Abstract
Borna disease virus (BDV) is a nonsegmented negative-strand RNA virus that belongs to the Mononegavirales. Unlike other animal viruses of this order, BDV replicates and transcribes in the nucleus of infected cells. Previous studies have shown that BDV uses RNA splicing machinery for its mRNA expression. In the present study, we identified spliced RNAs that use an alternative 3′ splice site, SA3, in BDV-infected cell lines as well as infected animal brain cells. Transient transfection analysis of cDNA clones of BDV RNA revealed that although SA3 is a favorable splice site in mammalian cells, utilization of SA3 is negatively regulated in infected cells. This negative splicing activity of the SA3 site is regulated by a putative cis-acting region, the exon splicing suppressor (ESS), within the polymerase exon of BDV. The BDV ESS contains similar motifs to other known ESSs present in viral and cellular genes. Furthermore, our results indicated that a functional polyadenylation signal just upstream of the BDV ESS is also involved in the regulation of alternative splicing of BDV. These observations represent the first documentation of complex RNA splicing in animal RNA viruses and also provide new insight into the mechanism of regulation of alternative splicing in animal viruses.
There are multiple posttranscriptional processes that are essential for expression of eukaryotic and viral genes. These include RNA capping, splicing, polyadenylation, and transport (1–6). Pre-mRNA splicing is often complex in mammalian cells, because the majority of pre-mRNAs have multiple introns, and these sometimes contain more than one 5′ and/or 3′ splice site. Alternative splicing of these pre-mRNAs involves the use of alternative 5′ or 3′ splice sites and exon skipping or inclusion, generating different pre-mRNAs potentially encoding multiple protein isoforms with distinct functions. A number of factors, including cis element in genomes and viral and cellular proteins, which affect splicing efficiency and splice site choice, have been identified (7–12). Splicing of HIV type 1 (HIV-1) tat RNA and bovine papillomavirus type 1 (BPV-1) early and late genes are regulated by suboptimal 3′ splice sites and exon splicing enhancer and suppressor elements (ESE and ESS), which bind to cellular regulatory factors (13–16). Such regulation is important to achieve the balance between alternatively spliced RNAs or spliced and unspliced RNAs. The proteins produced by regulated splicing are required at different stages of the virus replication or in different host cell environments and are involved in regulation of the viral life cycle. It has been demonstrated that reactivation of HIV-1 from the proviral latency requires a threshold level of Rev protein, which is generated by multiple splicing events (17, 18). Furthermore, the splicing level of NS1 mRNA of influenza virus regulates efficient transport of unspliced NS1 mRNA from the nucleus (19). In addition, the splicing pattern of the BPV-1 late mRNA depends on the differentiation stage of the infected cells (20, 21). Therefore, regulated splicing of viruses is a key step for accomplishing their life cycle in mammalian cells.
Borna disease virus (BDV) is a nonsegmented negative-strand RNA virus that belongs to the Mononegavirales (22, 23). Despite the similarity in genome organization to other members of this order, BDV has several clearly distinguishing features. One of the most striking characteristics of BDV is its localization for transcription. BDV replicates and transcribes in the nucleus of infected cells (24), whereas the other animal viruses of this order undergo their life cycle in the cell cytoplasm. Previous studies have demonstrated that BDV uses the RNA splicing machinery for gene expression, and the genome contains two introns (intron I and II; refs. 25 and 26). Transcripts that retain intron I serve as messages for expression of the gp18 matrix protein (M) of BDV, and those that retain intron II serve as messages for expression of the envelope glycoprotein (G). Transcripts that lack both introns serve as messages for expression of the Pol protein (L) of BDV (27). As in other viruses, splicing of the BDV genome is not 100% efficient. The 3′ splice sites of BDV have been shown to be suboptimal, and inefficiency of splicing is because of inaccessibility of the splice sites to the splicing complex (26). In view of the features of BDV biology, such as low-level production of infectious virus and viral persistence in infected cells, however, BDV must also regulate efficiency of gene expression by another mechanism posttranscriptionally. Furthermore, it is also likely that there are rare spliced RNAs that are conditionally expressed at different stages of the virus life cycle. Thus, research into the regulated RNA splicing of BDV should lead to a better understanding of the biological features of the virus. Here, we report spliced RNAs of BDV that use an alternative 3′ splice site. Our results demonstrate that a cis-acting splicing suppressor and a polyadenylation/termination (Pt) signal within the genome are involved in alternative splicing of BDV. These observations provide new insight into the mechanism responsible for regulation of alternative RNA splicing in animal viruses.
Materials and Methods
Cells and Viruses.
Madin–Darby canine kidney cell (MDCK), rat glioma cell (C6), human oligodendroglioma cell (OL), and COS-7 cell lines were maintained in DMEM containing 10% heat-inactivated FCS. Three persistently BDV-infected cell lines, MDCK/BDV (28), C6BV (29), and OL/BDV cells obtained by establishing a persistent BDV strain HuP2br (30) infection in OL cells, were maintained under the same conditions as the parental cell lines. These cells produced infectious BDV, and >90% of the cells were infected.
Animal Samples.
BDV-infected rat brain samples were obtained from 2 to 3 weeks after inoculating newborn Lewis rats intracerebrally with 20 μl of BDV stock prepared from a homogenized MDCK/BDV-infected cell line.
Thermostable Reverse Transcription (RT)–Time-Release PCR Analysis.
