Abstract
Recent work with microbial communities has demonstrated an adaptive response to artificial selection at the level of the ecosystem. The reasons for this response and the level at which adaptation occurs are unclear: does selection act implicitly on traits of individual species, or are higher-level traits genuinely being selected? If the ecosystem response is just the additive combination of the responses of the constituent species, then the ecosystem response could be predicted a priori, and the ecosystem-level selection process is superfluous. However, if the ecosystem response results from ecological interactions among species, then selection at a higher level is necessary. Here we perform artificial ecosystem selection experiments on an individual-based evolutionary simulation model of microbial ecology and observe a similar response to that seen with real ecosystems. We demonstrate that a significant fraction of artificially selected ecosystem responses cannot be accounted for by implicit lower-level selection of a single type of organism within the community, and that interactions among different types of organism contribute significantly to the response in the majority of cases. However, when the ecological problem posed by the artificial ecosystem selection process can be easily solved by a single dominant species, it often is.
Keywords: multilevel selection, microbial ecology, evolution, ecosystem selection
Recent work with microbial communities has demonstrated that artificial selection at the level of the ecosystem can lead to a sustained evolutionary response (1, 2). Statistically significant responses were observed in experiments where soil communities were selected for the dry weight of plant biomass they could support, and where pond-water communities were selected for their effect on the pH level of their liquid environment (2). Artificial ecosystem selection has also been used to create microbial communities capable of breaking down the environmental pollutant 3-chloroaniline (1). It has been argued that the published experiments have interesting implications for evolutionary theory, because they offer examples of multilevel selection (3, 4). In artificial selection scenarios, many of the problems that may prevent higher-level selection from being effective in nature are avoided, and various experimental results have demonstrated the power of group selection to drive adaptation in both single-species and multispecies groups (5–8). Recently, theoretical arguments have also been put forward to suggest circumstances where higher-level (or multilevel) selection can have a significant effect in nature (3, 4, 9). However, the presence in a functioning ecosystem of phenomena such as metabolic dependencies and nutrient cycling means that ecosystem selection is qualitatively different from classical group selection. The environment is an integral part of ecosystem dynamics.
It is unclear whether the existing laboratory results (1, 2) are due to the artificial ecosystem selection process implementing a form of parallel search for a particular species with the desired environmental effect, or whether the response to selection results from more complex synergistic interactions at the community level. It is possible that selecting at the ecosystem level is necessary to achieve the results reported, and that taking an intact community as the unit of selection allows selection to act on the dynamics of the ecosystem, i.e., allows evolutionary adaptation of nonadditive “phenotypic” traits that do not exist at the individual level. It is also possible that selecting at the community level simply casts the net wider in the search for a single species that provides the desired functional trait. We use a modeling approach to address this critical issue.
Population-based modeling has been used to explore multilevel selection in metacommunities, where it was shown that higher-level selection pressures could have a strong effect on ecological dynamics (10). Generalized Lotka–Volterra equations were used to model communities inhabiting semi-isolated patches, and patch-level selection pressures created by the metacommunity structure were shown to cause the local patch communities to diverge to different equilibria than would otherwise have been reached. More recently, population-based simulations of artificial ecosystem selection have been reported (11–13). Responses to artificial ecosystem selection for diversity (11) and for maximization of an arbitrary linear function of species composition (12) were found in a system modeling competition among different species, also based on sets of Lotka–Volterra population equations. However, Lotka–Volterra models do not allow direct mutualisms or metabolic dependencies, do not consider energy and material flows, and are deterministic, making them significantly removed from the stochastic, mutualistic, and thermodynamically constrained natural world. More importantly, the interaction matrices used (10–13) were fixed for the duration of each ecosystem “generation,” meaning there was no possibility of individual-level adaptation. This limitation on the effects of selection at the individual level is problematic for any study of multilevel selection, where the central questions concern the interaction between selection pressures at lower and higher levels. Although theoretical models that incorporate individual-level adaptation have been used to study frequency-dependent effects such as resource competition (14), no such models have been applied to study higher-level selection in the present context.
Here we present a set of artificial ecosystem selection experiments performed on simulated microbial microcosms held in isolated containers. The selected ecosystems are evolving microbial communities interacting with their abiotic environment, with selection performed on properties of the coupled biotic-abiotic system. The model allows individual-level adaptation and natural selection pressure generated by ecological interactions to be incorporated alongside artificially imposed ecosystem-level selection on the effect of the community on its abiotic environment. Artificial ecosystem selection experiments are performed by using a method similar to Swenson et al. (1, 2), by using properties of the environment as a target. A response similar to that seen in the laboratory experiments is observed and found to be robust to different ecosystem transmission methods, to the time for which the ecosystems are allowed to develop between selection events, and to the mutation rate of individuals. We further show whether the ecosystem response can be decomposed into the independent responses of individual species or is a genuine community-level property. (Although the concept of a species is not well defined for microbes, we use the term here to refer to a clonal group of individuals.)
