Abstract
Oxidation of protein thiolates is central to numerous redox-regulated processes. Bacillus subtilis OhrR is an organic peroxide sensor that represses expression of an inducible peroxiredoxin, OhrA. Here, we present evidence that oxidation of the sole cysteine residue in OhrR leads to a sulfenic acid-containing intermediate that retains DNA-binding activity: further reaction to generate either a mixed disulfide (S-thiolation) or a protein sulfenamide (sulfenyl-amide) derivative is essential for derepression. Protein S-thiolation protects OhrR from overoxidation and provides for a facile regeneration of active OhrR by thiol–disulfide exchange reactions. The sulfenamide can also be reduced by thiol–disulfide exchange reactions, although this process is much slower than for mixed disulfides. Recovery of oxidized OhrR from B. subtilis identifies three distinct S-thiolated species, including mixed disulfides with a novel 398-Da thiol, cysteine, and CoASH. Evidence for in vivo formation of the sulfenamide derivative is also presented.
Keywords: protein oxidation, redox regulation, S-thiolation, sulfenamide
Reactive oxygen species (ROS), such as H2O2, superoxide anion (O2•−), hydroxyl radical (•OH), and organic hydroperoxides (ROOH), can damage DNA, proteins, and membranes (1). ROS are a by-product of aerobic metabolism and are also generated by both animals and plants as part of their innate immune defenses (2, 3). In response, bacteria use transcription factors that sense ROS and regulate genes involved in their detoxification (4–6). Frequently, the sensing of ROS is mediated by oxidation of one or more protein thiolates.
One of the best-characterized bacterial sensors of ROS is Escherichia coli OxyR, which activates the expression of catalase, alkylhydroperoxide reductase (AhpCF), and thiol-disulfide homeostasis proteins in response to peroxide stress (6). OxyR contains a highly reactive Cys residue that forms an intramolecular disulfide bond in response to oxidative stress or depletion of reduced intracellular thiols (7, 8). OxyR can also be activated by other modifications of the active site thiol, including S-nitrosation, S-glutathionylation, and sulfenic acid formation (9), although the physiological relevance of these modifications remains controversial (6, 10).
In Bacillus subtilis, at least three transcription factors contribute to oxidative stress responses: σB, PerR, and OhrR (11). Oxidative stress induces the catalase KatB under control of σB general stress response (12). PerR, a ferric uptake regulator (Fur) family protein, regulates the expression of the major vegetative catalase (KatA), AhpCF, a Dps family protein (MrgA), and other peroxide stress proteins (4). OhrR represses ohrA, encoding a thiol-dependent peroxidase that detoxifies organic hydroperoxides by reduction to the corresponding alcohol (13). A paralog, ohrB, is under σB control (14).
OhrR is a member of the multiple antibiotic resistance (MarR/SlyA) family of winged-helix DNA-binding proteins (15). OhrR binds to an inverted repeat element overlapping the ohrA promoter and is inactivated by organic hydroperoxides (16). Oxidation of the sole cysteine (Cys15) of OhrR by cumene hydroperoxide (CHP) leads to the transient accumulation of a sulfenic acid (SOH) intermediate, followed by further protein oxidation: no intersubunit disulfide bond formation is observed (16). Oxidized OhrR is inactive for DNA binding, but activity is restored by treatment with the strong thiol reductant DTT. Because sulfenic acids, but not the higher oxidation states of sulfur, are reduced by DTT we suggested that sulfenic acid formation was sufficient for derepression (16). Because sulfenic acids are generally unstable (10), we speculated that in vivo the initially formed sulfenic acid might react with low molecular weight (LMW) intracellular thiols to form mixed disulfides (16).
Here, we have analyzed the regulatory thiolate switch in OhrR by reconstituting the relevant oxidation reactions in vitro and monitoring the fate of oxidized OhrR in cells. We demonstrate that, contrary to our starting hypothesis, the initially formed OhrR sulfenate is still active for DNA binding. In the absence of available thiols, this sulfenate condenses with a backbone amide to generate the protein sulfenamide and thereby triggers protein inactivation (and accounts for the previously detected DTT-reversible inactive species). In the presence of LMW thiols, the sulfenate rapidly generates a mixed disulfide. In cells, at least three distinct OhrR S-thiolated species are detected. S-thiolation and sulfenamide formation dissociate OhrR from operator DNA, protect against overoxidation, and provide pathways for regeneration of active repressor by thiol–disulfide exchange reactions.
Results
S-Thiolation Increases the Rate of OhrR Dissociation from Operator DNA.
Because B. subtilis contains Cys and reduced CoA (CoASH) as abundant LMW thiols (17, 18), we previously speculated that the initially formed Cys15-SOH of OhrR might react with these or other cellular thiols, resulting in S-thiolated OhrR (16). Indeed, when challenged with CHP in the presence of Cys, purified OhrR was quantitatively S-cysteinylated as monitored by MS [supporting information (SI) Figs. 6 and 7].
