Abstract
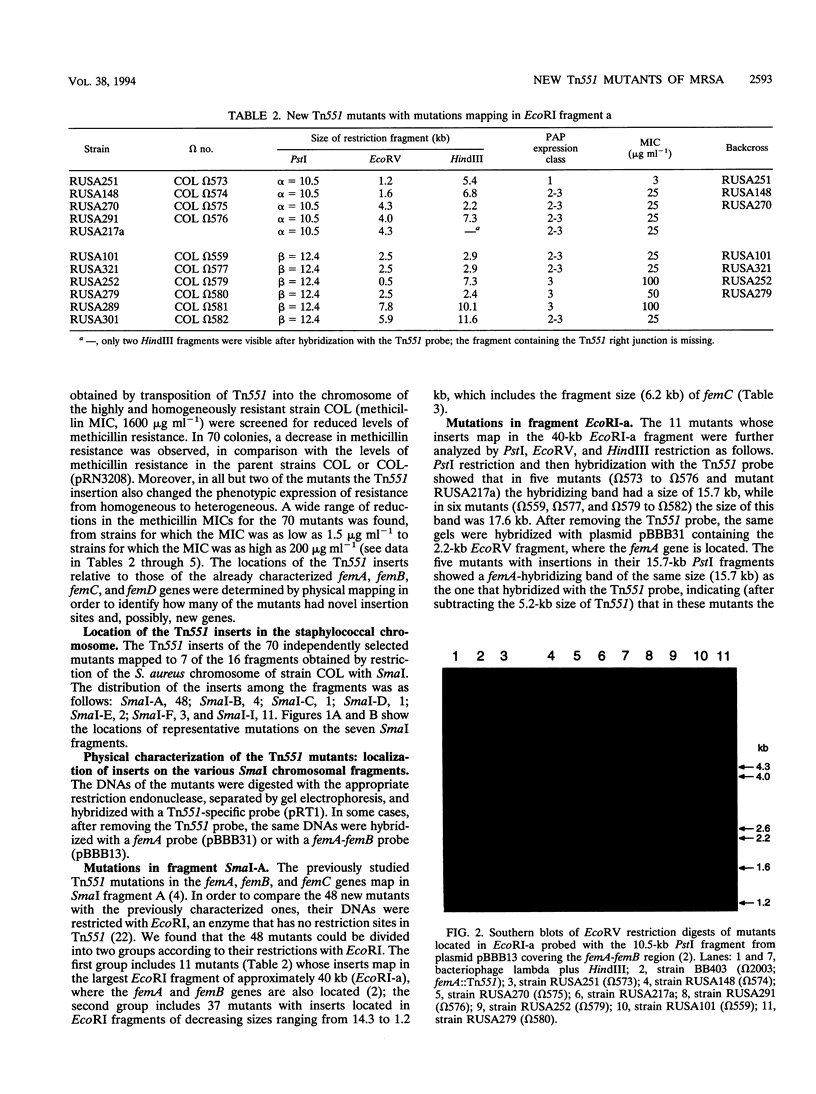
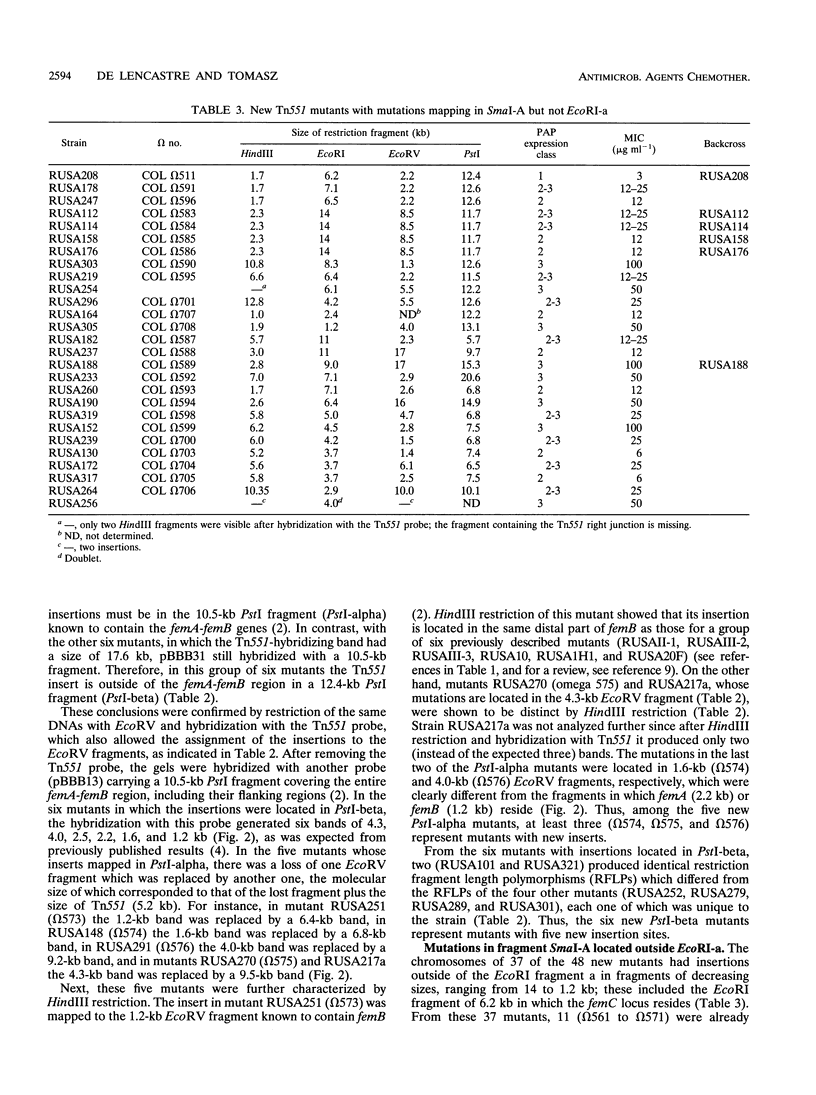
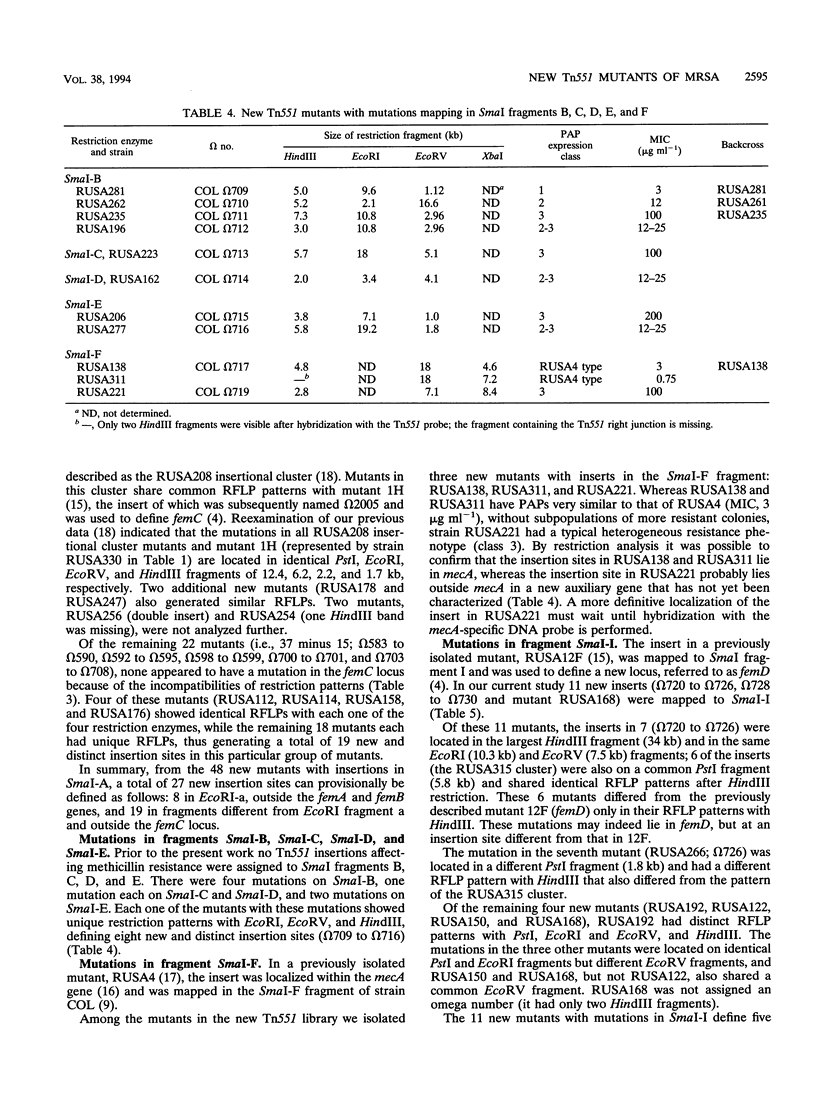
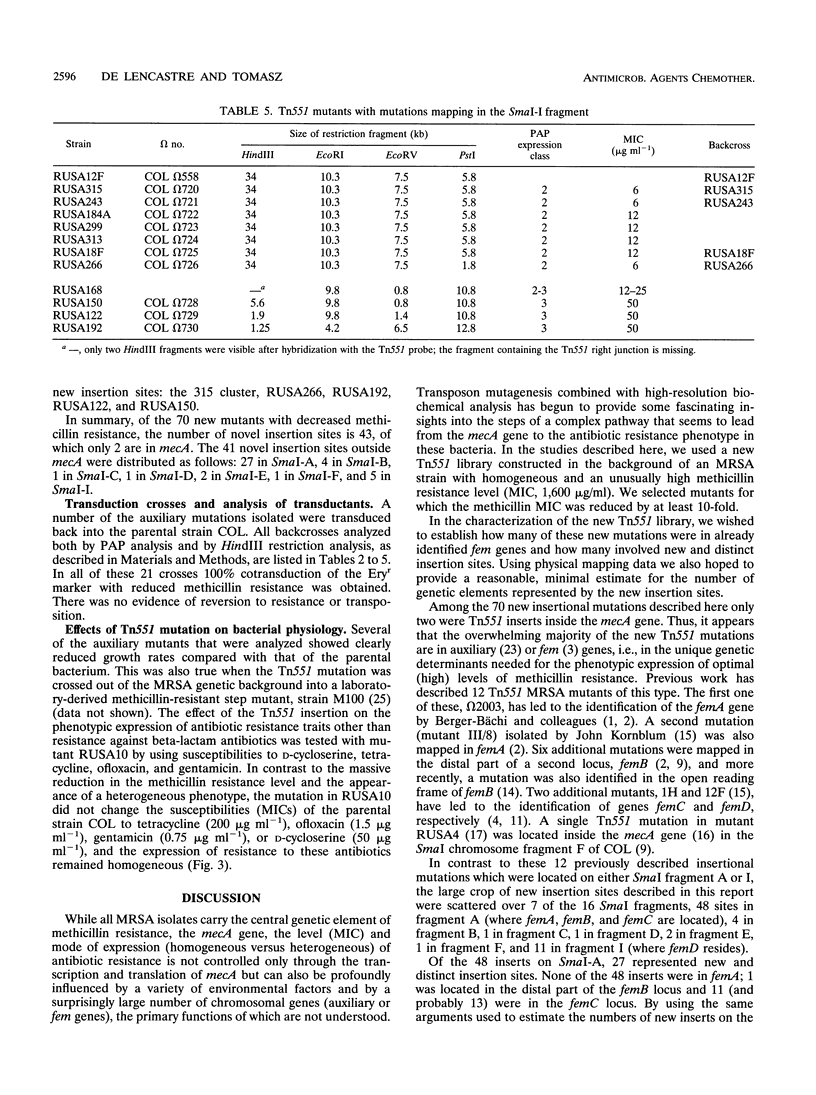
A new transposon library constructed in the background of the highly and homogeneously methicillin-resistant Staphylococcus aureus strain COL yielded 70 independent insertional mutants with reduced levels of antibiotic resistance. Restriction analysis with HindIII, EcoRV, EcoRI, and PstI and then Southern hybridization with probes for the transposon and for the femA-femB gene demonstrated that 41 of the 70 Tn551 mutants carried distinct and novel, as yet undescribed insertion sites, all of which were outside of the mecA gene and were also outside the already-characterized auxiliary genes femA, femB, femC, and femD. All previously described Tn551 mutations of this type were in genes located either on SmaI fragment A or SmaI fragment I. In contrast, inserts of the new library were located in 7 of the 16 SmaI chromosomal fragments, fragments A, B, C, D, E, F, and I. In all of the mutants, expression of methicillin resistance became heterogeneous, and the MIC for the majority of cells was reduced (1.5 to 200 micrograms ml-1) from the homogeneous methicillin MIC (1,600 micrograms ml-1) of the parental cells. Although identification of the exact number of genes inactivated through the new set of transposon inserts will require cloning and sequencing, a rough estimate of this number from mapping data suggests a minimum of at least 10 to 12 new genetic determinants, all of which are needed together with femA, femB, femC, and femD for the optimal expression of methicillin resistance.