Total RNAs were extracted from BDV-infected cultured cell lines, rat brain cells, or plasmid-transfected cells by using an RNA isolation kit (Nippon Gene, Toyama, Japan), and aliquots of 1 μg of total RNA were reverse transcribed with 50 μM of oligo(dT)20 primer. To prevent secondary structure formation of RNA templates during the RT step, we used the Thermoscript RT-PCR System (GIBCO/BRL), and RT was carried out for 1 h at 60°C.
The resulting cDNAs were used as templates for PCR amplification with the following primer pairs: S-1/A-2, S-1/A-12, S-1/A-9, and S-1/RVA-1. The sequences and nucleotide (nt) positions of primers used for this study are listed in Supplemental Table 1 (see www.pnas.org). PCR was performed in a total volume of 50 μl containing 3 μl of cDNA and 2.5 units of Taq polymerase [Amplitaq Gold (Perkin–Elmer)]. For efficient amplification of rare spliced RNAs in infected cells, we used the time-release PCR method (31). The reaction mixtures were preincubated at 94°C for 3 min followed by 35 cycles of PCR at 94°C for 1 min, 63°C for 1 min, and 74°C for 1 min 30 sec. Amplification products were analyzed by electrophoresis in 1.5% agarose gels.
RNase Protection Assay (RPA).
Aliquots of 10 μg of RNAs extracted from infected or transfected cells were hybridized with [32P]rUTP-labeled antisense riboprobe. To generate the riboprobe, an intronless BDV cDNA spanning splice site was amplified by PCR with S-20 and A-32 primers and inserted into pSPT19 (Boehringer Mannheim). The plasmid was transcribed by using T7 polymerase, and RPA was performed with an RPA III kit (Ambion, Austin, TX) according to the manufacturer's recommendations. The protected products were separated on 5% polyacrylamide/8 M urea gels for 1 h at a constant voltage of 300 V. After electrophoresis, the gels were dried on Whatman paper and then exposed to x-ray film for 1–3 days at −70°C. All cited percentages of spliced RNAs are averages of results obtained in at least three independent experiments.
Northern Blot Analysis.
Poly(A)+ RNA was purified from total RNA of OL and OL/BDV cell lines by using Oligotex-dT30 (Takara Shuzo, Kyoto). RNA samples were electrophoresed through a 1% agarose gel containing 2.2% formaldehyde and analyzed by Northern (RNA) blot hybridization with [32P]rUTP-labeled antisense riboprobe. To generate the riboprobe, BDV cDNA was amplified by RT-PCR and inserted into pSPT19. Hybridization was performed with the probe (1.0 × 106 cpm/ml) at 68°C overnight. After washing, specific reaction was detected to expose to x-ray film for 1–4 days at −70°C.
Plasmid Construction.
The BDV cDNA expression plasmid pCD2.1 used for transfection analysis was generated as follows. BDV cDNA was amplified with S-1.1 and A-10 primers (Supplemental Table 1; see www.pnas.org) by using RNA sample from OL/BDV cells. The PCR product was digested with EcoRI and SalI and cloned into pUC19. The pUC vector containing a 2.7-kb BDV cDNA clone was cut with EcoRI and filled with pCla I linker. The resulting vector was digested with ClaI and SalI, and then the fragment was inserted into HindIII-SalI sites of the expression vector pRVSV (32) to create pCD2.1. Plasmid pCD2.1 was used as a DNA template to create the other expression plasmids. To generate pCD2.2, pCD2.3, pCD2.4, pCD2.5, and pCD2.6, ApaI and XbaI sites in pCD2.1 were digested, and cDNA fragments amplified with S-11 and A-2.2 (pCD2.2), A-2.3 (pCD2.3), A-2.4 (pCD2.4), A-2.5 (pCD2.5), or A-2.6 (pCD2.6) primers (Supplemental Table 1) were inserted into these sites of the plasmid pCD2.1. To generate deletion clones of pCD2.5, the cDNA amplified with S-11 and A-25 primers was inserted into pCD2.1 lacking the ApaI and BamHI fragment. The resulting plasmid was cleaved by BamHI, and fragments amplified from pCD2.5 with S-5.1 (pCD2.5d1), S-5.2 (pCD2.5d2), or S-5.3 (pCD2.5d3), and A-28 primers were ligated into the BamHI sites. The pCD2.6d1 plasmid was created by insertion of the fragment from pCD2.6 amplified by S-6.1 and A-28 primers into pCD2.5, which was cleaved at the BamHI sites. To generate pCD2.6d2, the plasmid pCD2.6 was first digested with Sal I and Spl I, and then blunted and self ligated.
The expression plasmid pS1-FLAG was generated by amplification of BDV cDNA with S1-FF and S1-RF primers (Supplemental Table 1; see www.pnas.org) by using RNA sample from OL/BDV cells. The PCR product was digested with PstI and XbaI and cloned into pRVSV vector. The construction of an expression plasmid of BDV X protein, pcORFx1-FLAG, has been described elsewhere (33).
RACE Analysis.
The 3′ rapid amplification cDNA end (RACE) assay was carried out to determine the Pt sites of BDV transcripts. Total RNAs from BDV-infected cells were primed with oligo(dT)-adapter primer, RACE-A1, and RT was performed with Thermoscript (GIBCO/BRL) at 60°C for 1 h. The resulting first-strand cDNAs were used as templates for PCR with S-2 and RACE-A2 primer pair under the following conditions: 30 cycles of 94°C for 30 sec, 59°C for 30 sec, and 72°C for 40 sec. Nested amplification was performed by PCR with RACE-S3 and RACE-A2 under the following conditions: 25 cycles of 94°C for 20 sec, 59°C for 20 sec, and 72°C for 20 sec. The products of the nested PCR were cloned into pUC19 and sequenced.
