Abstract
The recent explosion in genomic sequencing has made available a wealth of data that can now be analyzed to identify protein antigens, potential targets for vaccine development. Here we present, in the context of Plasmodium falciparum, a strategy that rapidly identifies target antigens from large and complex genomes. Sixteen antigenic proteins recognized by volunteers immunized with radiation-attenuated P. falciparum sporozoites, but not by mock immunized controls, were identified. Several of these were more antigenic than previously identified and well characterized P. falciparum-derived protein antigens. The data suggest that immune responses to Plasmodium are dispersed on a relatively large number of parasite antigens. These studies have implications for our understanding of immunodominance and breadth of responses to complex pathogens.
The feasibility of a malaria vaccine is supported by experimental data demonstrating that protective immunity can be induced by exposure to intact parasite (1). In particular, animals or human volunteers immunized with radiation-attenuated Plasmodium spp. sporozoites can develop sterile immunity to subsequent challenge with infectious, nonattenuated, sporozoites (2). However, >5,000 proteins are expressed during the life cycle of the Plasmodium spp. parasite (3), and the protein antigens mediating the protective immunity induced by whole organism vaccination are largely unknown. Subunit vaccines currently in development are based on a single or few antigens and may therefore elicit too narrow a breadth of response, providing neither optimal protection nor protection on genetically diverse backgrounds. To duplicate the protection induced by whole organism vaccination (1, 2), we envision that a vaccine capable of targeting a large number of parasite-derived proteins by assembling their minimal CD8+ and CD4+ T cell epitopes into a multiepitope construct might be necessary.
Because of various factors related to antigen abundance and immunodominance, not all possible antigens are recognized by natural immune responses (4). Various approaches have been proposed for antigen identification (5–12). Herein, we report the development of a novel strategy that integrates bioinformatic predictions, HLA-supertype considerations (13), and in vitro cellular assays for the purpose of identifying potentially immunogenic protein antigens from the genomic sequences of complex pathogens. Traditionally, the use of these tools has been for the purpose of precisely identifying specific epitope sequences within a protein known to represent the target of cellular immunity. We hypothesized that these same tools could also be used on a broader scale as a means to efficiently identify new proteins that are strongly antigenic in the course of a natural infection, or in vaccinated individuals. The utility of this approach would be especially apparent in the context of a large and complex pathogen. We tested this hypothesis in the context of Plasmodium falciparum.
Methods
Study Subjects. Study subjects (n = 12; Table 4, which is published as supporting information on the PNAS web site, www.pnas.org) were healthy Caucasian male volunteers, aged 20–45 years (mean 34.8) recruited under a protocol approved by the Committee for the Protection of Human Subjects of the Naval Medical Research Center, the Office of the Special Assistant for Human Subject Protections at the Naval Bureau of Medicine and Surgery, and the Human Subjects Research Review Board of the Army Surgeon General, in accordance with the U.S. Navy regulation (SECNAVINST 3900.39B) governing the use of human subjects in medical research, and described in more detail in Supporting Text, which is published as supporting information on the PNAS web site.
Peptide Synthesis. Peptides for assessment of HLA binding antigenicity and immunogenicity were purchased commercially from Chiron Mimotopes (Chiron) initially as crude material. Quality control “spot checks” by mass spectrometry were used. In selected instances, for those antigens selected for additional deconvolution and HLA peptide binding assays, the peptides were resynthesized in-house, purified by reversed-phase HPLC, and the purity assessed by amino acid sequence and/or composition analysis. Purity was >95%.
HLA-Peptide Binding Assays. Assays to quantitatively measure peptide binding to class I and class II MHC molecules, based on the inhibition of binding of a high affinity radiolabeled peptide to purified MHC molecules, have been described in detail elsewhere (14). A brief description is also provided in Supporting Text.
Development of Predicted IC50 nM Value (PIC) Algorithms. The method of derivation of specific polynomial algorithm coefficients describing the average relative binding associated with each of the 20 naturally occurring amino acid residues, for each peptide position has been described by Gulukota et al. and others (15–17), and is described in more detail in Supporting Text.
IFN-γ Enzyme-Linked Immunospot (ELISPOT) Assay. The number of peptide-specific IFN-γ-secreting cells was determined by ex vivo IFN-γ ELISPOT, as described (18), and summarized in Supporting Text.
Statistical Analysis. All assays were performed in triplicate or quadruplicate. The significance of group differences were calculated by using the Student's t test, two-tailed (excel version 10.0, Microsoft). α < 0.05 was considered significant.
Results
Significant efforts in the last few years have been devoted by several groups toward the generation of procedures and algorithms that allow more effective and accurate prediction of MHC binding capacity. A methodology commonly used is the polynomial matrix, or linear coefficients, method. Polynomial or matrix methods to predict MHC binding (19) are based on the assumption that each of peptide side chains contributes independently to the binding affinity. Based on this assumption, a number of polynomial methods have been described. A common drawback of these methods is the fact that they provide only rank scores, and often limited to predictions for peptides of a specific size. As a result, it is difficult to combine predictions for different peptide sizes, or predictions for different MHC molecules.
The algorithms that we have developed are still based on the polynomial matrix method, but incorporate a number of additional mathematical transformations, including linear polynomial scaling, and further linear corrections based on minimizing the deviation of predicted values, to allow generation of allele-specific PIC. Because all predictions are expressed in terms of predicted nM KD values, this method, which is described in more detail in Materials and Methods, readily allows for combined predictions of ligands of different sizes (for example 9-mer versus 10-mers) or for different HLA molecules. In addition, by expressing algorithm scores in terms of IC50 nM values, scores can be related to affinity thresholds validated to be biologically relevant in the context of class I and class II HLA molecules (15, 20).
