Abstract
PTEN/MMAC1/TEP1 (PTEN, phosphatase deleted on chromosome ten; MMAC1, mutated in multiple advanced cancers; TEP1, tensin-like phosphatase) is a major human tumor suppressor gene whose suppressive activity operates on the phosphatidylinositol pathway. A single homologue of this gene, TEP1 (YNL128w), exists in the budding yeast Saccharomyces cerevisiae. Yeast strains deleted for TEP1 exhibit essentially no phenotype in haploids; however, diploids exhibit resistance to the phosphatidylinositol-3-phosphate kinase inhibitor wortmannin and to lithium ions. Although rates of cancer increase with age, neither tep1 haploids nor diploids have altered life spans. TEP1 RNA is present throughout the cell cycle, and levels are dramatically up-regulated during meiotic development. Although homozygous tep1 mutants initiate the meiotic program and form spores with wild-type kinetics, analysis of the spores produced in tep1 mutants indicates a specific defect in the trafficking or deposition of dityrosine, a major component of yeast spore walls, to the surface. Introduction of a common PTEN mutation found in human tumors into the analogous position in Tep1p produces a nonfunctional protein based on in vivo activity. These studies implicate Tep1p in a specific developmental trafficking or deposition event and suggest that Tep1p, like its mammalian counterpart, impinges on the phosphatidylinositol pathway.
Keywords: cancer, meiosis, Saccharomyces cerevisiae
The human tumor suppressor gene designated PTEN/MMAC1/TEP1 (PTEN, phosphatase deleted on chromosome ten; MMAC1, mutated in multiple advanced cancers; TEP1, tensin-like phosphatase) is either deleted or inactivated in a very high percentage of breast, endometrial, brain, and prostate cancers (1–3). Inactivation of the PTEN/MMAC/TEP1 gene (hereafter just PTEN) in mice has confirmed that it is a major tumor suppressor gene: several groups have independently shown that homozygous deletion of PTEN is embryonic lethal and that mice with only one functional copy of the gene are more likely to develop tumors (4–6).
Amino acid sequence analysis of the PTEN protein reveals similarity to protein phosphatases. However, although PTEN is capable of dephosphorylating peptide substrates in vitro, its most likely physiological substrates are the lipid phosphatidylinositol phosphates, in particular, phosphatidylinositol 3,4,5 triphosphate [PtdIns(3,4,5)P3] (7). Indeed, in the Cowden disease G129E mutation, the enzyme loses its ability to dephosphorylate the lipid substrate PtdIns(3,4,5)P3 but not peptide substrates (8, 9).
Further studies with the knockout mice have supported the hypothesis that the important function of PTEN is its lipid phosphatase activity. Embryonic fibroblasts from knockout strains have abnormally high levels of PtdIns(3,4,5)P3 and are resistant to apoptosis. Moreover, both the fibroblasts and tumor cells taken from the knockout mice, and several breast cancer cell lines, show extremely high levels of phosphorylated protein kinase B (PKB/Akt). PKB/Akt is a protein serine/threonine kinase that is activated by PtdIns(3,4,5)P3 and PtdIns(3,4)P2 (10). One of its functions is to suppress apoptosis in normal cells by phosphorylating, among other targets, the cell death regulator BAD, a member of the BCL-2 family (10). Constitutive activation of PKB/Akt is known to induce cellular transformation. Thus, PTEN functions at least in part to regulate the activity of PKB/Akt. However, the complete array of downstream targets of PTEN is still unclear.
Homologs of PTEN have been identified in Drosophila (11), Caenorhabditis elegans (12–14), and yeast (15). PTEN is essential for Drosophila (11), but mutations in the C. elegans PTEN result in a phenotype in the developmental process of dauer formation, which is elicited in response to starvation (12–14). In this study, we have analyzed the phenotype of yeast cells deleted for the PTEN homolog, TEP1 (YNL128w; 28% identical and 45% similar to human PTEN). In the absence of a functional TEP1 gene, yeast cells grow normally but homozygous diploids display resistance to lithium ions and wortmannin, a phosphatidylinositol 3-phosphate kinase (PI3-kinase) inhibitor, linking Tep1p to the phosphatidylinositol pathway. In addition, homozygous tep1 mutants display a specific defect in the developmental process of sporulation.
Materials and Methods
Yeast Strains and Media.
Routine growth and manipulation of Saccharomyces cerevisiae strains were performed (16). W303-1a (MATa leu2 his3 ura3 trp1), W303-1α (MATα leu2 his3 ura3 trp1), and JHY101 (W303 homozygous for tep1∷URA3) were used for the analysis of lithium, LY-294002, and wortmannin (Sigma) sensitivity, and for lifespan studies. Y1541 (MATa leu2-27 his4-260/Matα leu2-3,112 his4-280 trp1-1/trp1-1 ura3-1/ura3-1 ADE2/ade2 arg4∷kanR thr1-4/arg4∷kanR THR1 lys2/lys2) and Y1713 (Y1541 homozygous tep1∷LEU2) are derivatives of S288c; these strains were used for the analysis of sporulation. Yeast strain 334 (17) was used for analysis of protein. Yeast strains were transformed by using the lithium acetate procedure (18). All gene disruptions were performed by the one-step gene replacement described by Rothstein (19) and verified by PCR.