Results
Detection of a 3′ Splice Site and Alternative Splicing of BDV.
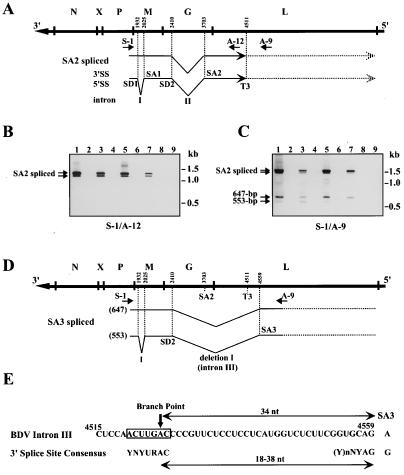
We first performed thermostable RT–time-release PCR to detect rare spliced RNAs in BDV-infected cultured cells and animal brains. The precise locations of PCR primers are indicated in Fig. 1A and are described in Supplemental Table 1 (www.pnas.org). By using the S-1/A-12 primer pair, we detected two fragments that could correspond to intron II [nt 2410–3703 of BDV genome (antigenome polarity)]-spliced RNAs, named SA2 spliced RNAs, at 1,297 and 1,203 bp, as previously reported (Fig. 1B, lanes 1, 3, 5 and 7) (25, 26). The S-1/A-9 primer pair amplified small faint bands at 647 and 553 bp on the gel, in addition to the fragments of 1,503 and 1,409 bp in all infected cells (Fig. 1C, lanes 1, 3, 5, and 7). Fragments corresponding to unspliced RNA species were also detected at 2,590 and 2,796 bp by using the S-1/A-12 and S-1/A-9 primer pairs, respectively, but the amplification of these longer products was inefficient under our conditions. To determine the sequence organization of the PCR DNAs, we cloned and sequenced the fragments amplified by the S-1/A-9 primer pair. Sequence analysis of several independent clones confirmed that the cloned fragments had the BDV genome sequence containing internal deletions. The DNA fragment of 1,503 bp lacked the intron II sequence, whereas the DNA fragment of 1,409 bp lacked both introns I (nt 1932–2025) and II sequences, confirming that these fragments were derived from SA2 spliced RNAs of BDV. The DNA fragment of 647 bp lacked the region between nt 2410 and 4559 (deletion I), and the fragment of 553 bp lacked both intron I and deletion I sequences (Fig. 1D). The deletion I sequence shared its 5′ end with intron II. Furthermore, inspection of the BDV antigenome polarity sequence at the boundary of deletion I revealed the presence of sequence motifs with similarity to the mammalian 3′ splice site consensus sequences (Fig. 1E) (34). The branchpoint sequence, in which six of seven nt positions matched the consensus branchpoint sequence, was found 34 nt from the possible 3′ splice site (Fig. 1E). The polypyrimidine tract also showed a suitable sequence, indicating that the deletion I sequence contained a 3′ splice site of BDV. Consequently, we designated deletion I as intron III of BDV and the 3′ splice site found at nt 4559 as SA3 (Fig. 1D).
Figure 1.
Detection of a 3′ splice site of BDV. (A) SA2 splicing of BDV. The outline of the BDV genome is indicated at the top. Arrows indicate approximate positions of primers used for RT-PCR. Positions of 5′ and 3′ splice site (SS) and a polyadenylation site (T3) are indicated. (B and C) Thermostable RT-time-release PCR of BDV RNA. RT-PCR was performed with S-1 and A-12 (B) or S-1 and A-9 (C) primer pairs. The images of agarose gels were captured electronically, and the pixels were inverted. Lanes: 1, MDCK/BDV; 2, MDCK; 3, C6BV; 4, C6; 5, OL/BDV; 6, OL; 7, BDV-infected rat brain; 8, uninfected rat brain; 9, MDCK/BDV RT(−). (D) Splicing patterns of BDV. Approximate location of a newly identified BDV intron and 3′ splice site (SA3) are shown. Lengths of PCR products are indicated at the left of the patterns in parentheses. (E) Comparison of the BDV intron III splice sequence with mammalian consensus sequence. A branchpoint nt is indicated by an arrow. Y, pyrimidine; R, purine; N, any base.
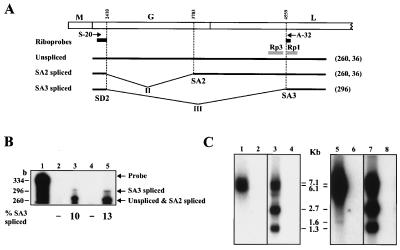
To confirm whether RNA species lacking intron III, named SA3 spliced RNAs, are generated during BDV infection, we performed RPA of RNA from BDV-infected cultured cells and rat brain cells by using an antisense riboprobe spanning nt 2150–4595 of BDV cDNA lacking the intron III sequence (Fig. 2A). By using this probe, fragments of 296 and 260 nt should be generated as protected RNAs that represent SA3 spliced and SA2 spliced or unspliced RNAs, respectively (Fig. 2A). As shown in Fig. 2B, SA3 spliced RNA was found as a faint band in both BDV-infected cultured cells and rat brain cells, whereas unspliced and SA2 spliced RNAs were detected as major protected signals (lanes 3 and 5). A 36-nt protected RNA corresponding to nt 4559–4595 was also found (data not shown). This observation demonstrated that the SA3 spliced RNAs do exist in BDV-infected cells, and that BDV uses alternative splicing for gene expression. Furthermore, RPA revealed that splicing efficiency of intron III was only around 10%, confirming the RT-PCR results shown in Fig. 1C.