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berger-Bächi B., Barberis-Maino L., Strässle A., Kayser F. H. FemA, a host-mediated factor essential for methicillin resistance in Staphylococcus aureus: molecular cloning and characterization. Mol Gen Genet. 1989 Oct;219(1-2):263–269. doi: 10.1007/BF00261186. [DOI] [PubMed] [Google Scholar]
- Berger-Bächi B. Insertional inactivation of staphylococcal methicillin resistance by Tn551. J Bacteriol. 1983 Apr;154(1):479–487. doi: 10.1128/jb.154.1.479-487.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger-Bächi B., Strässle A., Gustafson J. E., Kayser F. H. Mapping and characterization of multiple chromosomal factors involved in methicillin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 1992 Jul;36(7):1367–1373. doi: 10.1128/aac.36.7.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafson J., Strässle A., Hächler H., Kayser F. H., Berger-Bächi B. The femC locus of Staphylococcus aureus required for methicillin resistance includes the glutamine synthetase operon. J Bacteriol. 1994 Mar;176(5):1460–1467. doi: 10.1128/jb.176.5.1460-1467.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman B. J., Tomasz A. Expression of methicillin resistance in heterogeneous strains of Staphylococcus aureus. Antimicrob Agents Chemother. 1986 Jan;29(1):85–92. doi: 10.1128/aac.29.