An example of a combined simultaneous prediction of HLA-A*0201 binding for 1,735 different 9- and 10-mers, whose binding affinity for A*0201 were also experimentally determined, is shown in Fig. 2, which is published as supporting information on the PNAS web site, where PIC values are plotted as a function of the actual measured IC50 values (MIC). By using 500 nM as an affinity cutoff to define binding to HLA class I molecules (20), an algorithm score threshold of 100 nM allows the prediction of 52% of all peptides binding HLA-A*0201 with an affinity of 500 nM, or better. Notably, 75% of the peptides identified by the algorithm were indeed binders (true positives; conversely, only 25% are false positives). Increasing the threshold score to 200 nM allowed the selection of 78% of the A*0201 binders, and the percent of true positives only dropped to 69%. To date, we developed a panel of algorithms specific for a broad range of HLA class I and class II types (Table 5, which is published as supporting information on the PNAS web site).
We have described previously how a majority of HLA class I and II molecules can be classified in relatively few supertypes characterized by largely overlapping peptide repertoires (13). Peptides that are capable of binding to the majority of the most common molecules of a given supertype can be identified by in vitro peptide–MHC binding assays, and are designated as supertype binding peptides. A correlation between immunogenicity and the capacity to bind multiple molecules from a given HLA supertype has been documented in several studies (21–25), and high-affinity binding to the prototype allele of a given supertype has been shown to be highly predictive of supertype binding (17). Herein, we examined whether predictions based on prototype alleles could efficiently identify supertype binders. Accordingly, we analyzed Mage2/3, hepatitis B virus, hepatitis C virus, and Plasmodium protein sequences to examine how many of the previously identified supertype binders (15, 21–26) were identified by our predictive algorithms (100 nM was selected as the threshold predictive value; Fig. 2). Between 63% and 72% of known A1-, A2-, A3/A11-, A24-, and B7-supertype degenerate binders, and 90% of known DR-supertype binders, were identified (Table 1).
Table 1. Efficiency of predicting supertype binding by prototype allele algorithms.
|
Percent of supertype binders
predicted*
|
|||||||
|---|---|---|---|---|---|---|---|
| Source | Refs. | A2 | A11 | B7 | A1 | A24 | DR1 |
| HCV | 15, 21† | 88 (15 of 17)‡ | 71 (10 of 14) | 0 (0 of 2) | 50 (3 of 6) | 40 (2 of 5) | 100 (6 of 6) |
| P. falciparum§ | 15, 22, 23† | 82 (9 of 11) | 86 (6 of 7) | 0 (0 of 1) | 67 (2 of 3) | 78 (7 of 9) | 78 (7 of 9) |
| Mage2/3 | 15, 26† | 57 (8 of 14) | 100 (6 of 6) | 100 (4 of 4) | 80 (4 of 5) | 0 (0 of 1) | 100 (3 of 3) |
| HBV | 15, 25† | 63 (12 of 19) | 50 (6 of 12) | 75 (6 of 8) | 78 (7 of 9) | 65 (11 of 17) | 92 (11 of 12) |
| Total | 72 (44 of 61) | 72 (28 of 39) | 67 (10 of 15) | 70 (16 of 23) | 63 (20 of 32) | 90 (27 of 30) | |
A supertype binder is a peptide with the capacity to bind ≥50% of the MHC molecules in a given supertype.
J.S. and A.S., unpublished observations.
The numbers in parentheses indicate the number of supertype binders identified by the respective algorithm versus the total number of source-derived supertype binders.
Supertype peptides derived from the four previously well characterized P. falciparum antigens (PfCSP, PfSSP2, PfLSA, and PfEXP1).
Next, we proposed to screen potential protein antigens defined by using genomics and/or proteomic information for HLA-supertype peptides. Pools of algorithm-identified peptides could be then screened for reactivity with peripheral blood mononuclear cells (PBMC) of exposed/vaccinated individuals. Each pool is designed to include peptides representative of the six common class I and class II HLA-supertypes, and with ≈10 epitopes per supertype. Because the six most common HLA-supertypes afford coverage of >95% of the individuals in any population (13), both at the level of class I and class II molecules almost any individual should be capable of binding at least 20 different epitopes (10 epitopes for class I and class II, respectively). Thus, if an antigen is immunodominant and recognized in the process of natural infection (or vaccination or exposure, depending on the study design), this strategy would result in a “hit” even if the accuracy of prediction was as low as 10% (several independent studies have established an actual success rate on the order of 50–100%; refs. 17, 21–25, and 27). Because relatively large pools of peptides can be easily tested in assays such as ELISPOT, the method allows rapid identification of “hot spots,” or immunodominant protein antigens.
The recent completion of the genomic sequence of P. falciparum (3) and the elucidation of the P. falciparum proteome (28) by using multidimensional protein identification technology (MudPIT) (29) (which combines in-line high resolution liquid chromatography and tandem mass spectropscopy) provide a set of ORFs corresponding to potential P. falciparum target antigens, and evidence for expression of these genes in different stages of the parasite's life cycle. Therefore, to validate our hypothesis, we analyzed the P. falciparum genome sequence (3) and P. falciparum proteome (28) data. Tandem mass spectroscopy (MS/MS) spectra of peptide sequences generated by MudPIT analysis of P. falciparum sporozoite preparations were scanned against the P. falciparum genomic sequence database by using sequest software (30) to identify and prioritize a set of ORFs representing antigens potentially expressed in the sporozoite and intrahepatic stage of the parasite life cycle. Combined parasite/human (31, 32) or parasite/mosquito (Anopheles) (33) databases were also searched to account for spectra resulting from contaminating host or vector peptides.