Figure 2.
RNase protection and RNA blot assays of BDV RNA. (A) Schematic maps of BDV ORFs are shown at the top. The riboprobes for RPA and RNA blotting [Rp1 (nt 4560–4766) and Rp3 (nt 4075–4480)] are indicated by black and shaded bars, respectively. Expected sizes of protected fragments are indicated at the right of the patterns in parentheses. (B) RPA for SA3 splicing RNAs. Lanes: 1, RNase-untreated riboprobe; 2, uninfected rat brain; 3, BDV-infected rat brain, 4, OL cells; 5, OL/BDV cells. % of SA3 spliced RNAs is shown at the bottom of the lanes. (C) RNA blotting for SA3 spliced RNAs. Poly(A)+ RNAs from OL/BDV (lanes 1, 3, 5, and 7) and OL cells (lanes 2, 4, 6, and 8) were hybridized with Rp1 (lanes 1, 2, 5, and 6) or Rp3 (lanes 3, 4, 7, and 8) antisense probe. Lanes 5 to 8 represent an extended exposure (4 days) of the lanes 1 to 4. Appropriate sizes of the fragments are shown.
In addition, Northern blot analysis was carried out to confirm the generation of SA3 spliced mRNAs in OL/BDV cell line. After short exposure of the hybridized membrane, two bands, 7.1 and 6.1 kb, were detected by a riboprobe, Rp1, as previously described (Fig. 2 A and C, lane 1) (25, 26). To demonstrate rare RNA species that hybridized with Rp1, an extended exposure of the membrane was performed. After 4 days of exposure, an additional band was detected at 1.6 kb in the infected cells (Fig. 2C, lane 5). A faint signal is also found at around 2.3 kb by the longer exposure of the membrane. These bands were not seen with the hybridization by using riboprobe, Rp3, which are located in the intron III sequences (Fig. 1C, lanes 3 and 7), indicating that these rare RNA species resulted from the splicing of intron III.
SA3 Is a Favorable 3′ Splice Site but Is Less Active in BDV-Infected Cells.
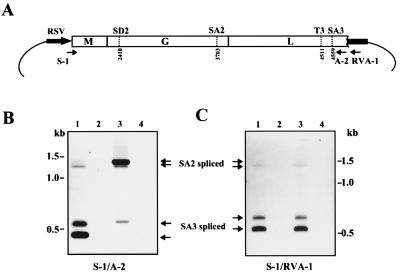
To characterize the splicing of intron III, called SA3 splicing, in more detail, a 2.7-kb BDV cDNA was cloned into an eukaryotic expression vector pRVSV (Fig. 3A). After transfection of COS-7 cells with pCD2.1, RNA was isolated at 48 h and reverse transcribed with oligo(dT) primer. BDV-specific cDNAs were then amplified with the S-1 and A-2 primer pair. COS-7 cells transfected with pCD2.1 produced both SA2 and SA3 spliced RNAs (Fig. 3B, lane 1), indicating that BDV intron III can be processed by the cellular RNA splicing machinery in the absence of BDV infection. Upper and lower bands in each spliced RNA correspond to RNA with and without intron I splicing, respectively. Furthermore, SA3 splicing was found as predominant product in the plasmid-transfected cells, whereas faint signals were observed for SA2 splicing (Fig. 3B, lane 1). This might have been because amplification of the smaller product was more efficient than that of the longer fragment. It is also possible that a weak detection of SA2 spliced RNAs in the transfected cells is caused by preferential recognition of SA3 in the alternative splicing of the viral RNAs. Despite this, however, the SA2 spliced RNAs were present as the major product in OL/BDV cells (Fig. 3B, lane 3; see also Fig. 1). This experiment revealed that although the SA3 site functions as a favorable splice signal within the genome, SA3 splicing is inefficient in persistently BDV-infected cells. To investigate whether this suppressive expression of SA3 splicing in the infected cells depends on the presence of viral proteins and/or transfected cell types, pCD2.1 was transfected into OL or OL/BDV cells, and RT-PCR was performed with a vector-specific primer, RVA-1. As shown in Fig. 3C, splicing of intron III in infected cells was as efficient as that observed in uninfected cells (lanes 1 and 3), suggesting that SA3 splicing is suppressed in a virus protein-independent manner in persistently BDV-infected cells.
Figure 3.
SA3 splicing is inefficient in persistently BDV-infected cells. (A) Schematic representation of BDV cDNA expression plasmid pCD2.1. Positions of splice sites and polyadenylation signal are indicated. Arrows show primers for RT-PCR. (B and C) Splicing analysis of cDNA expression plasmid. Arrows indicate SA2 or SA3 spliced products in the cells. Primer pairs used for analysis are indicated at the bottom. Lanes: (B) 1, pCD2.1-transfected COS-7 cells; 2, mock-transfected COS-7 cells; 3, OL/BDV cells; 4, OL cells. (C) 1, pCD2.1-transfected OL cells; 2, mock-transfected OL cells; 3, pCD2.1-transfected OL/BDV cells; 4, mock-transfected OL/BDV cells.
Detection of Negative Regulatory Activity of SA3 Splicing in the BDV Genome.