Plasmids.
TEP1 was isolated from yeast genomic DNA by PCR (5′ primer ACCGAATTCGAGTTCATAAATGATGAT, 3′ primer TACGGATCCTACTCCTTGGTTCTCATA). The resulting product was digested with EcoRI and XbaI, and inserted into the corresponding sites of pUC19 (New England Biolabs) to generate TEP1-JLH19. TEP1 was independently amplified from yeast genomic DNA by using primers complementary to flanking sequences (P85, 5′-GGATACCAGACTTTGGATTC; P86, 5′-CAATGCCTTTCAACCTACTCC). The amplified product was digested with BamHI and XbaI, and the resulting 1.86-kbp fragment was inserted into the corresponding sites of centromere-based pUN55 (20), generating pME1344. Sequence analysis confirmed that the wild-type gene was obtained. The XbaI–EcoRI fragment of TEP1-JLH19, where the XbaI site was filled in with the Klenow fragment of DNA polymerase I, was inserted into the SmaI and EcoRI sites of pUN15 (20) to generate B26-TEP1. The XbaI–EcoRI fragment of TEP1-JLH19, where the EcoRI site was filled in with the Klenow fragment of DNA polymerase I, was likewise inserted into the SmaI and XbaI sites of pUN105 (20) to create B34-TEP1. The 2-μ derivative, pME1519, was created by inserting the EcoRI–SalI (from the polylinker in the vector) TEP1 fragment from pME1344 into the corresponding sites of Yep352 (21).
The tep1∷URA3 deletion allele was constructed by excising the sequences between ClaI (−140) and SacI (1244) of TEP1-JLH19 and inserting a 1.2-kb fragment of URA3 to generate TEP1-KO3. TEP1-KO3 was digested with SphI for targeting to the TEP1 locus before transformation into yeast. The tep1∷LEU2 deletion allele was constructed in two steps. First, the EcoRI–XbaI TEP1 fragment from pME1344 was inserted into the corresponding sites of pHSS6 (22); the resulting plasmid was designated ME1394. The sequences between EcoRV and HindIII of TEP1 in pME1394 then were replaced with the HpaI–HindIII LEU2 fragment from Yep351 (21) to create pME1397. Plasmid ME1397 was digested with NotI before transformation into yeast.
To generate the tep1-G199E allele, the QuickChange Site-Directed Mutagenesis Kit (Stratagene) was used as recommended. The primers were P87, 5′-GT CGA ATG GGG AAA GAA GCT AGA CCA TAC TAT TG and P88, 5′-GT TAT CAT ACC AGA TGC TTC TTT CCC CAT TCG AC, where the bolded nucleotides represent the introduced mutation. Plasmid ME1344 and TEP1-INV (see below) were used as template, and the resulting plasmids were sequenced to confirm that only the indicated mutation was generated. TEP1-INV was obtained from Invitrogen and contains a Gal-driven TEP1 gene that has been modified at the 3′ end by addition of the V5 epitope tag.
Lifespan Determination.
To determine lifespan, cells were taken from logarithmically growing liquid cultures and plated at low density on yeast extract/peptone/dextrose. The cells were incubated at 30°C for 3–4 h. After incubation, the daughter cells were isolated as buds that had emerged from mother cells. The daughter cells were moved with an Olympic micromanipulator to uninhabited parts of the plate. All future buds produced by these daughter cells were removed by micromanipulation. The plates were grown at 30°C during working hours and moved to 4°C overnight. The positions of mother cells were noted to distinguish mother cells from daughter cells during symmetric division. On a rare occasion, a cell would be damaged by the micromanipulator and lyse immediately; this cell would be excluded from the data set. Micromanipulation was performed by using the Olympus BX-40 microscope under ×10 magnification. Time between readings increased from 1 h to 4 h toward the end of the experiment. Determinations of the significance of differences in mean lifespan between two strains were performed by using the nonparametric Wilcoxen signed rank test with a confidence level greater than 99% (23).
Analysis of Meiosis and Sporulation.
Cells were grown in yeast extract/peptone/acetate medium and sporulated as described (24). Aliquots from triplicate cultures were removed and fixed in 3.7% formaldehyde at indicated times after transfer to sporulation medium. Fixed cells were stained with the DNA-specific dye 4′,6-diamidino-2-phenyl-indole (DAPI) and examined by using fluorescent microscopy (25). Cells with two staining bodies were scored as having completed meiosis I, and cells with three or four staining bodies were scored as having completed meiosis II. The fixed cells also were examined by phase-contrast microscopy to assay ascus formation. A minimum of 600 cells was examined for each time point.
Quantitative dityrosine measurements were performed from triplicate cultures at the indicated times after transfer to sporulation medium as described (26). Cell fractionation was performed as described (26).
Fluorescence observations were made with a Nikon microscope equipped with custom filters (330WB80 exciter filter, 400CLP dichroic mirror, 400EFLP emission filter). For most observations, a drop of cells was mixed on a glass slide with a drop of 25% ammonium hydroxide (thus alkalizing the suspension for maximum fluorescence intensity) and a coverslip was added. The cells were photographed immediately with Ektachrome 400 film (Eastman Kodak) or Tri-X 400 black/white film.
Protein Analysis.