A panel of 27 ORFs representative of putative P. falciparum proteins was investigated. These proteins were among the first proteins identified by a P. falciparum proteomic analysis presented in more detail elsewhere (28). The panel of selected ORFs was fairly diverse in its characteristics and included a range of expression levels, stage specificity, and membrane association (Table 2). Their size ranged between 96 and 4,544 aa (mean, 1,252), and the percentage of P. falciparum sporozoite sequence coverage (i.e., the percentage of the protein sequence covered by identified peptides) ranged from 0.5% to 49.5%. This panel included 10 antigens expressed only in sporozoites, and 17 antigens common to other stages of the parasite life cycle. Seventeen of the ORFs were identified only in soluble MudPIT fractions, one was identified only in insoluble fractions, and nine were identified in both soluble and insoluble fractions. The relevance of this feature relies in the fact that the insoluble fractions are thought to be enriched with membrane or organelle associated proteins (29). Eight of the 27 ORFs had predicted signal sequences, and 10 had predicted transmembrane domains. However, no correlation between the presence of predicted transmembrane regions and recovery from the insoluble fraction was noted. The frequency of recognition in the P. falciparum proteome data set ranged between 16 peptide hits from six different sporozoite runs to single peptide hits. Although mass spectroscopy data cannot be taken as a quantitative measure of protein expression, the number of peptide hits is indicative of the abundance of each protein in the sporozoite preparations (29).
Table 2. Summary of characteristics of the selected panel of 27 putative P. falciparum antigens.
|
Peptides∥
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Antigen | Protein* | Amino acids† | Percent sequence coverage (PfSpz) | Sporozoite specific‡ | Fraction | SP§ | TM¶ | Stringent criteria | Relaxed criteria | MudPIT runs |
| 1 | PF10_0179** | 1,904 | 0.6 | Yes | Soluble | 0 | 0 | 1 | 1 | 1 |
| 2 | PFL0800c | 208 | 49.5 | Yes | Soluble and insoluble | 1 | 0 | 14 | 16 | 6 |
| 3 | PFI0165c | 2,404 | 1.3 | No | Soluble and insoluble | 0 | 0 | 8 | 11 | 2 |
| 4 | PFC0210c | 396 | 10.1 | Yes | Soluble | 1 | 1 | 2 | 2 | 2 |
| 5 | PF14_0372 | 400 | 3.0 | No | Soluble and insoluble | 1 | 2 | 4 | 6 | 4 |
| 6 | PF11_0341 | 1,062 | 5.5 | No | Soluble | 0 | 1 | 0 | 6 | 3 |
| 7 | mal4T2c4.plt1 | 104 | NA | ? | Soluble | - | - | ? | ? | 0 |
| 8 | PF13_0278 | 954 | 1.6 | No | Soluble | 0 | 0 | 2 | 5 | 1 |
| 9 | PFE0765w | 2,133 | 0.7 | No | Soluble | 0 | 0 | 1 | 2 | 1 |
| 10 | PF11_0479 | 3,029 | 2.4 | No | Soluble and insoluble | 0 | 0 | 4 | 8 | 4 |
| 11 | PFC0450w | 109 | 16.5 | Yes | Soluble | 1 | 2 | 0 | 1 | 1 |
| 12 | PF14_0074 | 395 | 26.6 | No | Soluble | 0 | 0 | 8 | 8 | 3 |
| 13 | PFC0700c | 307 | NA | ? | Soluble | 0 | 1 | 0 | 0 | 0 |
| 14 | PF14_0291 | 1,234 | 1.0 | Yes | Insoluble | 0 | 0 | 0 | 1 | 1 |
| 15 | MAL8P1.78 | 100 | 48.8 | Yes | Soluble and insoluble | 0 | 0 | 9 | 9 | 4 |
| 16 | MAL13P1.218 | 253 | 3.4 | Yes | Soluble | 0 | 0 | 0 | 1 | 1 |
| 17 | PFD0425w | 984 | 25.3 | Yes | Soluble | 1 | 0 | 15 | 18 | 4 |
| 18 | PF13_0320 | 1,791 | 1.4 | No | Soluble and insoluble | 0 | 21 | 3 | 6 | 3 |
| 19 | PF11_0435 | 1,815 | 0.8 | Yes | Soluble | 0 | 6 | 1 | 1 | 1 |
| 20 | PFI0260 | 4,544 | 2.2 | No | Soluble and insoluble | 0 | 0 | 9 | 13 | 6 |
| 21 | PF11_0226 | 2,024 | 0.8 | No | Soluble and insoluble | 1 | 0 | 3 | 5 | 1 |
| 22 | PFA0515w | 1,471 | 3.3 | No | Soluble | 0 | 0 | 1 | 8 | 2 |
| 23 | MAL6P1.252 | 2,647 | 1.7 | No | Soluble | 0 | 1 | 3 | 3 | 1 |
| 24 | PF14_0236 | 1,781 | 3.4 | No | Soluble and insoluble | 0 | 0 | 2 | 6 | 3 |
| 25 | PF14_0751 | 188 | 2.1 | Yes | Soluble | 1 | 1 | 0 | 1 | 1 |
| 26 | PF14_0648 | 96 | 0.5 | No | Soluble | 0 | 0 | 2 | 2 | 1 |
| 27 | PF11_0456 | 1,464 | 4.0 | No | Soluble | 0 | 0 | 0 | 5 | 2 |
| CSP | PFC0210c | 397 | 10.1 | Yes | Soluble | 1 | 1 | 2 | ||
| SSP2 | PF13_0201 | 574 | 36.1 | Yes | Soluble | 1 | 0 | 24 | ||
| EXP-1 | PF11_0224 | 162 | 72.9 | No | Soluble and insoluble | 1 | 1 | 6 | ||
| LSA-1 | X56203 | 1,909 | NA | No | NA | 1 | 1 | NA | ||
NA, not applicable.
Gene accession number.
The number of amino acids in the translated gene product of initial gene loci predictions, based on an incomplete and heterogeneous P. falciparum genomic sequence database (09/05/01 version).
“Yes” indicates sequences found only in sporozoite preparations, and “No” indicates sequences found in both the sporozoite preparations and preparations from different parasite life stages.