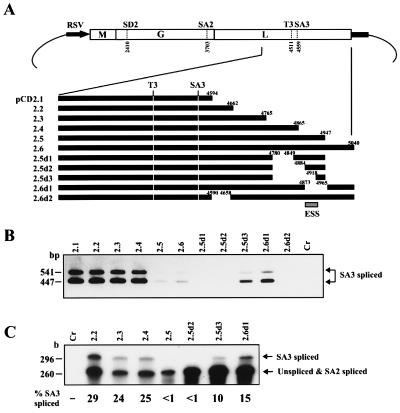
The results described above suggested the presence of elements with negative regulatory effects on SA3 splicing within the BDV genome. As previous studies have demonstrated that most of the viruses that use RNA splicing machinery contain cis-acting ESE or ESS regions for their regulated splicing (13–16), we next tried to identify the negative regulatory activity and to determine the approximate region corresponding to the activity within the BDV genome. We generated several plasmids (Fig. 4A) and used the cDNA expression system in COS-7 cells. Forty-eight hours after transfection, RNAs were isolated, and the efficiency of SA3 splicing was analyzed by RT-PCR and RPA. As shown in Fig. 4B, the cells transfected with pCD2.1, 2.2, 2.3, and 2.4 plasmids, which have 35-, 103-, 206-, and 306-nt flanking sequences in the 3′ portion from the SA3 site, respectively, showed efficient SA3 splicing. In contrast, the efficiency of SA3 splicing was markedly reduced in cells transfected with plasmids pCD2.5 and 2.6, which contain 388 and 481 nt flanking the 3′ portion of the SA3 site, respectively. In addition, two deletion forms of pCD2.5, pCD2.5d1, and 2.5d2, which lacked 68 (nt 4781–4848) and 103 nt (nt 4781–4883) also led to no detectable SA3 splicing signals. The suppression of SA3 spliced RNA was, however, released in cells transfected with pCD2.5d3 and 2.6d1, the plasmids lacking 137 (nt 4781–4917) and 91 nt (nt 4874–4964), respectively, although the levels of SA3 splicing did not reach that obtained in cells transfected with pCD2.1. We repeated this experiment at least six times and obtained similar results in each experiment. Furthermore, RPA also confirmed the efficiency of SA3 splicing in the cells transfected with these clones (Fig. 4C). Almost 30% SA3 splicing was found in the pCD2.2-transfected cells, whereas pCD2.5 showed less than 1% SA3 splicing. The pCD2.6d1, however, recovered the SA3 splicing to 15%. These observations demonstrated that the BDV genome indeed contains elements with negative regulatory activity on SA3 splicing and that the region at around 350 nt downstream of the SA3 site, at least nt 4873–4917, has an ESS activity of BDV (Fig. 4A).
Figure 4.
The BDV genome contains elements with negative regulatory effects on SA3 splicing. (A) A series of cDNA expression plasmid. Deletion sites are indicated by nt numbers. The putative ESS position is shown by a box. (B) RT-PCR analysis of deletion mutants. The mutants were transfected into COS-7 cells and SA3 splicing was detected by RT-PCR by using S-1/A-2 primer pairs. (C) RPA of deletion mutants. RNAs from the mutant-transfected cells were hybridized with the riboprobe shown in Fig. 2. Percent of SA3 splicing was indicated. Mutants used for the analysis are shown at the top of the lanes. Cr, mock-transfected COS-7 cells.
To elucidate the negative splicing activity of the BDV ESS region, we analyzed the sequences surrounding the region and compared the sequences with other known splicing suppressor elements. Interestingly, the BDV ESS contained two putative elements (Fig. 5A). Region 1 included three motifs, CUAGA (nt 4860), GGAUCC (nt 4868) and UUAG (nt 4891), similar to core sequences of the ESS element present in the first and second tat-coding exons of HIV-1 (ESS2 and ESS3; refs. 13, 14, and 35) within a small 35-nt sequence. Two of these motifs, CUAGA and UUAG, exactly matched the core motifs of the HIV-1 ESS, and the other, GGAUCC, was one nt different from the HIV-1 motif. Furthermore, we found a purine-rich region (region 2; nt 4896–4929), which was 70% A or G, immediately downstream of the UUAG motif (Fig. 5A). Previous studies revealed that purine-rich splicing elements are found in several cellular exons (9, 11, 36) and also demonstrated that an AG-rich sequence of BPV-1 ESS is required for strong suppression of BPV-1 late gene splicing (15, 16).
Figure 5.
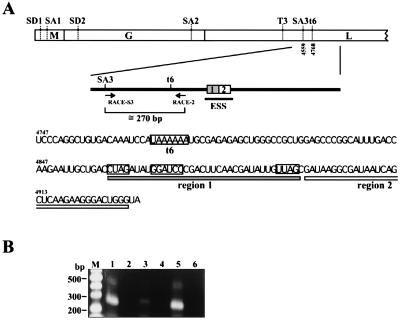
Detection of a functional polyadenylation signal upstream of the ESS region. (A) Sequences of the BDV ESS region. Schematic map of BDV ORFs is shown at the top. Splice and polyadenylation sites of BDV are shown by bars. Primers for 3′RACE analysis and putative elements of BDV ESS are also indicated. A polyadenylation signal (t6) and possible motifs for ESS are boxed. (B) 3′RACE analysis of BDV. RNAs from BDV-infected cells were subjected to 3′RACE analysis by using the primers shown in A. lanes: M, marker; 1, MDCK/BDV cells; 2, MDCK cells; 3, C6BV cells; 4, C6 cells; 5, OL/BDV cells; 6, OL cells.
A Possible Role of Polyadenylation in Alternative Splicing of BDV.