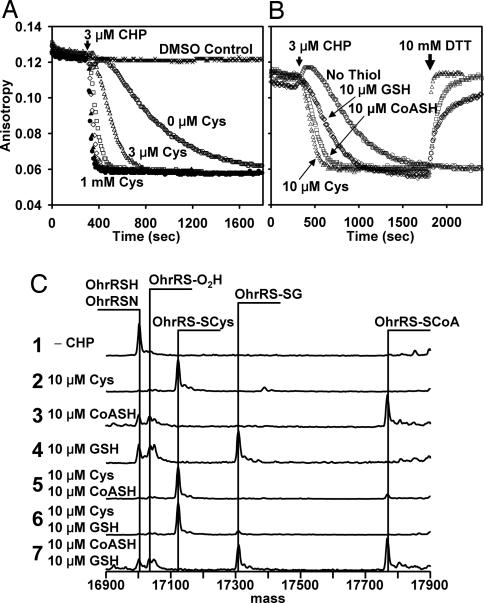
To investigate the effects of S-cysteinylation on the activity of OhrR, we monitored DNA binding in real time by using a fluorescence anisotropy (FA)-based DNA-binding assay. The DNA-binding activity of reduced OhrR was quite stable, even in air-saturated buffers (Fig. 1A, DMSO control). Addition of CHP in the absence of Cys led to the gradual loss of DNA-binding activity as a function of time. Significantly, the half-time for OhrR inactivation by CHP decreased from ≈9 min in the absence of Cys to <20 s with 1 mM Cys (Fig. 1A). Even 3 μM Cys led to a significant rate enhancement, and half-maximal dissociation rates were achieved with between 30 and 100 μM Cys (Fig. 1A). The dependence of protein inactivation on Cys concentration suggests that S-cysteinylation of OhrR (rather than sulfenic acid formation or protein dissociation) is rate-limiting in reactions with <100 μM Cys. Intracellular levels of Cys in B. subtilis are typically between 0.15 and 0.5 mM (17, 19), sufficient to mediate efficient S-cysteinylation.
Fig. 1.
Kinetic analysis of OhrR inactivation. (A) DNA-binding activity of purified OhrR (300 nM) was monitored by FA with labeled DNA (50 nM). Cysteine accelerates the CHP-dependent dissociation of OhrR from operator DNA. Samples were treated at the indicated time with a 10-fold molar excess (3 μM) of CHP in the presence of 0, 3 μM, 10 μM, 30 μM, 100 μM, or 1 mM Cys (in order of increasing dissociation rate). (B) OhrR was oxidized by CHP in either the presence or absence of the indicated LMW thiols. At the time indicated, 10 mM DTT was added to the thiol-containing reactions and the regain of DNA-binding activity was monitored. (C) In vitro oxidation products of OhrR were analyzed by ESI-MS. Purified OhrR (300 nM; trace 1) was treated with 3 μM CHP for 30 min in the presence of LMW thiols as indicated. Protein mass changes were monitored by ESI-MS.
Rates of protein S-thiolation can be strongly influenced by steric constraints and the chemical environment of the reactive Cys. To monitor the rate of OhrR S-thiolation by various LMW thiols, we exposed OhrR to CHP in the presence of low levels (10 μM) of Cys, CoASH, or glutathione (GSH) and monitored protein activity (Fig. 1B). Cys was most efficient at triggering rapid protein dissociation, whereas CoASH and GSH were somewhat less effective. As expected, addition of 10 mM DTT rapidly restored DNA-binding activity to S-thiolated OhrR (Fig. 1B). In contrast, OhrR oxidized in the absence of thiol exhibited a very slow reactivation rate (see below).
To monitor protein S-thiolation directly, we used electrospray ionization (ESI)-MS (Fig. 1C). Quantitative S-thiolation was achieved with 10 μM Cys, but not CoASH or GSH (quantitative S-thiolation was observed with 100 μM CoASH or 100 μM GSH; data not shown). In competition studies, S-thiolation with Cys was more efficient than that with either CoASH or GSH. Both Cys and CoASH are abundant LMW thiols in B. subtilis, but this organism lacks GSH (17, 19). Despite the relatively slow rate of S-glutathionylation of OhrR, when OhrR-expressing E. coli cells (which contain ≈5 mM GSH) were exposed to CHP the S-glutathionylated species could be easily detected by MS (SI Fig. 8).
Free Cys Mediates the Spontaneous Reactivation of OhrR.
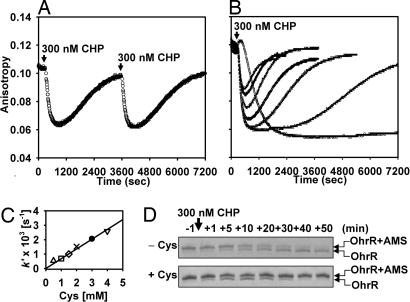
The preceding kinetic analyses were conducted by using a 10-fold molar excess of CHP over OhrR monomer. When we instead added one molar equivalent of CHP, we observed a spontaneous reactivation of OhrR in Cys-containing buffers (Fig. 2A). Under these conditions, the DNA-binding activity of OhrR is rapidly lost (kinact by CHP ≈2 × 104 M−1·s−1) followed by a slow recovery. To verify that the increasing anisotropy value corresponds to OhrR reactivation, CHP was again added and the inactivation/reactivation cycle repeated with virtually identical kinetics. As expected for a simple thiol–disulfide exchange reaction, the rate of OhrR reactivation linearly depended on Cys concentration (kreact = 0.68 M−1·s−1; Fig. 2 B and C).
Fig. 2.
Kinetics of OhrR oxidation and re-reduction in the presence of Cys. (A) OhrR (300 nM) and DNA were incubated in buffer with 1 mM Cys, and then 1 molar equivalent CHP was added at 5 min and 1 h. (arrows). (B) OhrR oxidation and re-reduction rates were measured as a function of Cys concentration. In order of increasing reactivation rate (and decreasing extent of net inactivation), the concentration of Cys was 0, 0.5, 1, 1.5, 2, 3, and 4 mM. (C) Pseudofirst-order rate constants (k′) for reactivation of OhrR were plotted against Cys concentration, where k′ = kreact[Cys]. (D) The AMS reactivity of Cys15 was monitored by SDS/PAGE in the absence or presence of 2 mM Cys. In the absence of Cys, OhrR is oxidized to an AMS-insensitive species, whereas in the presence of Cys this species appears transiently. See SI Fig. 9 for an image of the entire gel.
High concentrations of Cys both increased the rate of OhrR reactivation and impeded the complete inactivation by added oxidant. The latter effect is complex, but likely reflects both an increased reactivation rate and competition between free Cys and the OhrR Cys15 thiolate for limited CHP. In the presence of ≈104-fold molar excess of Cys (3 mM), the maximal extent of OhrR inactivation is reduced by half, suggesting an ≈104-fold greater reactivity of CHP with OhrR (k ≈2 × 104 M−1·s−1) than with free Cys (estimated k ≈2 M−1·s−1). This estimated rate of free Cys oxidation by CHP is comparable to that for H2O2 (≈10 M−1·s−1) (10). In contrast, 1 mM Cys completely inhibited the inactivation of OhrR by a 100-fold molar excess of H2O2 (30 μM) (data not shown), likely by serving as an alternate target for oxidation.
