Abstract
Resistance to ceftazidime, detected in isolates of Escherichia coli 5518 and Enterobacter gergoviae 3773 from our hospital, was transferred, together with resistance to aminoglycosides, trimethoprim, sulfonamide, and other beta-lactam antibiotics, by conjugation to E. coli JP559. Both E. coli transconjugants were resistant to ampicillin, all cephalosporins, and aztreonam but remained susceptible to cefoxitin and imipenem. The enzymes of the two transconjugant strains readily hydrolyzed cephalosporins in a spectrophotometric assay. Hybridization results suggested that the extended-spectrum beta-lactamase produced by E. coli 5518 was a non-TEM, non-SHV enzyme, the origin of which is currently unknown. The beta-lactamase produced by E. gergoviae 3773 was of the SHV type and was further proved to be SHV-2 by DNA sequencing. Thus, extended-spectrum beta-lactamases are occurring in China as well as in other parts of the world.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arakawa Y., Ohta M., Kido N., Fujii Y., Komatsu T., Kato N. Close evolutionary relationship between the chromosomally encoded beta-lactamase gene of Klebsiella pneumoniae and the TEM beta-lactamase gene mediated by R plasmids. FEBS Lett. 1986 Oct 20;207(1):69–74. doi: 10.1016/0014-5793(86)80014-x. [DOI] [PubMed] [Google Scholar]
- Barthélémy M., Péduzzi J., Ben Yaghlane H., Labia R. Single amino acid substitution between SHV-1 beta-lactamase and cefotaxime-hydrolyzing SHV-2 enzyme. FEBS Lett. 1988 Apr 11;231(1):217–220. doi: 10.1016/0014-5793(88)80734-8. [DOI] [PubMed] [Google Scholar]
- Barthélémy M., Péduzzi J., Bernard H., Tancrède C., Labia R. Close amino acid sequence relationship between the new plasmid-mediated extended-spectrum beta-lactamase MEN-1 and chromosomally encoded enzymes of Klebsiella oxytoca. Biochim Biophys Acta. 1992 Jul 13;1122(1):15–22. doi: 10.1016/0167-4838(92)90121-s. [DOI] [PubMed] [Google Scholar]
- Garbarg-Chenon A., Godard V., Labia R., Nicolas J. C. Nucleotide sequence of SHV-2 beta-lactamase gene. Antimicrob Agents Chemother. 1990 Jul;34(7):1444–1446. doi: 10.1128/aac.34.7.1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall L. M., Livermore D. M., Gur D., Akova M., Akalin H. E. OXA-11, an extended-spectrum variant of OXA-10 (PSE-2) beta-lactamase from Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1993 Aug;37(8):1637–1644. doi: 10.1128/aac.37.8.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzberg O. Refined crystal structure of beta-lactamase from Staphylococcus aureus PC1 at 2.0 A resolution. J Mol Biol. 1991 Feb 20;217(4):701–719. doi: 10.1016/0022-2836(91)90527-d. [DOI] [PubMed] [Google Scholar]
- Huletsky A., Couture F., Levesque R. C. Nucleotide sequence and phylogeny of SHV-2 beta-lactamase. Antimicrob Agents Chemother. 1990 Sep;34(9):1725–1732. doi: 10.1128/aac.34.9.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huovinen S., Huovinén P., Jacoby G. A. Detection of plasmid-mediated beta-lactamases with DNA probes. Antimicrob Agents Chemother. 1988 Feb;32(2):175–179. doi: 10.1128/aac.32.2.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacoby G. A., Medeiros A. A. More extended-spectrum beta-lactamases. Antimicrob Agents Chemother. 1991 Sep;35(9):1697–1704. doi: 10.1128/aac.35.9.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacoby G. A., Medeiros A. A., O'Brien T. F., Pinto M. E., Jiang H. Broad-spectrum, transmissible beta-lactamases. N Engl J Med. 1988 Sep 15;319(11):723–724. doi: 10.1056/NEJM198809153191114. [DOI] [PubMed] [Google Scholar]