Sequence analysis revealed a more interesting finding regarding regulation of alternative splicing of BDV. Just upstream of the ESS region, we found a putative Pt signal (previously referred to as t6; ref. 22), the utilization of which has not yet been identified in BDV-infected cells (Fig. 5A). The sequence of the t6 exactly matched the Pt signal consensus sequence of BDV (22). Therefore, if there are BDV pre-mRNAs that are terminated at the t6 signal, the pre-mRNAs could efficiently splice the intron III sequence, because the ESS region could be separated from the pre-mRNAs by stuttering of RNA-dependent RNA polymerase before accomplishment of splicing. To test this hypothesis, we investigated utilization of the t6 signal during BDV infection. To detect poly(A)+ mRNAs that terminated at the t6 signal, we performed 3′RACE by using total RNAs from BDV-infected cells. An oligo(dT)-adapter primer was used as an RT primer, and specific fragments were amplified by nested PCR. By using the primer pair, mRNAs that are terminated at the t6 signal should yield fragments of around 270 bp. As shown in Fig. 5B, we detected amplified products of the expected size in all BDV-infected cell lines (lanes 1, 3, and 5), and sequence analysis of the products confirmed polyadenylation at the t6 site with 20–25 adenylate residues (data not shown). This result revealed that the t6 is a functional Pt signal within the BDV genome.
The SA3 Spliced RNAs Can Form ORFs.
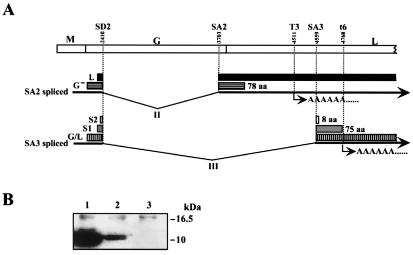
Sequence analysis of SA3 spliced RNAs revealed that the splicing could form predicted ORFs in all three frames. Frame 1 generated the largest ORF, which encodes the first 58 amino acids of G protein fused to L protein lacking one-quarter of the N terminus. Frames 2 and 3 generated ORFs encoding putative products of 75 (S1) and 8 (S2) amino acids, respectively (Fig. 6A). Interestingly, S1 ORF shared a start codon (nt 2393) with the L protein, which was formed by SA2 splicing (Fig. 6A; ref. 27). The AUG codon is flanked by a strong context for protein translational initiation (37), and the termination codon of the S1 ORF overlapped the t6 signal (Fig. 6A). To demonstrate whether the SA3 spliced RNA has a potential for production of polypeptides, we constructed an SA3 spliced cDNA expression plasmid, pS1-FLAG, which tags FLAG epitope in the C terminus of the predicted S1 ORF. After transfection of COS-7 cells, synthesis of the S1 protein was analyzed by Western blotting by using anti-FLAG antibody. As shown in Fig. 6B, the SA3 spliced cDNA clearly generated a protein (lane 2), with molecular weight that is almost the same as that of the X protein, indicating that the SA3 spliced RNA has a potential to translate a protein from the AUG signal of S1 ORF.
Figure 6.
The SA3 spliced RNA forms ORFs. (A) Schematic diagram of the new predicted BDV ORFs generated by SA3 splicing. Splice and polyadenylation sites in the BDV genome are indicated. ORFs and amino acid numbers of G-, S1 and S2 ORFs predicted by the BDV genome sequence are indicated. (B) Translation of predicted S1 protein in cDNA transfected COS-7 cells. Lanes: 1, pcORFx1-FLAG; 2, pS1-FLAG; 3, Mock. Appropriate sizes of molecular weight are shown.
Discussion
In this study, we found spliced RNAs of BDV that use an alternative 3′ splice site. This splice site, SA3, was indeed used in BDV-infected cell lines as well as infected rat brain cells, demonstrating alternative RNA splicing of BDV. Although the influenza virus is known as the only other RNA virus that uses alternative splicing for gene expression (38), the nonsegmented genome and alternative 3′ splice site of BDV indicated a manner and complexity of alternative splicing in this virus. Interestingly, cDNA expression analysis indicated that although the SA3 could be a favorable splice site in mammalian cells, utilization of SA3 is inefficient in infected cells. This analysis also revealed that viral proteins do not participate in the regulation of the SA3 splicing. It is still possible that the plasmid sequences at the 5′ and 3′ ends of the plasmid-derived RNA may contribute to some effects on the splicing (i.e., increasing the stability or splicing rate of the RNA) in the transfected cells. Our results demonstrated, however, that BDV contains elements that negatively regulate SA3 splicing activity within the genome. This suppression of SA3 splicing was controlled by a putative ESS, located at around 350 bp downstream of the SA3 site in an exon of BDV L protein. Recent studies on splicing suppressors or ESSs in eukaryotic pre-mRNAs have revealed that each ESS is located downstream of a suboptimal 3′ splice site in a regulated exon of its pre-mRNA and plays a negative role in splicing of the upstream intron (7, 8, 13–16, 39, 40). These ESSs are frequently located downstream of a juxtaposed ESE and form a bipartite splicing regulatory element in that exon. In BDV, however, we could not identify ESE activity in a juxtaposed upstream sequence of the ESS, because efficient SA3 splicing was detected in cDNA clones containing a series of deletions in upstream sequences of the ESS region (Fig. 4). This observation suggested that a juxtaposed ESE is not necessarily required for the ESS function. Therefore, it is likely that the BDV ESS directly inhibits the function of the SA3 site.