Cells were grown in synthetic complete medium lacking uracil and containing 1% galactose to induce expression of the TEP1 gene. Cells were collected, resuspended in 50 mM NaH2PO4, 300 mM NaCl, frozen in liquid nitrogen, and disrupted by glass bead agitation. Cell lysates were separated on 10% PAGE and immunoblots were performed with anti-V5 antibody (Invitrogen) as recommended.
Results and Discussion
TEP1 Is Not an Essential Gene in S. cerevisiae.
To determine the consequences of inactivation of TEP1 in yeast cells, diploid strains (W303 and S288C derivatives) were transformed with a tep1 deletion. The resulting transformants were sporulated and tetrads dissected. Spore viability was excellent, and tep1 segregants were recovered with the expected frequency (2:2). tep1 haploids displayed wild-type growth rate and mated normally. Neither haploid, nor diploid knockout strains derived from the haploids, showed any abnormal sensitivity to high or low temperatures. We conclude that TEP1 is not an essential gene in budding yeast.
Homozygous tep1 Diploids Are Resistant to Lithium Ions and the PI3-Kinase Inhibitor, Wortmannin.
For both W303A and S288C haploid tep1 strains, extensive screening of more than 50 different growth conditions failed to show any strong phenotype relative to wild-type yeast. Only a very slight resistance to lithium ions and to the fungal natural product wortmannin, an inhibitor of PI3-kinase (27), was detected. Although these weak phenotypes could be broadly interpreted as connecting TEP1 with the phosphatidylinositol signaling pathway—a connection now well established for its mammalian homologue PTEN—they were not useful for genetic screens aimed at characterizing its function further.
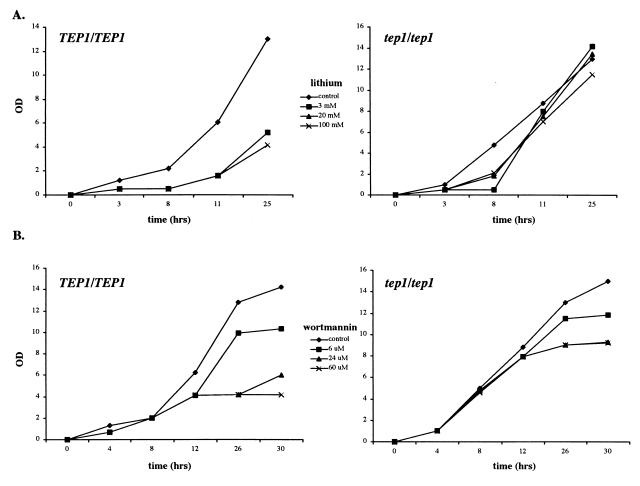
In contrast to the weak effect of lithium ions and wortmannin on haploid mutants, a significant effect was observed in diploids (Fig. 1). W303A diploids show pronounced growth inhibition at lithium concentrations of 3, 20, and 100 mM (Fig. 1A). The homozygous TEP1 knockout displays resistance to the same conditions. Wortmannin severely inhibits growth of the wild-type strain (although resistant mutants do arise spontaneously); the tep1/tep1 diploid is less sensitive, especially for concentrations of wortmannin in the 20 μM range (Fig. 1B). Interestingly, both wild-type and tep1 diploids are insensitive to another PI3-kinase inhibitor, LY-294002 (data not shown); this insensitivity may be because of an import failure.
Figure 1.
tep1 mutants are resistant to lithium ions and wortmannin. (A) Homozygous diploid TEP1 and Δtep1 strain (W303A) growth curves in the presence of lithium chloride in yeast extract/peptone/dextrose at 0 (control; ⧫), 3 mM (■), 20 mM (▴), and 100 mM (×). (B) Homozygous diploid TEP1 and Δtep1 strain (W303A) growth curves in the presence of wortmannin in YEPD + 2% DMSO at 0 (control; ⧫), 6 μM (■), 24 μM (▴), and 60 μM (×). Growth at 30°C was measured by optical density at 600 nm.
TEP1 Is Not a Lifespan Gene in S. cerevisiae.
Rates of cancer increase dramatically with age. Compared with the information we have about many biological processes, we know little about what determines the lifespan of an animal (28). C. elegans has proven to be a very convenient organism for lifespan studies, because its full genome sequence is available and its lifespan is ≈19 days at 20°C. Its dauer larva stage is long-lived, reproductively immature, and resistant to starvation and depravation. A number of genes that regulate dauer formation have been identified and assembled into a regulatory pathway. Mutation in these genes has been divided into two main classes: daf-d types that prevent dauer development and daf-c types that are mandatory for entry into the dauer state (29). The age-1 gene (daf-c type) encodes a PI3-kinase catalytic subunit. Mutations in daf-18 (daf-d type) suppress the increased longevity caused by mutations in daf-2 (daf-c type) and age-1 genes (29, 30). Recently several groups have shown that daf-18 is a homolog of PTEN (about 40% identity) and that PTEN acts to antagonize PI3-kinase function in vivo (13). Furthermore, Sun and colleagues (14) have shown that daf-18 dramatically shortens C. elegans lifespan, both in wild-type and a daf-2 mutant background that normally prolongs lifespan.