The oxidation state of Cys15 can be visualized by monitoring susceptibility to 4-acetamido-4′-maleimidylstilbene-2,2′-disulfonic acid (AMS) modification. In the presence of CHP and Cys there is a transient appearance of an alkylation-resistant form of OhrR (the S-cysteinylated protein) (Fig. 2D). These studies indicate that OhrR is functioning as a thiol-dependent peroxidase, albeit a very inefficient one. The catalytic cycle is limited by the slow rate of reactivation by Cys with 1 mM Cys (t1/2 ≈17 min; Fig. 2A). In contrast, thiol-dependent peroxidases of the Ohr family have catalytic turnover rates of >10 s−1 (20, 21).
The Rate of OhrR Oxidation Is Much Greater Than the Rate of DNA Dissociation in the Absence of Cysteine.
The preceding kinetic analyses indicate that OhrR rapidly dissociates from DNA when the reactive Cys15 thiolate (pKa ≈5.2; ref. 22) is S-cysteinylated. Because S-cysteinylation results from reaction of an initially formed Cys15 sulfenic acid with free Cys, oxidation to the sulfenate must itself be rapid. Yet, the functional inactivation of OhrR by CHP is slow in the absence of LMW thiols (Fig. 1A).
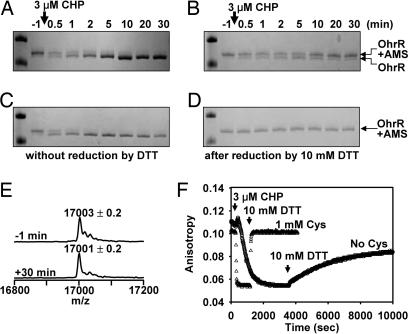
To determine the oxidation rate of Cys15 of OhrR directly, we monitored AMS reactivity after CHP treatment. The oxidation of Cys15 (t1/2 ≈30 s; Fig. 3A) was much faster than the rate of DNA dissociation (t1/2 ≈9 min; Fig. 1A). Importantly, the rate of thiol oxidation was similar in both the presence and absence of Cys (Fig. 3 A vs. C), despite the fact that the rate of DNA dissociation differed by >20-fold (Fig. 1A). A similar result was seen when AMS reactivity was monitored in reactions with 300 nM CHP (Fig. 2 D vs. B), although in this case the oxidation rate was slower (because of the lower concentration of CHP and the fact that CHP is no longer in excess relative to OhrR). Note that the oxidation rate of Cys15 to an AMS-resistant state (estimated kox ≈104 M−1·s−1) is comparable to the rate of functional inactivation of OhrR (Figs. 1A and 2A), consistent with the hypothesis that oxidation by CHP (rather than S-thiolation) is rate-limiting in the presence of 1 mM Cys.
Fig. 3.
Kinetics of OhrR oxidation and re-reduction in the absence of Cys. (A–D) AMS reactivity of OhrR after CHP treatment was monitored as a function of time. (A) OhrR oxidized in the absence of Cys. The accumulation of the faster migrating band as a function of time indicates a loss of AMS reactivity. (B) Samples shown in A were treated with DTT for 60 min before AMS alkylation. Oxidized OhrR was largely restored to an AMS-reactive form, consistent with the OhrR sulfenamide as the major product. (C and D) Shown are the parallel reactions in the presence of 1 mM Cys. Only the region of the gel corresponding to the OhrR-AMS and unalkylated OhrR bands is shown (see SI Fig. 9 for an image of the entire gel). (E) The molecular mass of OhrR was monitored by ESI-MS before and 30 min after CHP (3 μM) treatment (see SI Fig. 10 for the time series data). (F) Comparison of OhrR inactivation (by CHP) and reactivation (by DTT) in reactions with or without Cys.
Oxidization of OhrR in the Absence of LMW Thiols Leads to a Protein Sulfenamide.
To identify the functionally inactivated product of OhrR in the absence of LMW thiols, we recovered protein by trichloroacetic acid (TCA) precipitation after CHP addition (from reactions identical to those shown in Fig. 1) and monitored mass changes by ESI-MS (Fig. 3E and SI Fig. 10). We anticipated that Cys15-SH of OhrR would be oxidized to the sulfenic and possibly to the sulfinic and sulfonic acids as observed (16). However, the major peak exhibited a mass (17,001 Da) similar to that of reduced OhrR (17,003 Da) with only a minor peak corresponding to the sulfinic acid derivative (Fig. 3E). This 2-Da loss of mass was localized to Cys15 by ESI-MS-MS of the corresponding tryptic peptide (SI Fig. 11). Together with the observation that this species is resistant to AMS modification (Fig. 3A), these results suggest formation of a cyclic sulfenamide, a protein modification first documented in protein tyrosine phosphatase 1B (23, 24). Thus, oxidative inactivation of OhrR (in the absence of LMW thiols) likely proceeds via the rapid formation of a sulfenic acid followed by a slow cyclization in which Cys15-SOH reacts with the neighboring backbone amide (from Phe16). This condensation is essentially complete within 5–10 min (see SI Fig. 10).
When 10 mM DTT was added to reactions containing S-thiolated OhrR, there was a rapid reactivation of DNA-binding activity (Figs. 1B and 3F). In contrast, DTT treatment of OhrR oxidized in the absence of Cys led to a slow reactivation of ≈2/3 of the DNA-binding activity (Fig. 3F). Because OhrR oxidized in the absence of Cys is found primarily in the sulfenamide form (with a small amount of sulfinic acid; Fig. 3E), and sulfinic acids cannot be reduced by DTT, this result indicates that DTT can reduce the protein sulfenamide as also shown for protein tyrosine phosphatase 1B (23). Similarly, the reactivation of OhrR oxidized in the presence of 10 μM GSH exhibited complex kinetics: there was a rapid reactivation of approximately half of the DNA-binding activity (presumably corresponding to the S-glutathionylated fraction; Fig. 1C) and a slow phase that likely reflects the much slower reduction of the sulfenamide (Fig. 1B).