Sequence analysis of BDV ESS revealed that the region contained two putative elements that have motifs similar to the other ESS elements present in viral and cellular genes. Previous studies have shown that the core sequence necessary for HIV-1 tat ESS2 activity is CUAGA, and that the core sequences necessary for tat ESS3 activity are AGAUCC and UUAG (14, 35). Region 1 of BDV ESS contained all of the three motifs within a 35-nt sequence (Fig. 5A). A recent study revealed that these motifs in HIV-1 ESSs are important for binding to heterogeneous nuclear ribonucleoprotein A1 (hnRNP A1) that is widely expressed in mammalian cells and are required for the negative regulation of HIV-1 tat splicing (41). Furthermore, sequence comparison of several other potential ESS elements demonstrated that most contain a UAG motif, and a UAG motif is necessary for binding hnRNP A1 to regulate viral or cellular splicing or RNA transcription (41). Region 1 of BDV also contains two UAG motifs. Therefore, it is likely that the hnRNP A1 binds the cis-acting region of BDV and suppresses the SA3 splicing of its pre-mRNAs. In addition, a purine-rich sequence in the BDV ESS region 2 was similar to those observed in BPV-1 ESS and other cellular ESEs. Previous studies demonstrated that AG-rich elements present in cellular exons selectively bind to serine/arginine-rich (SR) proteins and enhance splicing efficiency of several cellular pre-mRNAs (9, 11, 36). In contrast, however, it has also been reported that the AG-rich sequence of BPV-1 ESS is important for suppression of splicing of the late mRNAs. A recent study suggested that the AG-rich region of BPV-1 could limit the amounts of SR proteins available for 5′ and 3′ splice sites interaction during pre-mRNA splicing and suppress efficient splicing of the mRNAs (16). It is also possible that the AG-rich element in BDV ESS binds to SR proteins and suppresses activated SA3 splicing.
Despite the sequence similarity between BDV ESS and other viral ESSs, the presence of a functional Pt signal, t6, upstream of the ESS revealed a mechanism for regulated alternative splicing of BDV. The pre-mRNAs that are terminated at the t6 signal could splice the intron III sequence efficiently, because the pre-mRNAs could not carry the ESS region in the sequences. On the other hand, SA3 splicing could be inefficient in the t6 readthrough pre-mRNAs. Strongly supporting this hypothesis, 3′RACE analysis clearly demonstrated that t6 is a functional signal during viral transcription. Although alternative Pt or readthrough mechanisms of RNA viruses have not yet been defined, alternative Pt of BDV pre-mRNAs could have an initial role in regulation of SA3 splicing, because the splicing reaction must involve 3′-end-processing of pre-mRNAs (2, 6).
The results presented here suggested that coupling of polyadenylation and splicing suppression are involved in alternative gene expression of the nonsegmented negative-strand RNA virus. Further studies with new techniques such as reverse-genetic systems are required to identify the detailed mechanism of RNA virus pre-mRNA splicing and its regulation in mammalian cells. Furthermore, the regulated alternative splicing of BDV suggested the functional importance of the proteins encoded by the SA3 spliced RNA in the life cycle of the virus. In a recent study, it has been demonstrated that AUG of the predicted S1 ORF is used to initiate translation of BDV L with SA2 splicing (27). This observation strongly supports the translational potential of the S1 ORF in infected cells that was demonstrated in the SA3 spliced cDNA-transfected cells. Further experiments are currently in progress to identify the proteins encoded by SA3 spliced RNAs in infected cells by using specific antibodies to the proteins produced from Escherichia coli and baculovirus systems.
Supplementary Material
Acknowledgments
This work was supported in part by the Special Coordination Funds for the Science and Technology from Science and Technology Agency (S.T.A.).
Abbreviations
- BDV
Borna disease virus
- RPA
RNase protection assay
- ESS
exon splicing suppressor
- ESE
exon splicing enhancer
- Pt
polyadenylation/termination
- MDCK
Madin–Darby canine kidney cells
- OL
oligodendroglioma cell
- RACE
rapid amplification of cDNA ends
- BPV-1
bovine papillomavirus type 1
- RT
reverse transcription
References
- 1.Bisaillon M, Lemay G. Virology. 1997;236:1–7. doi: 10.1006/viro.1997.8698. [DOI] [PubMed] [Google Scholar]
- 2.Sharp P A. Cell. 1994;77:805–815. doi: 10.1016/0092-8674(94)90130-9. [DOI] [PubMed] [Google Scholar]
- 3.Colgan D F, Manley J L. Genes Dev. 1997;11:2755–2766. doi: 10.1101/gad.11.21.2755. [DOI] [PubMed] [Google Scholar]
- 4.Gorlich D, Mattaj I W. Science. 1996;271:1513–1518. doi: 10.1126/science.271.5255.1513. [DOI] [PubMed] [Google Scholar]
- 5.Nakielny S, Dreyfuss G. Curr Opin Cell Biol. 1997;9:420–429. doi: 10.1016/s0955-0674(97)80016-6. [DOI] [PubMed] [Google Scholar]