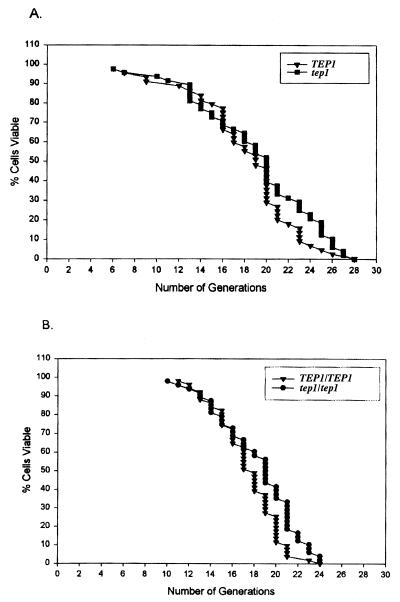
Yeast possess a finite lifespan that is similar in many attributes and implication to that of higher eukaryotes. To further characterize the yeast TEP1 gene, we determined the lifespan of a tep1 deletion strain (Fig. 2). TEP1 deletion had no effect on the lifespan of either haploid or diploid yeast strains. The average lifespan for TEP1 haploid cells was 18.25 divisions and 18.27 divisions for the TEP1 knockout. Wild-type diploid cells showed an average lifespan of 17.43 divisions; homozygous null diploid cells had an average of 17.85 divisions. We conclude that TEP1 does not directly participate in the aging process of S. cerevisiae.
Figure 2.
tep1 mutants do not have an altered lifespan. Lifespan analysis of TEP1 and Δtep1 (W303A) haploids (A) and diploids (B) according to ref. 22.
TEP1 mRNA Expression Is Markedly Elevated in Midsporulation.
The yeast developmental state referred to as sporulation, which consists of meiosis and spore morphogenesis, generates four haploid spores encapsulated into an ascus. During this developmental process, many genes are switched on and off in a carefully timed program. The most comprehensive gene expression analysis has been performed by Chu et al. (31) who used DNA microarrays containing nearly every yeast gene to assay changes in expression during sporulation. At least seven distinct temporal patterns of induction were observed, and the transcription factor Ndt80 was shown to be important for induction of a large group of genes at the end of meiotic prophase. One of these genes is TEP1, whose mRNA increases in abundance more than 15-fold during midsporulation. Consistent with TEP1 being up-regulated during meiosis, examination of its promoter region shows at least two consensus Ndt80 binding sites (31).
Diploids Homozygous for a tep1 Deletion Complete Meiosis and Form Spores.
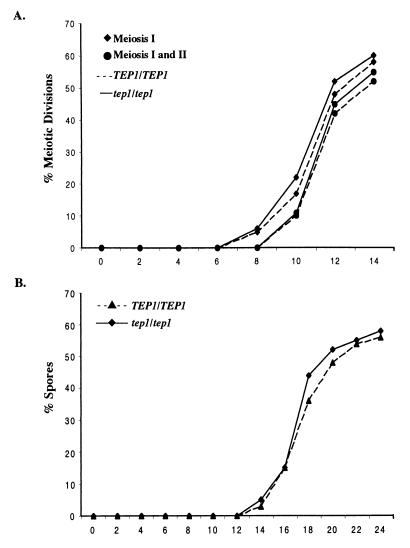
The failure to find any striking requirement for TEP1 in mitotically dividing cells and the up-regulation of TEP1 RNA in meiosis (31) prompted us to analyze the role of TEP1 in sporulation. Meiotic nuclear division and asci formation were examined in isogenic TEP1 and tep1 diploids induced to undergo meiosis. The percentage of cells completing meiosis I and meiosis II was determined by staining fixed cells with the DNA-specific dye DAPI. In the tep1 mutant, binucleate and tetranucleate cells appeared at the same time as in the corresponding TEP1 strain (Fig. 3A). Furthermore, the percentages of cells completing meiosis I and II were comparable in the two strains. Examination of asci formation by phase-contrast and fluorescence microscopy also indicated that spore maturation occurred with similar kinetics and levels in TEP1 and tep1 diploids (Fig. 3). Tetrad dissection of the resulting asci revealed that the absence of TEP1 had no affect on spore viability (data not shown). However, we did observe, in the W303A strain background, a significant percentage of damaged asci and released spores in the tep1 diploid (data not shown).
Figure 3.
Meiotic development in tep1 mutants. (A) The percentage of cells completing the meiosis I division (⧫) and both the meiosis I and II divisions (●) in homozygous Δtep1 (—) and TEP1 (- - -) strains monitored by fluorescence microscopy plotted against time (h) in sporulation medium. (B) The percentage of asci, monitored by phase contrast microscopy, in tep1 (⧫) and TEP1 (▴) strains plotted against time.
Spore Wall Formation Is Altered in tep1 Mutants.
Morphogenesis of spores initially requires the synthesis of an internal membrane surrounding the haploid nuclei generated from the meiotic divisions. Visualization of GFP-Spo14p was used to monitor spore membrane formation (26), which revealed that the process occurred normally in the tep1 mutant (data not shown). Late in meiosis, components of the spore wall are deposited in the luminal space of the prospore membrane (32). The outermost layer of the spore wall consists of dityrosine (33). Yeast cells synthesize the fluorescent molecule dityrosine only during ascus maturation. Qualitatively, dityrosine fluorescence can be monitored under UV light from sporulating cell patches on plates containing nitrocellulose filters (34). Such analysis revealed that tep1 mutants have reduced, although detectable, levels of dityrosine (data not shown).