We can also distinguish between reversible and irreversible protein oxidation by using AMS alkylation. In reactions containing 1 mM Cys, the protein was quantitatively S-cysteinylated (Fig. 3C and SI Fig. 6) and DTT fully restored the AMS reactivity of Cys15 (Fig. 3D). In reactions lacking free Cys, there was a complete loss of AMS reactivity after 30 min of treatment (Fig. 3A). Treatment with DTT restored most, but not all, of the AMS reactivity (Fig. 3B). This finding is consistent with the ability of DTT to regenerate reduced Cys15 from the sulfenamide, but not from the sulfinic acid, derivative.
The Transiently Formed OhrR Sulfenate Is Active for DNA Binding.
The disparity between the rapid rate of OhrR oxidation (Fig. 3A) and the slow loss of DNA-binding activity (in the absence of thiols; Fig. 1A) is inconsistent with our previous hypothesis that sulfenate formation is sufficient for derepression (16). Instead, the kinetic analyses above suggest that the initially formed sulfenate retains DNA-binding activity and that protein dissociation is triggered by either S-thiolation or sulfenamide formation.
The transient formation of a Cys15 sulfenate was previously documented by chemical modification with 7-chloro-4-nitrobenzo-2-oxa-1,3-diazole (NBD-Cl): 30 s after CHP addition, the sulfenate ester of NBD was the major reaction product (with decreasing levels at later time points) (16). This sulfenate intermediate is also detected by the rapid loss of AMS reactivity upon CHP addition (Fig. 3A). Consistent with the presence of a protein sulfenate, TCA precipitation of partially oxidized samples (containing both the SH and SOH forms of OhrR) leads to the appearance of disulfide-linked dimers (see SI Fig. 9 for discussion). In the presence of LMW thiols, the sulfenate intermediate is rapidly trapped as the mixed disulfide and the protein inactivated (Fig. 1). In contrast, in the absence of thiols there is a prominent shoulder on the DNA dissociation curve (Fig. 1 A and B; reactions with no thiol added) corresponding to a 2- to 3-min period after CHP addition during which OhrR retains activity. In these reactions, the loss of DNA-binding activity is correlated with the accumulation of the cyclic sulfenamide (SI Fig. 10) rather than the loss of AMS reactivity (Fig. 3A). Together, these results suggest that the OhrR sulfenate (Cys15-SOH) retains DNA-binding activity and that further processing is necessary for protein inactivation.
Oxidation of OhrR in Vivo Leads to Both S-Thiolated and Sulfenamide Species.
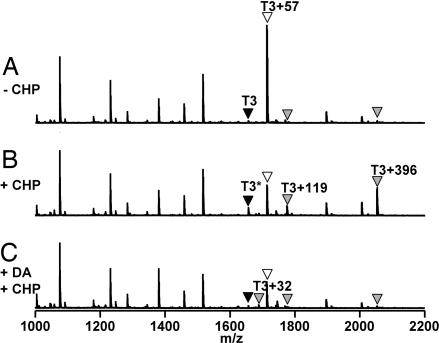
For in vivo analyses, we constructed a B. subtilis strain expressing a fully functional, epitope-tagged OhrR–FLAG protein. We purified OhrR–FLAG from B. subtilis cells before and after treatment with CHP. Iodoacetamide (IA) was used to alkylate Cys15 in reduced OhrR before analysis by trypsin digestion and MALDI-TOF MS. In the untreated sample, OhrR was fully reduced and Cys15 reacted with IA to generate the carboxyamidomethyl derivative (Cys15 CAM) (Fig. 4A). However, after ≈2 min of CHP treatment the amount of OhrR accessible to IA modification was greatly decreased and several other peaks with increased intensity were identified (Fig. 4B).
Fig. 4.
In vivo oxidation products of OhrR. OhrR-FLAG was recovered from B. subtilis and alkylated with IA, and the oxidation state of Cys15 was monitored by MALDI-TOF MS analysis of tryptic digests. (A) In untreated cells, the T3 tryptic peptide is fully alkylated by IA to generate the CysCAM derivative (T3+57 Da; white triangle). (B) In cells treated with 100 μM CHP for ≈2 min, oxidation leads to the appearance of an S-cysteinylated peak (T3+119 Da), an S-thiolated peak (T3+396 Da), and an alkylation-resistant peak (T3∗) with mass similar to fully reduced (CysSH) T3 (black triangle). (C) In cells treated with diamide for 1 min followed by 100 μM CHP, the yield of S-thiolated products is greatly decreased and the Cys15-SO2H derivative (T3+32) is now detected. Note that the intensities of other OhrR tryptic peptides (the peaks between 1,000 and 1,600 m/z) are not affected by oxidation.
Unexpectedly, oxidation of OhrR led to three different S-thiolated species (corresponding to the mixed disulfides with Cys, CoASH, and a novel 398-Da LMW thiol). The highest intensity oxidation-dependent peak (m/z = 2,052.9 Da), as detected in several replicate MS experiments and using different ionization modes, corresponded to the Cys15-containing tryptic peptide (T3; Leu10–Arg23) with an additional 396 Da. Because disulfide bond formation leads to a loss of 2-Da mass, this finding is consistent with S-thiolation with a 398-Da thiol. Further fragmentation by MS/MS confirmed that the T3+396 peptide was modified on Cys15 (SI Fig. 12). MS experiments also revealed the formation of S-cysteinylated OhrR (m/z = 1,775.8 Da = T3 + 119 Da). We did not detect an adduct with CoASH in this experiment (Fig. 4B), likely because CoA-modified peptides ionize poorly in the positive ion mode (25). In negative ion mode, a small peak corresponding to the CoA adduct of the T3 peptide (m/z = 2,419.9 Da) was detected (SI Fig. 13). As expected, these S-thiolated peaks were not detected in samples treated with DTT before analysis (SI Fig. 13 and data not shown) nor in cells treated with diamide, a thiol-oxidizing agent that depletes intracellular thiols, before CHP addition (Fig. 4C).