- 6.Zhao J, Hyman L, Moore C. Microbiol Mol Biol Rev. 1999;63:405–445. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caputi M, Casari G, Guenzi S, Tagliabue R, Sidoli A, Melo C A, Baralle F E. Nucleic Acids Res. 1994;22:1018–1022. doi: 10.1093/nar/22.6.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Del Gatto F, Breathnach R. Mol Cell Biol. 1995;15:4825–4834. doi: 10.1128/mcb.15.9.4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lavigueur A, La Branche H, Kornblihtt A R, Chabot B. Genes Dev. 1993;7:2405–2417. doi: 10.1101/gad.7.12a.2405. [DOI] [PubMed] [Google Scholar]
- 10.Ramchatesingh J, Zahler A M, Neugebauer K M, Roth M B, Cooper T A. Mol Cell Biol. 1995;15:4898–4907. doi: 10.1128/mcb.15.9.4898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun Q, Mayeda A, Hampson R K, Krainer A R, Rottman F M. Genes Dev. 1993;7:2598–2608. doi: 10.1101/gad.7.12b.2598. [DOI] [PubMed] [Google Scholar]
- 12.Tanaka K, Watakabe A, Shimura Y. Mol Cell Biol. 1994;14:1347–1354. doi: 10.1128/mcb.14.2.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Amendt B A, Si Z H, Stoltzfus C M. Mol Cell Biol. 1995;15:4606–4615. doi: 10.1128/mcb.15.8.4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Si Z, Amendt B A, Stoltzfus C M. Nucleic Acids Res. 1997;25:861–867. doi: 10.1093/nar/25.4.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng Z M, He P, Baker C C. J Virol. 1996;70:4691–4699. doi: 10.1128/jvi.70.7.4691-4699.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng Z M, He P J, Baker C C. J Virol. 1999;73:29–36. doi: 10.1128/jvi.73.1.29-36.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pomerantz R J, Trono D, Feinberg M B, Baltimore D. Cell. 1990;61:1271–1276. doi: 10.1016/0092-8674(90)90691-7. [DOI] [PubMed] [Google Scholar]
- 18.Pomerantz R J, Seshamma T, Trono D. J Virol. 1992;66:1809–1813. doi: 10.1128/jvi.66.3.1809-1813.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alonso-Caplen F V, Krug R M. Mol Cell Biol. 1991;11:1092–1098. doi: 10.1128/mcb.11.2.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barksdale S K, Baker C C. J Virol. 1993;67:5605–5616. doi: 10.1128/jvi.67.9.5605-5616.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barksdale S, Baker C C. J Virol. 1995;69:6553–6556. doi: 10.1128/jvi.69.10.6553-6556.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Briese T, Schneemann A, Lewis A J, Park Y S, Kim S, Ludwig H, Lipkin W I. Proc Natl Acad Sci USA. 1994;91:4362–4366. doi: 10.1073/pnas.91.10.4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cubitt B, Oldstone C, de la Torre J C. J Virol. 1994;68:1382–1396. doi: 10.1128/jvi.68.3.1382-1396.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Briese T, de la Torre J C, Lewis A, Ludwig H, Lipkin W I. Proc Natl Acad Sci USA. 1992;89:11486–11489. doi: 10.1073/pnas.89.23.11486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cubitt B, Oldstone C, Valcarcel J, Carlos de la Torre J. Virus Res. 1994;34:69–79. doi: 10.1016/0168-1702(94)90120-1. [DOI] [PubMed] [Google Scholar]
- 26.Schneider P A, Schneemann A, Lipkin W I. J Virol. 1994;68:5007–5012. doi: 10.1128/jvi.68.8.5007-5012.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walker M P, Jordan I, Briese T, Fischer N, Lipkin W I. J Virol. 2000;74:4425–4428. doi: 10.1128/jvi.74.9.4425-4428.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thierer J, Riehle H, Grebenstein O, Binz T, Herzog S, Thiedemann N, Stitz L, Rott R, Lottspeich F, Niemann H. J Gen Virol. 1992;73:413–416. doi: 10.1099/0022-1317-73-2-413. [DOI] [PubMed] [Google Scholar]
- 29.Carbone K M, Rubin S A, Sierra-Honigmann A M, Lederman H M. J Virol. 1993;67:1453–1460. doi: 10.1128/jvi.67.3.1453-1460.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakamura Y, Takahashi H, Shoya Y, Nakaya T, Watanabe M, Tomonaga K, Iwahashi K, Ameno K, Momiyama N, et al. J Virol. 2000;74:4601–4611. doi: 10.1128/jvi.74.10.4601-4611.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kebelmann-Betzing C, Seeger K, Dragon S, Schmitt G, Moricke A, Schild T A, Henze G, Beyermann B. BioTechniques. 1998;24:154–158. doi: 10.2144/98241pf01. [DOI] [PubMed] [Google Scholar]
- 32.Sakai H, Siomi H, Shida H, Shibata R, Kiyomasu T, Adachi A. J Virol. 1990;64:5833–5839. doi: 10.1128/jvi.64.12.5833-5839.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malik T H, Kobayashi T, Ghosh M, Kishi M, Lai P K. Virology. 1999;258:65–72. doi: 10.1006/viro.1999.9715. [DOI] [PubMed] [Google Scholar]
- 34.Green M R. Annu Rev Genet. 1986;20:671–708. doi: 10.1146/annurev.ge.20.120186.003323. [DOI] [PubMed] [Google Scholar]
- 35.Si Z H, Rauch D, Stoltzfus C M. Mol Cell Biol. 1998;18:5404–5413. doi: 10.1128/mcb.18.9.5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Staknis D, Reed R. Mol Cell Biol. 1994;14:7670–7682. doi: 10.1128/mcb.14.11.7670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kozak M. J Cell Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lamb R A, Lai C J, Choppin P W. Proc Natl Acad Sci USA. 1981;78:4170–4174. doi: 10.1073/pnas.78.7.4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Staffa A, Acheson N H, Cochrane A. J Biol Chem. 1997;272:33394–33401. doi: 10.1074/jbc.272.52.33394. [DOI] [PubMed] [Google Scholar]
- 40.Wentz M P, Moore B E, Cloyd M W, Berget S M, Donehower L A. J Virol. 1997;71:8542–8551. doi: 10.1128/jvi.71.11.8542-8551.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Caputi M, Mayeda A, Krainer A R, Zahler A M. EMBO J. 1999;18:4060–4067. doi: 10.1093/emboj/18.14.4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.