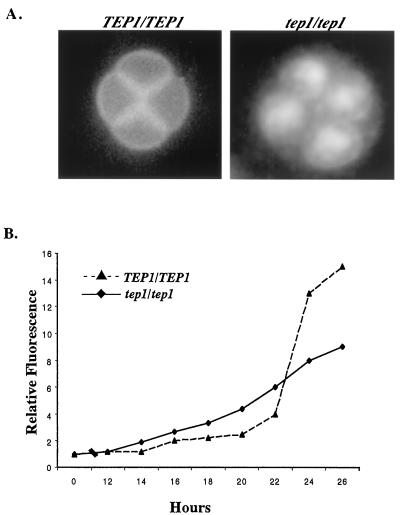
The defect in dityrosine fluorescence in tep1mutant is also apparent on microscopic examination of DAPI-stained, sporulating W303A cells (33, 35). Twenty-four hours after introduction into sporulation medium, wild-type cells have formed the characteristic tetrads with bright sharp spore wall fluorescent boundaries; the dityrosine fluorescence obscures the DAPI staining of the nuclei in these spores (Fig. 4A). In contrast, the tep1/tep1 spores show no defined wall fluorescence but rather, the spore cytoplasm is filled with fluorescent material and despite this the DAPI-stained nuclei are clearly visible (Fig. 4A).
Figure 4.
Dityrosine is altered in tep1 mutants. (A) Fluorescent microscopy of TEP1 and tep1 sporulating cells. After 24 h in sporulation medium, cells were stained with DAPI and observed for natural dityrosine fluorescence simultaneously in the emission range of 420–460 nm. In TEP1 cells, strong surface dityrosine ring fluorescence can be seen outlining the four spores. DAPI staining is not observable in these cells because the surface dityrosine fluorescence masks it. In Δtep1 strains, the localized dityrosine fluorescence is replaced by a dispersed intracellular signal and DAPI staining is seen. (B) Relative fluorescence intensity at 315 nm of cell wall extracts extracted from tep1 (⧫) and TEP1 (▴) strains plotted against time in sporulation medium.
Normal asci, with mature functional spore walls, exhibit resistance to heat shock, ether, and enzymatic digestion (24). Analysis of the effect that reduced dityrosine has on spore wall function was assessed by monitoring resistance to heat shock, ether, and glusulase digestion. No differences were observed on cell viability and growth when TEP1 and tep1 spores were subjected to these treatments (data not shown). Hence, reduced dityrosine does not affect spore functionality of tep1 mutants.
Dityrosine deposition also was monitored quantitatively by extracting cell wall components from sporulating cultures of TEP1 and tep1 strains. The fluorescence excitation spectrum of dityrosine was monitored in the extracts as described (26). The excitation spectrum obtained from sporulating TEP1 and tep1 diploids was characteristic of dityrosine with a peak of fluorescence at 315 nm (33). Consistent with the qualitative analysis, these measurements indicated that tep1 mutants displayed a 2-fold reduction in dityrosine levels (Fig. 4B). Interestingly, examination of dityrosine at various times throughout the meiotic program revealed that dityrosine accumulated earlier in the mutant compared with wild type, although the final level of dityrosine was lower (Fig. 4B).
Cellular fractionation was performed to determine whether the defect observed in dityrosine fluorescence was a consequence of poor deposition in the spore wall or a defect in the synthesis of the fluorescent molecule itself. Spectrophotometric analysis of the soluble cellular fraction showed more fluorescence characteristic of dityrosine in the mutant compared with the wild type. In contrast, more dityrosine fluorescence was detected in the wild type in the particulate and whole-cell extract. These results suggest that the defect in dityrosine is a consequence of faulty trafficking or deposition and not in the synthesis of the fluorescent molecule.
Analysis of the tep1-G199E Mutant.
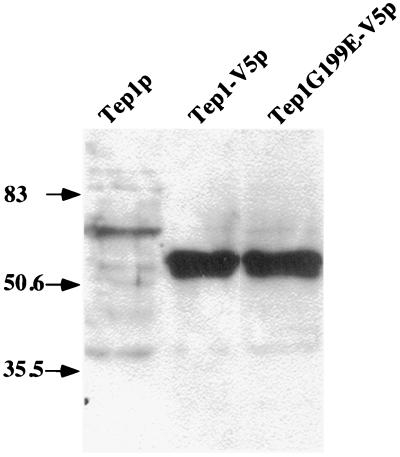
To test the hypothesis that lipid phosphatase activity is important for Tep1p function in yeast, we used complementation assays to determine the functionality of tagged versions of wild-type and mutant Tep1 proteins. We mutagenized the conserved glycine residue at position 199, which corresponds to residue 129 in human PTEN, to a glutamate. The corresponding human mutant protein retains the capability to dephosphorylate peptide substrates but fails to dephosphorylate phosphatidylinositol molecules in vitro and consequently fails to trigger apoptosis (8). Strains harboring tagged wild-type Tep1p was functional based on complementation of the dityrosine defect. In contrast, yeast strains harboring tep1-G199E as the only source of Tep1p, display the same defect in dityrosine deposition as strains harboring a complete deletion of the gene, supporting the hypothesis that the essential function of TEP1, like its human counterpart, is its lipid phosphatase activity. The stability of the mutant protein was verified by Western blot analysis (Fig. 5).
