual resistance to beta-lactam antibiotics. J Antimicrob Chemother. 1987 Dec;20(6):919–921. doi: 10.1093/jac/20.6.919. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G. Nucleotide sequence of the ampicillin resistance gene of Escherichia coli plasmid pBR322. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3737–3741. doi: 10.1073/pnas.75.8.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshita S., Sato M., Toba M., Masahashi W., Hashimoto-Gotoh T. High-copy-number and low-copy-number plasmid vectors for lacZ alpha-complementation and chloramphenicol- or kanamycin-resistance selection. Gene. 1987;61(1):63–74. doi: 10.1016/0378-1119(87)90365-9. [DOI] [PubMed] [Google Scholar]
- Vuye A., Verschraegen G., Claeys G. Plasmid-mediated beta-lactamases in clinical isolates of Klebsiella pneumoniae and Escherichia coli resistant to ceftazidime. Antimicrob Agents Chemother. 1989 May;33(5):757–761. doi: 10.1128/aac.33.5.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M., Iyobe S., Inoue M., Mitsuhashi S. Transferable imipenem resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1991 Jan;35(1):147–151. doi: 10.1128/aac.35.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodford N., Payne D. J., Johnson A. P., Weinbren M. J., Perinpanayagam R. M., George R. C., Cookson B. D., Amyes S. G. Transferable cephalosporin resistance not inhibited by clavulanate in Escherichia coli. Lancet. 1990 Jul 28;336(8709):253–253. doi: 10.1016/0140-6736(90)91784-8. [DOI] [PubMed] [Google Scholar]
- Yamamoto T., Murayama S. Y., Sawai T. Cloning and expression of the gene(s) for cephalosporinase production of Citrobacter freundii. Mol Gen Genet. 1983;190(1):85–91. doi: 10.1007/BF00330328. [DOI] [PubMed] [Google Scholar]
- Yang Y. J., Wu P. J., Livermore D. M. Biochemical characterization of a beta-lactamase that hydrolyzes penems and carbapenems from two Serratia marcescens isolates. Antimicrob Agents Chemother. 1990 May;34(5):755–758. doi: 10.1128/aac.34.5.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Yotsuji A., Minami S., Inoue M., Mitsuhashi S. Properties of novel beta-lactamase produced by Bacteroides fragilis. Antimicrob Agents Chemother. 1983 Dec;24(6):925–929. doi: 10.1128/aac.24.6.925. [DOI] [PMC free article] [PubMed] [Google Scholar]