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henze U., Sidow T., Wecke J., Labischinski H., Berger-Bächi B. Influence of femB on methicillin resistance and peptidoglycan metabolism in Staphylococcus aureus. J Bacteriol. 1993 Mar;175(6):1612–1620. doi: 10.1128/jb.175.6.1612-1620.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hächler H., Kayser F. H., Berger-Bächi B. Homology of a transferable tetracycline resistance determinant of Clostridium difficile with Streptococcus (Enterococcus) faecalis transposon Tn916. Antimicrob Agents Chemother. 1987 Jul;31(7):1033–1038. doi: 10.1128/aac.31.7.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornblum J., Hartman B. J., Novick R. P., Tomasz A. Conversion of a homogeneously methicillin-resistant strain of Staphylococcus aureus to heterogeneous resistance by Tn551-mediated insertional inactivation. Eur J Clin Microbiol. 1986 Dec;5(6):714–718. doi: 10.1007/BF02013311. [DOI] [PubMed] [Google Scholar]
- Matthews P., Tomasz A. Insertional inactivation of the mec gene in a transposon mutant of a methicillin-resistant clinical isolate of Staphylococcus aureus. Antimicrob Agents Chemother. 1990 Sep;34(9):1777–1779. doi: 10.1128/aac.34.9.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami K., Tomasz A. Involvement of multiple genetic determinants in high-level methicillin resistance in Staphylococcus aureus. J Bacteriol. 1989 Feb;171(2):874–879. doi: 10.1128/jb.171.2.874-879.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornelas-Soares A., de Lencastre H., de Jonge B., Gage D., Chang Y. S., Tomasz A. The peptidoglycan composition of a Staphylococcus aureus mutant selected for reduced methicillin resistance. J Biol Chem. 1993 Dec 15;268(35):26268–26272. [PubMed] [Google Scholar]