Absence (0) or presence (1) of predicted N-terminal signal peptide (SP), based on the SignalP algorithm (see http://plasmodb.org).
Absence (0) or presence (1-21) of predicted transmembrane domains (TM), based on the TMHMM algorithm (see http://plasmodb.org).
The number of peptides identified by using either the stringent or relaxed criteria, respectively, as described in the text.
Sequences can be accessed via PlasmoDB, the official database of the malaria parasite genome project, at (http://plasmodb.org).
Four known and well characterized P. falciparum preerythrocytic-stage antigens [circumsporozoite protein (CSP), sporozoite surface protein 2 (SSP2), exported protein 1 (LSA-1), and exported protein 1 (EXP-1)] were analyzed in parallel by using the same methodology. These antigens vary in molecular mass between 23 kDa (EXP-1) and 230 kDa (LSA-1). CSP and SSP2 are expressed only in the sporozoite stage, LSA-1 is liver stage-specific, and EXP-1 is expressed in both the liver and erythrocytic stages of the parasite life cycle. In the course of proteomic analysis (28), CSP, SSP2, and EXP were identified by between 2 (CSP) and 24 (SSP2) peptide hits, and were associated with between 10.1% (CSP) and 72.9% (EXP-1) sequence coverage (28) (Table 2). LSA-1 was not identified during this analysis because it is expressed only during the liver stage of the P. falciparum life cycle (34), and MudPIT studies were not carried out with P. falciparum liver stage parasite preparations (because of limitations in the availability of liver-stage parasite material). Two of the three known antigens (CSP and SSP2) were detected only in the soluble MudPIT fractions, whereas EXP-1 was detected in both soluble and insoluble (membrane protein) fractions (28).
The panel of candidate antigens was originally identified by searching against an incomplete and heterogeneous database. When searched against the final P. falciparum genomic sequence database (2) by using refined gene model predictions, and taking into consideration sequence information from the Anopheles (vector) (33) and human (host) (31, 32) databases, 19 of the 27 putative antigens could be identified by using stringent selection criteria, six could be identified only if the selection criteria were relaxed (no tryptic end requirement, cross-correlation score for +2 spectra lowered by 0.05) (28), and one (antigen 7) could not be found in the database, indicating that it either is not a gene or the gene modeling algorithm missed it. For another antigen (antigen 13), the peptide used to originally identify the protein is still found in the final P. falciparum proteome data set with a good cross-correlation score, meaning that the experimental MS/MS spectrum matched the theoretical spectrum of the identifying peptide fairly well, but its DeltaCn parameter (the normalized difference in cross-correlation scores between top scoring peptides for a given antigen) was very low (0.02). Thus, it cannot be definitively established from the sequest result whether the peptide/protein was in the P. falciparum sporozoite preparations analyzed. Overall, the panel of 27 ORFs can be considered representative of a wide range of potential target antigens with different characteristics of expression and subcellular localization. Comparison of the antigenicity of these proteins should therefore allow one to determine those characteristics that may be considered appropriate predictors of antigenicity, and whether proteomic data can be used successfully to identify and prioritize candidate vaccine antigens.
Amino acid sequences from the 27 ORFs were scanned with HLA-A1, -A2, -A3, -B7, and -DR supertype prototype PIC algorithms. A total of 3,241 sequences were identified (range = 14–435; mean = 120 sequences per antigen). A set of 1,142 peptides was synthesized (range = 13–50; mean = 42; see Table 6, which is published as supporting information on the PNAS web site), where only the top 10 scoring candidates per supertype per antigen were selected for larger ORFs. Control sets of peptides from the four known and well characterized P. falciparum antigens (CSP, SSP2, LSA1, and EXP1) were predicted according to the same criteria applied to the panel of 27 unknown ORFs, and synthesized. Analysis revealed that the predictive algorithms were highly effective in identifying already known and validated epitopes from those antigens. Specifically, the majority (≈81%) of known HLA-degenerate epitopes from those antigens (Table 7, which is published as supporting information on the PNAS web site) were identified by using the ImmunoSense algorithms: 87.1% (27 of 31) of previously identified class I (HLA-A2, -A3/A11, -A1, -A24) epitopes and 63.6% (7 of 11) of class II (DR1) epitopes.
In our previous analysis of known P. falciparum antigens, we considered, as a selection criterion, a high degree of conservancy of the predicted epitopes among sequences of different P. falciparum isolates. Because only a single P. falciparum genomic sequence is available at this time, this criterion could not be used in our analysis; however, it could be considered as additional sequences become available.
Predicted supertype epitopes were tested for their capacity to induce and recall IFN-γ immune responses by using PBMC from volunteers immunized with irradiated P. falciparum sporozoites (n = 8), or control volunteers mock immunized in parallel (n = 4) (Table 4). Sporozoite-immune individuals develop a vigorous and multifaceted immune response directed against multiple antigens, and it is presumed that the entire repertoire of sporozoite-induced T cell specificities is represented in those individuals. Peptides were tested as pools containing 1 μg/ml each peptide, with each antigen represented by a separate pool, in IFN-γ ELISPOT assays (18). Positive and negative control epitopes from well characterized non-P. falciparum antigens (cytomegalovirus, influenza, Epstein–Barr virus, HIV) were also included.
Considering a stimulation index (ratio test response/control) > 2.0 as positive, 16 of the 27 previously untested proteins were reproducibly recognized as antigens by at least two of the eight irradiated sporozoite immunized volunteers tested in two or more assays, but not by any of the four mock immunized controls (Table 3). Nine of the 27 antigens (nos. 2, 5, 18, 3, 22, 20, 11, 21, and 13) were recognized by at least 50% of irradiated sporozoite volunteers and classified as highly antigenic. Three antigens (nos. 1, 12, and 17) were recognized by 37.5% of volunteers and classified as intermediately antigenic. Four antigens were recognized by two volunteers, and classified as weakly antigenic. Finally, 11 of the 27 unknown antigens induced IFN-γ responses of sufficient magnitude to meet our criteria of positivity. Positive control peptides derived from cytomegalovirus, influenza, and Epstein–Barr virus were recognized by 87.5% of the volunteers tested.