Figure 5.
Immunoblot of Tep1-V5p and mutant variant. Whole-cell extracts were prepared as described in Materials and Methods. Two times the amount of extract was loaded for the cells harboring Tep1p compared with Tep1-V5p and Tep1G199E-V5p. The positions of the prestained molecular mass markers are indicated by arrows and are expressed in kDa.
Overexpression of TEP1 Fails to Complement the Mutant Defect, Although Displaying No Effect on Wild Type.
In the course of our studies, we noticed that, whereas TEP1 on a centromere-based plasmid complemented the dityrosine defect of tep1 mutants, the same sequences on a 2-μ-based plasmid, which exists at approximately 40 copies per cell (36), did not. To eliminate the possibility that sequences were altered in subcloning steps to produce a nonfunctional gene, we performed two experiments. First, we transplaced the fragment from the 2-μ vector back to the centromere-based plasmid and confirmed that it was able to complement the tep1 mutant defect. Second, we cotransformed TEP1 2 μ and centromere-based plasmids into wild-type and tep1 mutants and found that the sequences still failed to rescue the mutant phenotype, while having no deleterious affect on wild type. In addition, when tep1 mutants harboring TEP1 2 μ were propagated, a percentage of cells could overcome the effect and were now rescued. (Recombination and/or loss or damage of the plasmid leading to copy reduction could account for these observations.) One interpretation of these results is that in the absence of TEP1 other pathways are altered to compensate for its inactivation.
Conclusions
In this study, we have analyzed the requirement of TEP1, the yeast homolog of PTEN, in the life and development of the budding yeast, S. cerevisiae. We find that TEP1 is dispensable for growth, and that haploid yeast strains harboring TEP1 deletions display only subtle phenotypes with respect to lithium and wortmannin resistance. In diploids, lithium and wortmannin resistance is more striking. Both lithium and wortmannin have been shown to affect yeast growth by inhibiting enzymes in the phosphatidylinositol signaling pathway.
We also find that TEP1 plays a role in the trafficking of dityrosine to the outside of the spore wall during the diploid-specific process of sporulation. Formation and accumulation of dityrosine in the yeast outer spore wall is believed to be a multistep process (33). Accumulation of dityrosine (up to 20% of the total protein hydrolysate) makes the spores resistant to enzymatic digestion and to chemical and physical damage. Enzymes involved in the incorporation of the dityrosine precursors into the spore wall or in the epimerization of the LL-dityrosine precursor have not yet been identified. Two developmentally regulated genes expressed in midsporulation, DIT1 and DIT2, are required for production of the dityrosine precursor. DIT1 is of unknown function; DIT2 is a member of the cytochrome P450 superfamily and is believed to catalyze the oxidation of tyrosine to form the orthobiphenyl linkage that forms dityrosine and renders it stable to hydrolysis (37). Our studies have shown that TEP1, which is also preferentially expressed in midsporulation, is required for proper transport (and/or deposition) of dityrosine into the outer wall. Emr and colleagues (38) have shown that the yeast phosphatidylinositol signaling pathway is involved in vesicular transport, which might be required to shuttle dityrosine (or a protein containing it) to the spore wall during spore maturation. Taken together, these results implicate TEP1 in the phosphatidylinositol pathway in yeast.
The defect observed in tep1 mutants in spore morphogenesis is consistent with a large body of evidence that indicates that phosphoinositides are important for cellular differentiation (39). Interestingly, phosphoinositides are in vivo activators of phospholipase D (PLD; ref. 40), which is essential for sporulation in yeast (25, 41). However, we have found no genetic interactions between yeast PLD and TEP1 (J.E., unpublished observations). Nonetheless, the maturation of the spore wall and its connection to the meiotic cell is analogous to the interaction of epithelial cells with the extracellular matrix. Perturbation of these cell contacts are important in carcinogenesis and suggest that analysis of TEP1 in yeast will be directly applicable to understanding the role of its homolog PTEN as a tumor suppressor in humans.
Surprisingly, as one scales the evolutionary ladder, PTEN plays an increasingly important role. This is emphasized by the findings that inactivation of PTEN in mice results in embryonic lethality and a propensity for the development of tumors in the heterozygous state (4–6). The Drosophila PTEN homolog also is required for viability (11). In C. elegans, however, the PTEN homolog is only required for the dauer response to starvation (12–14). Analogous to dauer formation, sporulation in yeast is a developmental state that is activated in response to starvation. The PTEN homolog in yeast plays a role in this process, although it is not essential for the formation of viable spores. One explanation for the subtle effect observed in yeast is offered by the observation that overexpression of TEP1 does not suppress the tep1 phenotype. The most likely explanation for this finding is that there may be other phosphatases that use the same substrate in yeast and/or that disruption of one enzyme can be compensated for by alterations in other activities. Genetic screens should uncover these pathways and provide a molecular understanding of the functional consequences of altered phosphatidylinositol pathways in vivo.