- Oshida T., Tomasz A. Isolation and characterization of a Tn551-autolysis mutant of Staphylococcus aureus. J Bacteriol. 1992 Aug;174(15):4952–4959. doi: 10.1128/jb.174.15.4952-4959.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattee P. A. Distribution of Tn551 insertion sites responsible for auxotrophy on the Staphylococcus aureus chromosome. J Bacteriol. 1981 Jan;145(1):479–488. doi: 10.1128/jb.145.1.479-488.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw J. H., Clewell D. B. Complete nucleotide sequence of macrolide-lincosamide-streptogramin B-resistance transposon Tn917 in Streptococcus faecalis. J Bacteriol. 1985 Nov;164(2):782–796. doi: 10.1128/jb.164.2.782-796.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasz A., Nachman S., Leaf H. Stable classes of phenotypic expression in methicillin-resistant clinical isolates of staphylococci. Antimicrob Agents Chemother. 1991 Jan;35(1):124–129. doi: 10.1128/aac.35.1.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonin E., Tomasz A. Beta-lactam-specific resistant mutants of Staphylococcus aureus. Antimicrob Agents Chemother. 1986 Oct;30(4):577–583. doi: 10.1128/aac.30.4.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jonge B. L., Chang Y. S., Gage D., Tomasz A. Peptidoglycan composition in heterogeneous Tn551 mutants of a methicillin-resistant Staphylococcus aureus strain. J Biol Chem. 1992 Jun 5;267(16):11255–11259. [PubMed] [Google Scholar]