In addition to S-thiolation, a second consequence of protein oxidation was the retention of a peak (T3∗) apparently identical in mass to the unmodifed, fully reduced T3 peptide (Fig. 4B). Because OhrR was quantitatively alkylated by IA in extracts from cells that had not been exposed to CHP, the T3∗ peptide is evidence for a form of OhrR in which Cys15 is in an alkylation-resistant state. The most likely possibility, based on our in vitro studies, is a protein sulfenamide. Although this peptide did not show a loss of 2 Da, as expected for sulfenamide formation, results with authentic OhrR sulfenamide (prepared in vitro; Fig. 3E) indicate that this mass change is not detected by MALDI-TOF analyses of tryptic digests, even when it is present as judged by ESI-MS. Note that a stable sulfenic acid form of OhrR (Cys15-SOH) was not detected under these conditions, although this is the likely precursor of both the mixed disulfide and sulfenamide products (10). Moreover, the sulfinic acid derivative is detected only in cells depleted of LMW thiols by diamide treatment and then challenged with CHP (Fig. 4C), or when significantly higher amounts of CHP (1 mM) are used (data not shown). Thus, we conclude that the major fate of the initially formed sulfenate, both in vivo and in vitro, is further processing to either the S-thiolated or sulfenamide products (Fig. 5).
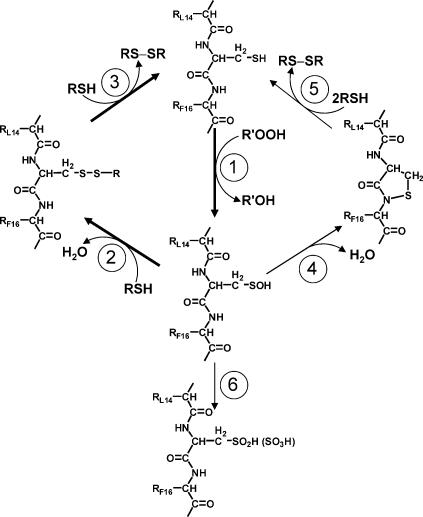
Fig. 5.
Mechanism of sensing organic hydroperoxides by OhrR. OhrR is normally present in the cell in the fully reduced state and (①) reacts rapidly with organic hydroperoxides (R′-OOH) to generate a sulfenic acid intermediate. The OhrR sulfenate is still active for DNA-binding (see text), but can be rapidly S-thiolated (②), which inactivates OhrR and allows derepression of its target gene (encoding the OhrA peroxiredoxin). Re-reduction, by either spontaneous or enzyme-catalyzed thiol-disulfide exchange reactions regenerates the active repressor (③). The sulfenate can also spontaneously react to generate the sulfenamide (④), which accounts for the slow loss of DNA-binding activity observed in vitro in the absence of thiols. Sulfenamides can be reduced by thiol-disulfide exchange reactions (⑤) as described (23, 24). When intracellular thiols are depleted, overoxidation of Cys15 to the sulfinic and sulfonic acid derivatives can occur (⑥).
Discussion
Reactive protein thiolates play a central role in the redox regulation of numerous proteins. Recent work has highlighted the versatility of thiol-based redox switches (6, 26). In addition to their well characterized roles in the reversible formation of protein disulfides, reactive thiolates can also participate in other pathways including (i) mixed-disulfide bond formation with LMW thiols, (ii) overoxidation to sulfinic and sulfonic acids, and (iii) condensation to the protein sulfenamide.
Oxidation of reactive protein thiolates to the sulfenic acid (Fig. 5, reaction 1) is often followed by condensation with another protein thiol to generate an intramolecular or intermolecular protein disulfide (Fig. 5, reaction 2). Well characterized examples include oxidation of the E. coli OxyR and yeast Yap1 transcription factors (6, 26). A similar mechanism is used by Xanthomonas campestris OhrR, which forms an intersubunit disulfide bond between Cys22 (equivalent to B. subtilis OhrR Cys15) and Cys127 (27). Indeed, most OhrR orthologs contain two Cys residues that may participate in their regulatory cycle. In contrast, OhrR proteins in various Bacillus spp., Oceanobacillus iheyensis, Streptomyces coelicolor, Sinorhizobium meliloti, and Rhodopseudomonas palustris all contain a single Cys residue and presumably function by pathways analogous to those documented here for the B. subtilis protein.
For those proteins lacking a suitably positioned protein thiol, the initially formed Cys sulfenate may instead form a mixed disulfide with a LMW thiol. Protein S-glutathionylation is a frequently observed modification that regulates the activity of numerous proteins (28). S-cysteinylation is much less common and has not previously been documented in bacterial cells, likely because most bacteria do not contain Cys as the major intracellular thiol (17, 29). However, protein S-cysteinylation has been reported for extracellular proteins in humans such as serum albumin and transthyretin (30). Disulfide bond formation between proteins and CoASH is not frequently observed, although it clearly occurs during the catalytic cycle of CoA disulfide reductase (31). CoASH is an abundant LMW thiol in Staphylococcus aureus and other Gram-positive bacteria (17, 32), Borrelia burgdorferi (33), and several thermophilic Archaea (34). Thus, it is likely that other instances of protein S-thiolation by CoASH will be discovered. Indeed, 45% of CoASH in Bacillus megaterium spores is found as mixed disulfides with protein (35).