Table 3. Summary of irradiated sporozoite immune reactivities against a subset of the panel of 27 P. falciparum proteins and four known antigens.
| Source | Antigen | No. volunteers responding | Percent volunteers responding | SI* | SFC† |
|---|---|---|---|---|---|
| Test Plasmodium proteins | 2 | 8 | 100 | 3.3 ± 1.3 | 122.7 ± 90.6 |
| 5 | 7 | 87.5 | 2.8 ± 1.1 | 101.8 ± 74.0 | |
| 18 | 6 | 75 | 2.2 ± 0.2 | 58.4 ± 24.4 | |
| 3 | 6 | 75 | 2.6 ± 0.4 | 119.1 ± 69.5 | |
| 22 | 5 | 62.5 | 2.9 ± 0.9 | 108.4 ± 78.1 | |
| 20 | 4 | 50 | 2.5 ± 0.5 | 74.8 ± 40.1 | |
| 11 | 4 | 50 | 3.1 ± 0.8 | 81.3 ± 47.9 | |
| 21 | 4 | 50 | 2.3 ± 0.3 | 48.2 ± 16.5 | |
| 13 | 4 | 50 | 2.9 ± 1.1 | 92.2 ± 50.1 | |
| 1 | 3 | 37.5 | 2.4 ± 0.2 | 61.4 ± 42.0 | |
| 17 | 3 | 37.5 | 2.4 ± 0.3 | 57.6 ± 34.5 | |
| 12 | 3 | 37.5 | 2.2 ± 0.3 | 48.2 ± 16.5 | |
| 16 | 2 | 25 | 2.2 ± 0.3 | 27.2 ± 23.3 | |
| 15 | 2 | 25 | 2.5 ± 0.7 | 28.8 ± 20.0 | |
| 19 | 2 | 25 | 2.7 ± 0.6 | 31.3 ± 22.6 | |
| 9 | 2 | 25 | 2.5 ± 0.2 | 32.0 ± 28.3 | |
| Range | 2-8 | 25-100 | 2.0-5.9 | 10.0-254.0 | |
| Mean | 4.1 | 50.8 | 2.6 | 80.3 | |
| Known Plasmodium antigens | CSP | 3 | 37.5 | 2.7 ± 0.6 | 41.6 ± 20.1 |
| SSP2 | 5 | 62.5 | 2.9 ± 1.0 | 97.2 ± 82.0 | |
| LSA1 | 2 | 25 | 3.2 ± 1.4 | 34.2 ± 29.0 | |
| EXP1 | 1 | 12.5 | 3.4 | 41.3 | |
| Range | 1-5 | 12.5-62.5 | 2.0-4.2 | 12.3-230.0 | |
| Mean | 2.75 | 34.4 | 2.9 | 65.5 | |
| Control epitopes‡ | CMV, EBV, influenza | 7 | 87.5§ | 4.0 ± 2.2 | 59.0 ± 30.5 |
CMV, cytomegalovirus; EBV, Epstein-Barr virus.
SI (stimulation index) indicates fold over background response to media and negative control peptide (HLA-A2 restricted HIV gag protein, residues 77-85), mean ± SD.
Net SFC indicates counts above background counts to media and negative control peptide (HLA-A2 restricted HIV gag, residues 77-85), mean ± SD.
Positive control peptides derived from CMV phosphoprotein, residues 495-503 (HLA-A2); influenza virus matrix protein, residues 58-66 (HLA-A2); influenza virus nucleoprotein, residues 265-273 (HLA-A3); and EBV EBNA3, residues 339-347 (HLA-B8).
Positive responses to control epitopes were detected in all four mock-immunized volunteers.
The magnitude of these responses is relatively low, but not substantially different from those observed with the pool of positive control peptides. The specificity of these responses is further demonstrated by the fact that no response was observed for all antigens in mock-immunized controls, and all individuals in the case of 11 of the experimental P. falciparum proteins analyzed.
Pools of predicted epitopes from the known antigens (CSP, SSP2, LSA1, and EXP1) were also recognized by irradiated sporozoite volunteers, though the frequency of response to those pools was somewhat lower than that to pools of peptides representing previously validated epitopes derived from the same antigens (22, 23) (Table 3). It is particularly noteworthy that the reactivity against several of the newly identified antigens exceeded the reactivities observed against well-characterized antigens, suggesting that some of the novel antigens identified by using our strategy may represent better candidates for vaccine development. For example, responses to antigen 2 were significantly better than responses to EXP-1, LSA-1 or CSP, as evidenced by the number of responding volunteers (8 of 8 vs. 1 of 8, 2 of 8, and 3 of 8; P = 0.001, 0.007, and 0.026), and number of spot-forming cells (SFCs) (122.7 vs. 41.3, 34.2, and 41.6). Responses were also better than to PfSSP2, but those differences were not significant. Similarly, responses to antigen 5 were significantly better than responses to EXP-1 or LSA-1 (number of responding volunteers: 7 of 8 vs. 1 of 8, 2 of 8; P = 0.010, and 0.041; number of SFCs: 101.8 vs. 41.3 and 34.2), and better than responses to CSP or SSP2 (Table 3).