In mammals, PTEN dephosphorylates phosphatidylinositols, in particular PtdIns(3,4,5)P3, thus generating PtdIns(4,5)P2 (7). Although PtdIns(4,5)P2 is an important lipid that plays a key role in cell signaling, cytoskeletal and membrane trafficking in yeast as well as all organisms examined (42), to date, PtdIns(3,4,5)P3 has not been found in yeast. What, then, is Tep1p dephosphorylating? In addition to PtdIns(4,5)P2, yeast cells contain PtdIns(3)P, PtdIns(3,4)P2, and PtdIns(3,5)P2, at least two of which play important roles in vacuolar trafficking (38). No changes in levels of any of these lipids are observed in tep1 mutants (J.C. and G.A.P., unpublished observation). This may be because of the inability to detect localized changes in particular phosphatidylinositol species. Alternatively, alteration of pathways that compensate for the absence of TEP1 could restore the important phosphatidylinositols to near normal levels, thus no differences can be detected. As the full range of PTEN substrates has not been characterized, it remains possible that PTEN and/or its homologs also could act on the soluble inositols, which recently have been shown to be important players in nuclear signaling (43). The possibility that dephosphorylation of protein substrates is also important for the biological action of PTEN and/or Tep1 still has not been ruled out either. Studies of the function of Tep1 in budding yeast should aid in understanding the role(s) of PTEN in mammalian cells, even if their physiological effects are quite different.
Acknowledgments
We thank S. Chu, J. Haber, J. Gray, and S. Rudge for advice and encouragement. Funding was provided by: CaPCURE and the Ellison Medical Foundation (to G.A.P.); Carol M. Baldwin Breast Cancer Research Foundation and American Cancer Society RPG-99-122-01-MBC (to J.E.); and National Institutes of Health/National Cancer Institute Grant RO1 CA77695 (to H.S.).
Abbreviations
- PTEN
phosphatase deleted on chromosome ten
- MMAC1
mutated in multiple advanced cancers
- TEP1
tensin-like phosphatase
- PI3-kinase
phosphatidylinositol-3-kinase
- PtdIns(3,4,5)P3
phosphatidylinositol 3,4,5 triphosphate
- DAPI
4′,6-diamidino-2-phenyl-indole
References
- 1.Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang S I, Puc J, Miliaresis C, Rodgers L, McCombie R, et al. Science. 1997;275:1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 2.Li D M, Sun H. Cancer Res. 1997;57:2124–2129. [PubMed] [Google Scholar]
- 3.Steck P A, Pershouse M A, Jasser S A, Yung W K, Lin H, Ligon A H, Langford L A, Baumgard M L, Hattier T, Davis T, et al. Nat Genet. 1997;15:356–362. doi: 10.1038/ng0497-356. [DOI] [PubMed] [Google Scholar]
- 4.Podsypanina K, Ellenson L H, Nemes A, Gu J, Tamura M, Yamada K M, Cordon-Cardo C, Catoretti G, Fisher P E, Parsons R. Proc Natl Acad Sci USA. 1999;96:1563–1568. doi: 10.1073/pnas.96.4.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suzuki A, de la Pompa J L, Stambolic V, Elia A J, Sasaki T, del Barco Barrantes I, Ho A, Wakeham A, Itie A, Khoo W, et al. Curr Biol. 1998;8:1169–1178. doi: 10.1016/s0960-9822(07)00488-5. [DOI] [PubMed] [Google Scholar]
- 6.Cristofano A D, Pesce B, Cordon-Cardo C, Pandolfi P P. Nat Genet. 1998;19:348–355. doi: 10.1038/1235. [DOI] [PubMed] [Google Scholar]
- 7.Maehama T, Dixon J E. J Biol Chem. 1998;273:13375–13378. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- 8.Myers M P, Pass I, Batty I H, Van der Kaay J, Stolarov J P, Hemmings B A, Wigler M H, Downes C P, Tonks N K. Proc Natl Acad Sci USA. 1998;95:13513–13518. doi: 10.1073/pnas.95.23.13513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Furnari F B, Huang H J, Cavenee W K. Cancer Res. 1998;58:5002–5008. [PubMed] [Google Scholar]
- 10.Cantley L C, Neel B G. Proc Natl Acad Sci USA. 1999;96:4240–4245. doi: 10.1073/pnas.96.8.4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang H, Potter C J, Tao W, Li D-M, Brogiolo W, Hafen E, Sun H, Xu T. Development (Cambridge, UK) 1999;126:5365–5372. doi: 10.1242/dev.126.23.5365. [DOI] [PubMed] [Google Scholar]
- 12.Ogg S, Ruvkun G. Mol Cell. 1998;2:887–893. doi: 10.1016/s1097-2765(00)80303-2. [DOI] [PubMed] [Google Scholar]
- 13.Gil E B, Malone L E, Liu L X, Johnson C D, Lees J A. Proc Natl Acad Sci USA. 1999;96:2925–2930. doi: 10.1073/pnas.96.6.2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mihaylova V T, Borland C Z, Manjarrez L, Stern M J, Sun H. Proc Natl Acad Sci USA. 1999;96:7427–7432. doi: 10.1073/pnas.96.13.7427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li L, Ernsting B R, Wishart M J, Lohse D L, Dixon J E. J Biol Chem. 1997;272:29403–29406. doi: 10.1074/jbc.272.47.29403. [DOI] [PubMed] [Google Scholar]