In the absence of an available thiol, the protein sulfenic acid can be further oxidized to the sulfinic (SO2H) and sulfonic (SO3H) acids (Fig. 5, reaction 6). Although generally considered as irreversible oxidation states, eukaryotes contain enzymes (e.g., sestrin and sulfiredoxin) that can reduce the sulfinic acid in 2-Cys peroxiredoxins to the thiol (36–38). Thus, in this specific case, the sulfinic acid can function as a reversible redox switch. Alternatively, the Cys sulfenate can condense with the adjacent backbone amide to generate a cyclic protein sulfenamide (also known as a sulfenyl-amide; see Fig. 5, reaction 4). This reaction product was only recently detected in the x-ray crystal structures of oxidized protein tyrosine phosphatase 1B (23, 24). The extent to which sulfenamides might occur in oxidized proteins, and the pathways and rates of formation and rereduction, are largely unexplored.
Here, we have documented a critical role of S-thiolation in regulation of B. subtilis OhrR. The formation of mixed disulfides between OhrR and free thiols greatly increases the rate of protein dissociation from operator DNA, prevents irreversible protein overoxidation to the sulfinic or sulfonic acids, and provides a facile pathway for the regeneration of active OhrR by thiol–disulfide exchange reactions (Fig. 5). This mechanism relies on the ability of the peroxidatic cysteine of OhrR (Cys15) to react rapidly with organic peroxides to form a sulfenic acid derivative (Fig. 5, reaction 1). In the presence of Cys, OhrR inactivation by CHP (kinact ≈2 × 104 M−1·s−1) is nearly as sensitive as the H2O2-dependent oxidation of B. subtilis PerR (kinact ≈105 M−1·s−1) (39) and E. coli OxyR (kox ≈2 × 105 M−1·s−1) (8). However, this should be considered a lower limit because CHP is not likely to be the physiologically relevant oxidant and protein oxidation (rather than S-thiolation) is expected to be rate-limiting in the cell (estimated >0.15 mM Cys). Indeed, X. campestris OhrR has been found to be highly sensitive to linoleic acid hydroperoxide, which may be prevalent in the soil environment (40).
Our analyses indicate that OhrR forms several different mixed disulfide species in vivo. MS analyses reveal formation of a mixed disulfide with a newly discovered 398-Da LMW thiol. A thiol of this mass has also been detected recently in Bacillus anthracis (41). In addition, OhrR forms mixed disulfides with Cys and CoASH. Of those thiols we were able to test in vitro, reaction occurs with Cys ≫ CoASH ≈ GSH. However, when expressed in E. coli, S-glutathionylated OhrR is readily detected upon oxidation (SI Fig. 8).
In addition to S-thiolation, we report the in vitro formation (in the absence of LMW thiols) of an OhrR protein sulfenamide (Fig. 3E, SI Fig. 6, and SI Fig. 10). The detection of an alkylation-resistant form of OhrR in immunoprecipitation studies suggests that the sulfenamide derivative also forms in the cell (Fig. 5; reaction 4). Like the S-thiolated species, the sulfenamide derivative is inactive for DNA binding, but activity can be restored (albeit slowly) by the strong thiol reductant DTT (Fig. 3 B and F). Previously, we reported that oxidation of OhrR (in the absence of Cys) led to formation of a relatively stable sulfenic acid derivative and, over time, further oxidation to the sulfinic and sulfonic acid species. Using a DNaseI footprinting assay we found that DNA-binding activity could be restored by DTT (16). Because DTT reduces sulfenates (42), but not sulfinic acids, we interpreted this as evidence of a stable (and inactive) sulfenate. In retrospect, the ability of DTT to restore DNA binding was likely indicative of sulfenamide formation and, as reported here, the sulfenic acid derivative is still active for DNA binding. Protein sulfenamides were not known at the time of these original experiments. Here, we document the in vitro kinetics of protein sulfenamide formation and re-reduction and provide experimental evidence for such species in vivo.
OhrR is related to the MarR/SlyA family of winged-helix-turn-helix DNA-binding proteins (15). The structure of an OhrR–ohrA operator complex reveals that Cys15 is hydrogen-bonded to Tyr29 and Tyr40 on the other subunit, and we therefore suggested that sulfenate formation might disrupt these critical hydrogen bonds (22). In light of the studies reported here, we now conclude that sulfenate formation does not alter protein conformation sufficiently to functionally inactivate the repressor, and that inactivation requires either mixed-disulfide bond or sulfenamide formation. Although the signaling mechanisms regulating repressor activity are likely to be diverse, recent results suggest that at least one key virulence regulator in S. aureus (MgrA) is regulated by oxidation at an active site similar to that described for B. subtilis OhrR (43). Thus, the regulatory mechanisms documented here may have broad relevance for stress responses in this and related human pathogens.
Experimental Procedures
Preparation of OhrR.
OhrR was purified after overexpression in E.coli BL21/DE3(pLysS) as described (16). OhrR concentrations were determined by using the calculated value of ε280 nm = 14,440 M−1·cm−1, and aliquots of 200 μM (monomer) stock were stored at −70°C in buffer A [20 mM Tris, pH 8.0/100 mM NaCl/5% (vol/vol) glycerol] containing 1 mM DTT and 1 mM EDTA. For in vitro oxidation experiments, DTT and EDTA were removed by using Bio-Spin 6 Chromatography columns (Bio-Rad, Hercules, CA).
FA.
A 6-carboxyfluorescein-(6F-)-labeled ohrA operator DNA fragment was generated by annealing 5′-6F-TACAATTAAATTGTATACAATTAAATTGTA-3′ (Integrated DNA Technologies, Coralville, IA) and its unlabeled complement. FA measurements (λex = 495 nm; slit width = 15 nm, λem = 520 nm; slit width = 20 nm, integration time = 1 s) were performed with 50 nM DNA and 300 nM OhrR (monomer) in 3 ml of 20 mM Tris (pH 8.0) containing 150 mM NaCl and 5% (vol/vol) glycerol. FA values were recorded automatically every 10 s with a LS55 luminescence spectrometer (PerkinElmer, Wellesley, MA). The g factor was 1.07 ± 0.01. Kinetic traces were analyzed by using a pseudofirst-order approximation where appropriate.