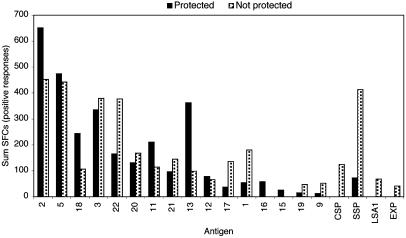
Overall, there was no significant difference for protected versus not protected volunteers with regard to either rate of response (42 of 128 vs. 32 of 128, P = 0.215) or magnitude of response as assessed by stimulation index (all antigens, 2.7 ± 0.9 vs. 2.5 + 0.6; high immune reactivity, 2.9 ± 1.0 vs. 2.6 ± 0.6; intermediate, 2.5 ± 0.4 vs. 2.3 ± 0.3; low, 2.2 vs. 2.3 ± 0.2; P = 0.259) or mean SFC (all antigens, 71.4 ± 51.8 vs. 90.5 ± 70.4; high immune reactivity, 89.0 ± 50.7 vs. 99.3 ± 80.4; intermediate, 29.1 ± 16.8 vs. 68.8 ± 26.9; low, 55.3 vs. 65.8 ± 31.8; P = 0.182). However, at the level of individual antigens, there was a trend for some antigens to be preferentially recognized by protected volunteers: for example, antigens 2 and 13 (Fig. 1). Interestingly, responses to the well characterized antigens, CSP, SSP2, LSA1, and EXP1, were detected predominantly in nonprotected volunteers (Fig. 1). Additional studies with increased sample sizes will be required to more finally establish whether individuals protected against P. falciparum sporozoite challenge selectively recognize specific antigens. Such studies will also establish whether protective immune responses in humans vaccinated with irradiated P. falciparum sporozoites are narrowly focused on a few immunodominant antigens and epitopes, or alternatively, are dispersed on a relatively large numbers of parasite antigens.
Fig. 1.
Antigen-specific reactivity in irradiated sporozoite immunized volunteers protected or not protected against sporozoite challenge. PBMCs derived from volunteers immunized with irradiated P. falciparum sporozoites and either protected or not protected against challenge with infectious sporozoites were tested by IFN-γ ELISPOT assay against peptide pools from putative P. falciparum proteins, as described in the text. Data are presented as summed SFC of all responses that met predefined criteria of positivity.
HLA-A2 peptide pools from antigens 2, 5, and 13, and HLA-A1 and HLA-DR peptide pools from antigens 2 and 5, are recognized by irradiated sporozoite volunteers who express the respective HLA alleles, but not by mock immunized controls.
Additionally, a comprehensive analysis of HLA binding against the HLA-A1, -A2, -A3/11, -A24, and -DR supertypes has been completed for selected antigens (antigens 2, 5, 12, and 13). Several degenerate binders have been identified for each supertype/antigen combination, and 50–70% of the predicted supertype peptides have been shown to have the capacity to bind to 50% or more of the HLA molecules from a given supertype (Tables 8 and 9, which are published as supporting information on the PNAS web site). These data represent an important validation of the predictive strategy used, and demonstrate that our predictive algorithms can be used to effectively predict pools of supertype binding peptides.
Further analysis revealed the extent to which the antigenicity results correlate with the proteomic data. Of the nine antigens associated with highest immune reactivity, six were identified by multiple peptide hits in multiple MudPIT runs (see Tables 2 and 3). However, the converse was not true in that some proteins identified by multiple hits in multiple runs were either weakly or nonantigenic (e.g., antigens 10, 15, and 24). Clearly, some antigens are preferentially recognized by the immune system for reasons that may not simply reflect the relative abundance of antigen. Furthermore, four of the six antigens identified by only a single peptide hit were moderately to highly antigenic (e.g., antigens 1, 11, 16, and 19). Finally, some but not all antigens that were identified as candidates according to the relaxed MudPIT criteria, but not according to the stringent criteria, were highly antigenic (e.g., antigens 6 and 22). When relative antigenicity (Table 3) was directly compared with relative abundance of expression in the P. falciparum life cycle (Table 2), no significant association was detected (r2 = 0.088; simple linear regression analysis). These data indicate that although genomics, proteomics, and in silico approaches can be used to prioritize ORFs or antigens for characterization, they are not sufficient on their own and must be considered in combination with other strategies, such as immunological screening, which take into account biological activity or function.
Discussion
The data presented herein have several important implications. First, we describe a novel strategy to mine genomic sequence databases for the identification and prioritization of novel target antigens and epitopes recognized by experimentally vaccinated, infected and/or naturally exposed humans, in the context of multiple genetic restrictions. This report provides an example of identifying antigens through epitope predictions; all other methods and reports described in the literature using algorithm predictions start from known antigens to identify epitopes. Our data support the potential of this approach to identify immunodominant antigens and to allow for prioritization among those antigens on the basis of immune reactivity. We have analyzed 27 proteins out of ≈1,000 estimated to be expressed in the sporozoite/liver stage (≈3% of all potential proteins), and identified nine highly antigenic antigens. Eleven of the 27 antigens were not recognized by any volunteer in any assay. In independent studies in our laboratory, seven of the 27 ORFs tested here were assessed in a humoral immunoscreening assay. Of those, four of four antigens classified with high immune reactivity and one of one antigen with intermediate immune reactivity were shown to be recognized by malaria-immune sera but not by malaria-naïve sera, in as many as five of eight experiments (P. Quinones-Casas, personal communication). Even given that the subset screened represents only a small fraction of putative preerythrocytic stage proteins, the number of antigens shown to be recognized by sporozoite-immune T cell responses are 3-fold more than the sum total of all antigens previously known. Extrapolating this data, it is possible that several hundred proteins are targets of the immune response to irradiated sporozoites in the general human population. We are currently undertaking a systematic study of additional protein antigens to expand the observations to a larger number of P. falciparum proteins. Of those that are recognized, antigens of the most interest for vaccine development would be those preferentially targeted by immune responses that efficiently kill or inhibit the function of the parasite. In that regard, antigens and epitopes targeted by CD8+ T cell response may be considered of the greatest interest, based on evidence in animal models implicating CD8+ T cells as critical mediators of protective immunity.