- de Jonge B. L., Chang Y. S., Gage D., Tomasz A. Peptidoglycan composition of a highly methicillin-resistant Staphylococcus aureus strain. The role of penicillin binding protein 2A. J Biol Chem. 1992 Jun 5;267(16):11248–11254. [PubMed] [Google Scholar]
- de Jonge B. L., Sidow T., Chang Y. S., Labischinski H., Berger-Bachi B., Gage D. A., Tomasz A. Altered muropeptide composition in Staphylococcus aureus strains with an inactivated femA locus. J Bacteriol. 1993 May;175(9):2779–2782. doi: 10.1128/jb.175.9.2779-2782.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lencastre H., Couto I., Santos I., Melo-Cristino J., Torres-Pereira A., Tomasz A. Methicillin-resistant Staphylococcus aureus disease in a Portuguese hospital: characterization of clonal types by a combination of DNA typing methods. Eur J Clin Microbiol Infect Dis. 1994 Jan;13(1):64–73. doi: 10.1007/BF02026129. [DOI] [PubMed] [Google Scholar]
- de Lencastre H., Sá Figueiredo A. M., Urban C., Rahal J., Tomasz A. Multiple mechanisms of methicillin resistance and improved methods for detection in clinical isolates of Staphylococcus aureus. Antimicrob Agents Chemother. 1991 Apr;35(4):632–639. doi: 10.1128/aac.35.4.632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lencastre H., de Jonge B. L., Matthews P. R., Tomasz A. Molecular aspects of methicillin resistance in Staphylococcus aureus. J Antimicrob Chemother. 1994 Jan;33(1):7–24. doi: 10.1093/jac/33.1.7. [DOI] [PubMed] [Google Scholar]