ESI-MS Analysis and AMS Modification Experiment.
For ESI-MS analysis, 1 ml of reaction was mixed with 110 μl of 100% TCA, and proteins were recovered by centrifugation at 16,100 × g for 10 min. After additional washing with 10% TCA, samples were dissolved in 30 μl of 2% acetic acid (vol/vol) and 50% methanol (vol/vol) and analyzed with an Esquire-LC ion trap mass spectrometer (Bruker, Billerica, MA). For each sample, at least three spectra were acquired and independently deconvoluted by using the Daltoniks DataAnalysis program (Bruker), and the average mass was presented as average ± SD. For SDS/PAGE analysis of AMS modification (Figs. 2D and 3 A and C), TCA-precipitated samples were resuspended with 200 mM Tris (pH 8.0) containing 1% SDS and 10 mM AMS and incubated for 2 h in the dark. To check the re-reduction of OhrR (Fig. 3 B and D), TCA-precipitated samples were incubated with 10 mM DTT in 200 mM Tris buffer (pH 8.0) containing 1% SDS for 1 h, recovered by 20% TCA precipitation, and then alkylated with AMS. Samples were resolved on 16% SDS/PAGE gels by using a Tris-Tricine buffer system.
Monitoring OhrR Oxidation in B. subtilis.
The ohrR ORF with its own promoter was PCR-amplified and cloned into the HindIII and EagI sites of pJL070 (44) to replace perR with ohrR. The resulting plasmid was used for transformation of HB2014 (13) to generate a strain containing ohrR-FLAG at the amyE locus designated HB9121. Cells of OD600 = 0.8 in 250 ml of LB medium were untreated (Fig. 4A) or treated with 100 μM CHP (Fig. 4B) for ≈2 min. For Fig. 4C cells were treated with 1 mM diamide for 1 min, a time sufficient for the depletion of intracellular Cys (18), before CHP treatment. OhrR-FLAG was recovered as described for PerR-FLAG (44) in the presence of 100 mM IA, digested with trypsin, and analyzed with a 4700 MALDI-TOF mass spectrometer (Applied Biosystems, Foster City, CA).
Supplementary Material
Acknowledgments
We thank the helpful staff of the Cornell Biotechnology Resource Center Proteomics and Mass Spectrometry Core Facility and Dr. A. Claiborne and R. Fahey for communicating results before publication. This work was supported by National Science Foundation Grant MCB-0640616 (to J.D.H.) and a Government of Thailand Scholarship (to S.S.).
Abbreviations
- AMS
4-acetamido-4′-maleimidylstilbene-2,2′-disulfonic acid
- IA
iodoacetamide
- TCA
trichloroacetic acid
- CHP
cumene hydroperoxide
- ROS
reactive oxygen species
- LMW
low molecular weight
- CoASH
reduced CoA
- FA
fluorescence anisotropy
- GSH
glutathione
- ESI
electrospray ionization.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0702081104/DC1.
References
- 1.Imlay JA. Annu Rev Microbiol. 2003;57:395–418. doi: 10.1146/annurev.micro.57.030502.090938. [DOI] [PubMed] [Google Scholar]
- 2.Fang FC. Nat Rev Microbiol. 2004;2:820–832. doi: 10.1038/nrmicro1004. [DOI] [PubMed] [Google Scholar]
- 3.Torres MA, Dangl JL. Curr Opin Plant Biol. 2005;8:397–403. doi: 10.1016/j.pbi.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 4.Mongkolsuk S, Helmann JD. Mol Microbiol. 2002;45:9–15. doi: 10.1046/j.1365-2958.2002.03015.x. [DOI] [PubMed] [Google Scholar]
- 5.Toledano MB, Delaunay A, Monceau L, Tacnet F. Trends Biochem Sci. 2004;29:351–357. doi: 10.1016/j.tibs.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 6.Paget MS, Buttner MJ. Annu Rev Genet. 2003;37:91–121. doi: 10.1146/annurev.genet.37.110801.142538. [DOI] [PubMed] [Google Scholar]
- 7.Lee C, Lee SM, Mukhopadhyay P, Kim SJ, Lee SC, Ahn WS, Yu MH, Storz G, Ryu SE. Nat Struct Mol Biol. 2004;11:1179–1185. doi: 10.1038/nsmb856. [DOI] [PubMed] [Google Scholar]
- 8.Aslund F, Zheng M, Beckwith J, Storz G. Proc Natl Acad Sci USA. 1999;96:6161–6165. doi: 10.1073/pnas.96.11.6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim SO, Merchant K, Nudelman R, Beyer WF, Jr, Keng T, DeAngelo J, Hausladen A, Stamler JS. Cell. 2002;109:383–396. doi: 10.1016/s0092-8674(02)00723-7. [DOI] [PubMed] [Google Scholar]
- 10.Poole LB, Karplus PA, Claiborne A. Annu Rev Pharmacol Toxicol. 2004;44:325–347. doi: 10.1146/annurev.pharmtox.44.101802.121735. [DOI] [PubMed] [Google Scholar]