Second, it was known previously that B cell responses against complex pathogens are directed against a large number of antigens. In the case of malaria, for example, immunoprecipitation with human sera followed by 2D gels revealed a large number of different antigens (35–37). However, it is commonly assumed that T cell responses are directed against a relatively few immunodominant epitopes. Our study represents the first example showing that T cell responses in humans immunized with irradiated sporozoites are dispersed over a relatively large number of parasite antigens, rather than narrowly focused on very few immunodominant proteins and epitopes. Moreover, our results support the concept that a multiantigen vaccine will likely be needed to induce high effective protective immunity against malaria, and that a multiepitope approach should mimic natural responses in individuals experimentally vaccinated or naturally exposed to P. falciparum. Although data from murine studies show that immune responses directed against single antigens can be protective in selected inbred strains, genetic restriction of immune responses and protective immunity is well established (38), and it is widely believed that immune responses against multiple target antigens or epitopes will be required to effectively immunized an outbred human population. Indeed, clinical trials conducted to date with vaccines based on single antigens have been disappointing (39).
Our data also suggest that strict immunodominance (4) (i.e., only one or a few antigens or epitopes dominating the responses) does not apply in the context of the complex Plasmodium spp. parasite. Rather, a very complex and multispecific response exists and, among those antigens recognized, defined patterns of immunodominance might be more quantitative than qualitative. We are expanding our studies to determine whether this situation extends to naturally occurring infections, and to vaccination and other experimental systems.
Overall our data provide the foundation for development of an antigen map of sporozoite/liver stages (immunosome), and for the development and optimization of multiantigen and multiepitope vaccines designed to mimic the complexity of responses elicited by natural infection.
Supplementary Material
Acknowledgments
We are indebted to a number of individuals and funding sources, as detailed in Supporting Text.
Abbreviations: PBMC, peripheral blood mononuclear cells; ELISPOT, enzyme-linked immunospot; CSP, circumsporozoite protein; EXP-1, exported protein 1; LSA-1, liver-stage antigen 1; MIC, measured IC50 nM value; MudPIT, multidimensional protein identification technology; SFC, spot-forming cells; SSP2, sporozoite surface protein 2; PIC, predicted IC50 nM value.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. PF10_0179, PFL0800c, PFI0165c, PFC0210c, PF14_0372, PF11_0341, mal4T2c4.p1t1, PF13_0278, PFE0765w, PF11_0479, PFC0450w, PF14_0074, PFC0700c, PF14_0291, MAL8P1.78, MAL13P1.218, PFD0425w, PF13_0320, PF11_0435, PFI0260, PF11_0226, PFA0515w, MAL6P1.252, PF14_0236, PF14_0751, PF14_0648, and PF11_0456).
References
- 1.Good, M. F. & Doolan, D. L. (1999) Curr. Opin. Immunol. 11, 412–419. [DOI] [PubMed] [Google Scholar]
- 2.Hoffman, S. L., Goh, L. M., Luke, T. C., Schneider, I., Le, T. P., Doolan, D. L., Sacci, J., de la Vega, P., Dowler, M., Paul, C., et al. (2002) J. Infect. Dis. 185, 1155–1164. [DOI] [PubMed] [Google Scholar]
- 3.Gardner, M. J., Hall, N., Fung, E., White, O., Berriman, M., Hyman, R. W., Carlton, J. M., Pain, A., Nelson, K. E., Bowman, S., et al. (2002) Nature 419, 498–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yewdell, J. W. & Bennink, J. R. (1999) Annu. Rev. Immunol. 17, 51–88. [DOI] [PubMed] [Google Scholar]
- 5.Kawakami, Y. & Rosenberg, S. A. (1997) Int. Rev. Immunol. 14, 173–192. [DOI] [PubMed] [Google Scholar]
- 6.Rotzschke, O., Falk, K., Deres, K., Schild, H., Norda, M., Metzger, J., Jung, G. & Rammensee, H. G. (1990) Nature 348, 252–254. [DOI] [PubMed] [Google Scholar]
- 7.Van Bleek, G. M. & Nathenson, S. G. (1990) Nature 348, 213–216. [DOI] [PubMed] [Google Scholar]
- 8.Hunt, D. F., Michel, H., Dickinson, T. A., Shabanowitz, J., Cox, A. L., Sakaguchi, K., Appella, E., Grey, H. M. & Sette, A. (1992) Science 256, 1817–1820. [DOI] [PubMed] [Google Scholar]
- 9.Cox, A. L., Skipper, J., Chen, Y., Henderson, R. A., Darrow, T. L., Shabanowitz, J., Engelhard, V. H., Hunt, D. F. & Slingluff, C. L., Jr. (1994) Science 264, 716–719. [DOI] [PubMed] [Google Scholar]
- 10.Kern, F., Bunde, T., Faulhaber, N., Kiecker, F., Khatamzas, E., Rudawski, I. M., Pruss, A., Gratama, J. W., Volkmer-Engert, R., Ewert, R., et al. (2002) J. Infect. Dis. 185, 1709–1716. [DOI] [PubMed] [Google Scholar]
- 11.Davenport, M. P. & Hill, A. V. (1996) Mol. Med. Today 2, 38–45. [DOI] [PubMed] [Google Scholar]