- Jelsch C., Lenfant F., Masson J. M., Samama J. P. Beta-lactamase TEM1 of E. coli. Crystal structure determination at 2.5 A resolution. FEBS Lett. 1992 Mar 9;299(2):135–142. doi: 10.1016/0014-5793(92)80232-6. [DOI] [PubMed] [Google Scholar]
- Kliebe C., Nies B. A., Meyer J. F., Tolxdorff-Neutzling R. M., Wiedemann B. Evolution of plasmid-coded resistance to broad-spectrum cephalosporins. Antimicrob Agents Chemother. 1985 Aug;28(2):302–307. doi: 10.1128/aac.28.2.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knothe H., Shah P., Krcmery V., Antal M., Mitsuhashi S. Transferable resistance to cefotaxime, cefoxitin, cefamandole and cefuroxime in clinical isolates of Klebsiella pneumoniae and Serratia marcescens. Infection. 1983 Nov-Dec;11(6):315–317. doi: 10.1007/BF01641355. [DOI] [PubMed] [Google Scholar]
- Lee K. Y., Hopkins J. D., Syvanen M. Direct involvement of IS26 in an antibiotic resistance operon. J Bacteriol. 1990 Jun;172(6):3229–3236. doi: 10.1128/jb.172.6.3229-3236.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindberg F., Lindquist S., Normark S. Inactivation of the ampD gene causes semiconstitutive overproduction of the inducible Citrobacter freundii beta-lactamase. J Bacteriol. 1987 May;169(5):1923–1928. doi: 10.1128/jb.169.5.1923-1928.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier J., Levesque R. C. Cloning of SHV-2, OHIO-1, and OXA-6 beta-lactamases and cloning and sequencing of SHV-1 beta-lactamase. Antimicrob Agents Chemother. 1990 Aug;34(8):1577–1583. doi: 10.1128/aac.34.8.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordmann P., Ronco E., Naas T., Duport C., Michel-Briand Y., Labia R. Characterization of a novel extended-spectrum beta-lactamase from Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1993 May;37(5):962–969. doi: 10.1128/aac.37.5.962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippon A., Labia R., Jacoby G. Extended-spectrum beta-lactamases. Antimicrob Agents Chemother. 1989 Aug;33(8):1131–1136. doi: 10.1128/aac.33.8.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn J. P., Miyashiro D., Sahm D., Flamm R., Bush K. Novel plasmid-mediated beta-lactamase (TEM-10) conferring selective resistance to ceftazidime and aztreonam in clinical isolates of Klebsiella pneumoniae. Antimicrob Agents Chemother. 1989 Sep;33(9):1451–1456. doi: 10.1128/aac.33.9.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders C. C., Sanders W. E., Jr Emergence of resistance during therapy with the newer beta-lactam antibiotics: role of inducible beta-lactamases and implications for the future. Rev Infect Dis. 1983 Jul-Aug;5(4):639–648. doi: 10.1093/clinids/5.4.639. [DOI] [PubMed] [Google Scholar]
- Sanders C. C., Sanders W. E., Jr Type I beta-lactamases of gram-negative bacteria: interactions with beta-lactam antibiotics. J Infect Dis. 1986 Nov;154(5):792–800. doi: 10.1093/infdis/154.5.792. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
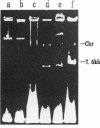
- Takahashi S., Nagano Y. Rapid procedure for isolation of plasmid DNA and application to epidemiological analysis. J Clin Microbiol. 1984 Oct;20(4):608–613. doi: 10.1128/jcm.20.4.608-613.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willetts N., Wilkins B. Processing of plasmid DNA during bacterial conjugation. Microbiol Rev. 1984 Mar;48(1):24–41. doi: 10.1128/mr.48.1.24-41.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y. J., Wu P. J., Livermore D. M. Biochemical characterization of a beta-lactamase that hydrolyzes penems and carbapenems from two Serratia marcescens isolates. Antimicrob Agents Chemother. 1990 May;34(5):755–758. doi: 10.1128/aac.34.5.755. [DOI] [PMC free article] [PubMed] [Google Scholar]