- 16.Rose M D, Winston F, Hieter P. Methods in Yeast Genetics, A Laboratory Course Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1990. [Google Scholar]
- 17.Hovland P, Flick J, Johnston M, Sclafani R A. Gene. 1989;83:57–64. doi: 10.1016/0378-1119(89)90403-4. [DOI] [PubMed] [Google Scholar]
- 18.Ito H, Fukada Y, Murata K, Kimura A. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rothstein R. In: Guide to Yeast Genetics and Molecular Biology. Guthrie C, Fink G R, editors. Vol. 194. San Diego: Academic; 1991. pp. 281–301. [Google Scholar]
- 20.Elledge S J, Davis R W. Gene. 1988;70:303–312. doi: 10.1016/0378-1119(88)90202-8. [DOI] [PubMed] [Google Scholar]
- 21.Hill J E, Myers A M, Koerner T J, Tzagoloff A. Yeast. 1986;2:163–167. doi: 10.1002/yea.320020304. [DOI] [PubMed] [Google Scholar]
- 22.Seifert H S, Chen E Y, So M, Heffron F. Proc Natl Acad Sci USA. 1986;83:735–739. doi: 10.1073/pnas.83.3.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kennedy B K, Jr, Austriaco N R, Jr, Guarente L. J Cell Biol. 1994;127:1985–1993. doi: 10.1083/jcb.127.6.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krisak L, Strich R, Winters R S, Hall J P, Mallory M J, Kreitzer D, Tuan R S, Winter E. Genes Dev. 1994;8:2151–2161. doi: 10.1101/gad.8.18.2151. [DOI] [PubMed] [Google Scholar]
- 25.Rose K, Rudge S A, Frohman M A, Morris A J, Engebrecht J. Proc Natl Acad Sci USA. 1995;92:12151–12155. doi: 10.1073/pnas.92.26.12151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rudge S A, Cavenagh M M, Kamath R, Sciorra V A, Morris A J, Kahn R A, Engebrecht J. Mol Biol Cell. 1998;9:2025–2036. doi: 10.1091/mbc.9.8.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cutle N S, Heitman J, Cardenas M E. J Biol Chem. 1997;272:27671–27677. doi: 10.1074/jbc.272.44.27671. [DOI] [PubMed] [Google Scholar]
- 28.Fench C H. Longevity, Senescence, and the Genome. Chicago: Univ. of Chicago Press; 1990. [Google Scholar]
- 29.Larsen P L, Albert P S, Riddle D L. Genetics. 1995;139:1567–1583. doi: 10.1093/genetics/139.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dorman J B, Albinder B, Shroyer T, Kenyon C. Genetics. 1995;141:1399–1406. doi: 10.1093/genetics/141.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chu S, DeRisi J, Eisen M, Mulholland J, Botstein D, Brown P O, Herskowitz I. Science. 1998;282:699–705. doi: 10.1126/science.282.5389.699. [DOI] [PubMed] [Google Scholar]
- 32.Byers B. In: The Molecular Biology of the Yeast Saccharomyces: Life Cycle and Inheritance. Strathern J N, Jones E W, Broach J R, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1981. pp. 59–96. [Google Scholar]
- 33.Briza P, Winkler G, Kalchhauser H, Breitenbach M. J Biol Chem. 1986;261:4288–4294. [PubMed] [Google Scholar]
- 34.Esposito R E, Dresser M, Breitenbach M. In: Guide to Yeast Genetics and Molecular Biology. Guthrie C, Fink G R, editors. Vol. 194. San Diego: Academic; 1990. pp. 110–131. [Google Scholar]
- 35.Smail E H, Briza P, Panagos A, Berenfeld L. Infect Immun. 1995;63:4078–4083. doi: 10.1128/iai.63.10.4078-4083.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Broach J R, Volkert F C. In: The Molecular and Cellular Biology of the Yeast Saccharomyces. Genome Dynamics, Protein Synthesis, and Energetics. Broach J R, Pringle J R, Jones E W, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1991. pp. 297–331. [Google Scholar]
- 37.Briza P, Breitenbach M, Ellinger A, Segall J. Genes Dev. 1990;4:1775–1789. doi: 10.1101/gad.4.10.1775. [DOI] [PubMed] [Google Scholar]
- 38.Burd C G, Babst M, Emr S D. Semin Cell Dev Biol. 1998;9:527–533. doi: 10.1006/scdb.1998.0255. [DOI] [PubMed] [Google Scholar]
- 39.Divecha N, Irvine R. Cell. 1995;80:269–278. doi: 10.1016/0092-8674(95)90409-3. [DOI] [PubMed] [Google Scholar]
- 40.Sciorra V A, Rudge S A, Prestwich G D, Frohman M A, Engebrecht J, Morris A J. EMBO J. 1999;20:5911–5921. doi: 10.1093/emboj/18.21.5911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rudge S A, Morris A J, Engebrecht J. J Cell Biol. 1998;140:2025–2036. doi: 10.1083/jcb.140.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martin T F J. Annu Rev Cell Dev Biol. 1998;14:231–264. doi: 10.1146/annurev.cellbio.14.1.231. [DOI] [PubMed] [Google Scholar]
- 43.Odom A R, Stahlberg A, Wente S R, York J D. Science. 2000;287:2026–2029. doi: 10.1126/science.287.5460.2026. [DOI] [PubMed] [Google Scholar]