- 12.Aidoo, M., Lalvani, A., Allsopp, C. E., Plebanski, M., Meisner, S. J., Krausa, P., Browning, M., Morris-Jones, S., Gotch, F., Fidock, D. A., et al. (1995) Lancet 345, 1003–1007. [DOI] [PubMed] [Google Scholar]
- 13.Sette, A. & Sidney, J. (1999) Immunogenetics 50, 201–212. [DOI] [PubMed] [Google Scholar]
- 14.Sidney, J., Southwood, S., Oseroff, C., Del Guercio, M. F., Sette, A. & Grey, H. (1998) in Current Protocols in Immunology (Wiley, New York), pp. 18.3.1–18.3.19. [DOI] [PubMed]
- 15.Southwood, S., Sidney, J., Kondo, A., del Guercio, M. F., Appella, E., Hoffman, S., Kubo, R. T., Chesnut, R. W., Grey, H. M. & Sette, A. (1998) J. Immunol. 160, 3363–3373. [PubMed] [Google Scholar]
- 16.Gulukota, K., Sidney, J., Sette, A. & DeLisi, C. (1997) J. Mol. Biol. 267, 1258–1267. [DOI] [PubMed] [Google Scholar]
- 17.Sidney, J., Southwood, S., Mann, D. L., Fernandez-Vina, M. A., Newman, M. J. & Sette, A. (2001) Hum. Immunol. 62, 1200–1216. [DOI] [PubMed] [Google Scholar]
- 18.Brice, G. T., Graber, N. L., Hoffman, S. L. & Doolan, D. L. (2001) J. Immunol. Methods 257, 55–69. [DOI] [PubMed] [Google Scholar]
- 19.Sette, A., Buus, S., Appella, E., Smith, J. A., Chesnut, R., Miles, C., Colon, S. M. & Grey, H. M. (1989) Proc. Natl. Acad. Sci. USA 86, 3296–3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sette, A., Vitiello, A., Reherman, B., Fowler, P., Nayersina, R., Kast, W. M., Melief, C. J., Oseroff, C., Yuan, L., Ruppert, J., et al. (1994) J. Immunol. 153, 5586–5592. [PubMed] [Google Scholar]
- 21.Chang, K. M., Gruener, N. H., Southwood, S., Sidney, J., Pape, G. R., Chisari, F. V. & Sette, A. (1999) J. Immunol. 162, 1156–1164. [PubMed] [Google Scholar]
- 22.Doolan, D. L., Hoffman, S. L., Southwood, S., Wentworth, P. A., Sidney, J., Chesnut, R. W., Keogh, E., Appella, E., Nutman, T. B., Lal, A. A., et al. (1997) Immunity 7, 97–112. [DOI] [PubMed] [Google Scholar]
- 23.Doolan, D. L., Southwood, S., Chesnut, R., Appella, E., Gomez, E., Richards, A., Higashimoto, Y. I., Maewal, A., Sidney, J., Gramzinski, R. A., et al. (2000) J. Immunol. 165, 1123–1137. [DOI] [PubMed] [Google Scholar]
- 24.Threlkeld, S. C., Wentworth, P. A., Kalams, S. A., Wilkes, B. M., Ruhl, D. J., Keogh, E., Sidney, J., Southwood, S., Walker, B. D. & Sette, A. (1997) J. Immunol. 159, 1648–1657. [PubMed] [Google Scholar]
- 25.Bertoni, R., Sidney, J., Fowler, P., Chesnut, R. W., Chisari, F. V. & Sette, A. (1997) J. Clin. Invest. 100, 503–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kawashima, I., Hudson, S. J., Tsai, V., Southwood, S., Takesako, K., Appella, E., Sette, A. & Celis, E. (1998) Hum. Immunol. 59, 1–14. [DOI] [PubMed] [Google Scholar]
- 27.Altfeld, M. A., Livingston, B., Reshamwala, N., Nguyen, P. T., Addo, M. M., Shea, A., Newman, M., Fikes, J., Sidney, J., Wentworth, P., et al. (2001) J. Virol. 75, 1301–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Florens, L., Washburn, M. P., Raine, J. D., Anthony, R. M., Grainger, M., Haynes, J. D., Moch, J. K., Muster, N., Sacci, J. B., Tabb, D. L., et al. (2002) Nature 419, 520–526. [DOI] [PubMed] [Google Scholar]
- 29.Washburn, M. P., Wolters, D. & Yates, J. R., III (2001) Nat. Biotechnol. 19, 242–247. [DOI] [PubMed] [Google Scholar]
- 30.Eng, J. K., McCormack, A. L. & Yates, J. R. (1994) J. Am. Soc. Mass Spectrom. 5, 976–989. [DOI] [PubMed] [Google Scholar]
- 31.Lander, E. S., Linton, L. M., Birren, B., Nusbaum, C., Zody, M. C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. (2001) Nature 409, 860–921. [DOI] [PubMed] [Google Scholar]
- 32.Venter, J. C., Adams, M. D., Myers, E. W., Li, P. W., Mural, R. J., Sutton, G. G., Smith, H. O., Yandell, M., Evans, C. A., Holt, R. A., et al. (2001) Science 291, 1304–1351. [DOI] [PubMed] [Google Scholar]
- 33.Holt, R. A., Subramanian, G. M., Halpern, A., Sutton, G. G., Charlab, R., Nusskern, D. R., Wincker, P., Clark, A. G., Ribeiro, J. M., Wides, R., et al. (2002) Science 298, 129–149.12364791 [Google Scholar]
- 34.Guerin-Marchand, C., Druilhe, P., Galey, B., Londono, A., Patarapotikul, J., Beaudoin, R. L., Dubeaux, C., Tartar, A., Mercereau-Puijalon, O. & Langsley, G. (1987) Nature 329, 164–167. [DOI] [PubMed] [Google Scholar]
- 35.Brown, G. V., Anders, R. F., Mitchell, G. F. & Heywood, P. F. (1982) Nature 297, 591–593. [DOI] [PubMed] [Google Scholar]
- 36.Brown, G. V., Coppel, R. L., Vrbova, H., Grumont, R. J. & Anders, R. F. (1982) Exp. Parasitol. 53, 279–284. [DOI] [PubMed] [Google Scholar]
- 37.Brown, G. V., Stace, J. D. & Anders, R. F. (1983) Am. J. Trop. Med. Hyg. 32, 1221–1228. [DOI] [PubMed] [Google Scholar]
- 38.Doolan, D. L., Sedegah, M., Hedstrom, R. C., Hobart, P., Charoenvit, Y. & Hoffman, S. L. (1996) J. Exp. Med. 183, 1739–1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Richie, T. L. & Saul, A. (2002) Nature 415, 694–701. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.