- 11.Helmann JD, Wu MF, Gaballa A, Kobel PA, Morshedi MM, Fawcett P, Paddon C. J Bacteriol. 2003;185:243–253. doi: 10.1128/JB.185.1.243-253.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Engelmann S, Lindner C, Hecker M. J Bacteriol. 1995;177:5598–5605. doi: 10.1128/jb.177.19.5598-5605.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuangthong M, Atichartpongkul S, Mongkolsuk S, Helmann JD. J Bacteriol. 2001;183:4134–4141. doi: 10.1128/JB.183.14.4134-4141.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Volker U, Andersen KK, Antelmann H, Devine KM, Hecker M. J Bacteriol. 1998;180:4212–4218. doi: 10.1128/jb.180.16.4212-4218.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ellison DW, Miller VL. Curr Opin Microbiol. 2006;9:153–159. doi: 10.1016/j.mib.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 16.Fuangthong M, Helmann JD. Proc Natl Acad Sci USA. 2002;99:6690–6695. doi: 10.1073/pnas.102483199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Newton GL, Arnold K, Price MS, Sherrill C, Delcardayre SB, Aharonowitz Y, Cohen G, Davies J, Fahey RC, Davis C. J Bacteriol. 1996;178:1990–1995. doi: 10.1128/jb.178.7.1990-1995.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gusarov I, Nudler E. Proc Natl Acad Sci USA. 2005;102:13855–13860. doi: 10.1073/pnas.0504307102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fahey RC, Brown WC, Adams WB, Worsham MB. J Bacteriol. 1978;133:1126–1129. doi: 10.1128/jb.133.3.1126-1129.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lesniak J, Barton WA, Nikolov DB. EMBO J. 2002;21:6649–6659. doi: 10.1093/emboj/cdf670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cussiol JR, Alves SV, de Oliveira MA, Netto LE. J Biol Chem. 2003;278:11570–11578. doi: 10.1074/jbc.M300252200. [DOI] [PubMed] [Google Scholar]
- 22.Hong M, Fuangthong M, Helmann JD, Brennan RG. Mol Cell. 2005;20:131–141. doi: 10.1016/j.molcel.2005.09.013. [DOI] [PubMed] [Google Scholar]
- 23.Salmeen A, Andersen JN, Myers MP, Meng TC, Hinks JA, Tonks NK, Barford D. Nature. 2003;423:769–773. doi: 10.1038/nature01680. [DOI] [PubMed] [Google Scholar]
- 24.van Montfort RL, Congreve M, Tisi D, Carr R, Jhoti H. Nature. 2003;423:773–777. doi: 10.1038/nature01681. [DOI] [PubMed] [Google Scholar]
- 25.Janek K, Wenschuh H, Bienert M, Krause E. Rapid Commun Mass Spectrom. 2001;15:1593–1599. doi: 10.1002/rcm.417. [DOI] [PubMed] [Google Scholar]
- 26.Barford D. Curr Opin Struct Biol. 2004;14:679–686. doi: 10.1016/j.sbi.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 27.Panmanee W, Vattanaviboon P, Poole LB, Mongkolsuk S. J Bacteriol. 2006;188:1389–1395. doi: 10.1128/JB.188.4.1389-1395.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Giustarini D, Rossi R, Milzani A, Colombo R, Dalle-Donne I. J Cell Mol Med. 2004;8:201–212. doi: 10.1111/j.1582-4934.2004.tb00275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hand CE, Honek JF. J Nat Prod. 2005;68:293–308. doi: 10.1021/np049685x. [DOI] [PubMed] [Google Scholar]
- 30.Ghezzi P. Biochem Soc Trans. 2005;33:1378–1381. doi: 10.1042/BST0331378. [DOI] [PubMed] [Google Scholar]
- 31.Mallett TC, Wallen JR, Karplus PA, Sakai H, Tsukihara T, Claiborne A. Biochemistry. 2006;45:11278–11289. doi: 10.1021/bi061139a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.delCardayre SB, Stock KP, Newton GL, Fahey RC, Davies JE. J Biol Chem. 1998;273:5744–5751. doi: 10.1074/jbc.273.10.5744. [DOI] [PubMed] [Google Scholar]
- 33.Boylan JA, Hummel CS, Benoit S, Garcia-Lara J, Treglown-Downey J, Crane EJ, 3rd, Gherardini FC. Mol Microbiol. 2006;59:475–486. doi: 10.1111/j.1365-2958.2005.04963.x. [DOI] [PubMed] [Google Scholar]
- 34.Hummel CS, Lancaster KM, Crane EJ., III FEMS Microbiol Lett. 2005;252:229–234. doi: 10.1016/j.femsle.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 35.Setlow B, Setlow P. Biochem Biophys Res Commun. 1977;75:45–50. doi: 10.1016/0006-291x(77)91286-4. [DOI] [PubMed] [Google Scholar]
- 36.Rhee SG, Kang SW, Jeong W, Chang TS, Yang KS, Woo HA. Curr Opin Cell Biol. 2005;17:183–189. doi: 10.1016/j.ceb.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 37.Jacob C, Holme AL, Fry FH. Org Biomol Chem. 2004;2:1953–1956. doi: 10.1039/b406180b. [DOI] [PubMed] [Google Scholar]
- 38.Budanov AV, Sablina AA, Feinstein E, Koonin EV, Chumakov PM. Science. 2004;304:596–600. doi: 10.1126/science.1095569. [DOI] [PubMed] [Google Scholar]
- 39.Lee JW, Helmann JD. Nature. 2006;440:363–367. doi: 10.1038/nature04537. [DOI] [PubMed] [Google Scholar]
- 40.Klomsiri C, Panmanee W, Dharmsthiti S, Vattanaviboon P, Mongkolsuk S. J Bacteriol. 2005;187:3277–3281. doi: 10.1128/JB.187.9.3277-3281.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nicely NI, Parsonage D, Paige C, Newton GL, Fahey RC, Leonardi R, Jackowski S, Mallett TC, Claiborne A. Biochemistry. 2007;46:3234–3245. doi: 10.1021/bi062299p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kumar RA, Koc A, Cerny RL, Gladyshev VN. J Biol Chem. 2002;277:37527–37535. doi: 10.1074/jbc.M203496200. [DOI] [PubMed] [Google Scholar]
- 43.Chen PR, Bae T, Williams WA, Duguid EM, Rice PA, Schneewind O, He C. Nat Chem Biol. 2006;2:591–595. doi: 10.1038/nchembio820. [DOI] [PubMed] [Google Scholar]
- 44.Lee JW, Helmann JD. J Biol Chem. 2006;281:23567–23578. doi: 10.1074/jbc.M603968200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.