To address the question of how artificial selection produces a response in the selected ecosystems, we ask two main questions. First of all, was selection above the level of a single species (clonal group) necessary to achieve the observed response? This question is answered by searching for a species in the selected community that is alone responsible for generating the desired ecosystem property; if such a species exists, it could potentially have been found by lower-level artificial selection methods. Second, if higher-level selection is shown to be necessary, we ask whether the observed response results from the additive combination of a number of species, each making an independent contribution to the net response, or whether the observed response results from ecological interactions among species. If the former, the same response could in theory have been produced by carefully picking a complementary set of species based on their individual properties, and selection at the level of the ecosystem is superfluous (despite being the mechanism by which the community was assembled in this case). If the latter, the observed performance is a nonadditive function of the actions of the constituent species of the community (i.e., the community response is not equal to the sum of its parts) and is therefore not decomposable. In such cases we have the strongest argument for higher-level selection acting on traits above the level of the individual.
Model Description.
The “Flask” model (15, 16) [see supporting information (SI) Text and SI Table 2] simulates a flask containing a neutral liquid matrix in which is suspended a microbial population. The composition of the liquid medium determines the environment of the microbes. Some of the chemicals present are “nutrients” that may be consumed as food and converted to biomass, whereas others are nonconsumable and form part of the abiotic environment. The environment is assumed to have properties such as temperature, pH, salinity, etc., that both affect and can be affected by microbial activity. Nonconsumable chemicals and physical properties of the flask environment are collectively referred to as “abiotic factors,” to distinguish them from nutrients.
There is a flow of liquid medium through each flask that occurs continuously at a prescribed rate. The inflow brings with it influxes of nutrients at fixed concentrations and steady inputs to abiotic factors, whereas the outflow removes fixed proportions of stored nutrients and abiotic factors. The liquid medium in each flask is assumed to be well mixed, so that in the absence of perturbation, the composition of the medium in each flask will reach a homogeneous steady state.
Microbes are modeled as simple organisms that consume and excrete nutrients and affect the levels of abiotic factors in their environment as a byproduct of metabolism. The precise ratios in which nutrients are consumed and excreted are genetically encoded for each individual, as are associated effects on abiotic factors and preferred abiotic conditions (i.e., the state of the abiotic environment in which growth rate is maximized). The amount of nutrients consumed by a microbe is constrained by availability, by a universal maximum consumption rate, and by the fit between the current state of the abiotic environment and the microbe's preferences. Microbes affect the environmental levels of abiotic factors as a by-product of their metabolic actions, in proportion to the amount of biomass created.
Microbes grow by converting consumed nutrients to biomass and reproduce by splitting when their biomass reaches a fixed threshold. All nutrients have an equal value, and microbes have a universal standard conversion efficiency, so that the only determinants of differential growth rates among microbes are their genetically specified metabolisms. A microbe that grows at the maximum possible rate will reproduce approximately every 12 timesteps, but nutrient limitation and adverse abiotic conditions commonly cause much slower growth rates; the growth and reproduction of microbes mean that nutrient limitation is the normal ecosystem condition. Mutation may occur during each reproduction event by selecting a new random allele with probability Pmut (experimentally varied in the range [0,0.1]) at each locus; otherwise each offspring microbe receives an identical copy of the parental genotype. Biomass is reduced at a fixed rate to represent the inevitable thermodynamic inefficiency of metabolism and the cost of maintaining cellular machinery. Microbes die if their biomass drops below a fixed threshold, which can happen in sustained periods of nutrient limitation. They may also die “from natural causes,” with a low probability at each timestep. This mechanism is a catch-all for death by predation, senescence, etc., and serves to thin out the microbial population in an unbiased way, thus promoting continuing competition and individual-level selection. When a microbe dies, it is assumed that its remaining biomass is washed out and lost from the system.
Growth of a population occurs only as a result of individual growth and reproduction and is not specified a priori as in more traditional population ecology models such as Lotka–Volterra systems. Flask ecosystem carrying capacities are determined by nutrient supply and typically measure in the hundreds for the parameters used. These quantities are small by comparison with real-world microbial populations, a constraint enforced by the demands of computational tractability, but the small population sizes (and high mutation rates) are reasonable if each individual in the model is considered to represent many genetically similar real-world individuals. The shared environment creates individual-level selection pressure on metabolic requirements and environmental preferences, but the nature of this selection pressure changes over time as microbial activity alters the environment. Ecological and evolutionary dynamics of these model ecosystems are discussed elsewhere (16).
Artificial Selection Target.
The “phenotypic” ecosystem trait used for artificial selection is based on the levels of the abiotic factors in the flask environment. Basing the fitness of flask ecosystems on properties of the environment rather than the biotic population avoids any prespecification of the type of population that will provide a good solution to the evolutionary problem. An arbitrary target state of the abiotic environment is assigned, with the deviation error of the actual abiotic state of a flask from this target constituting its performance score, Φ:
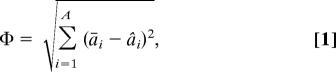 |
where āi is the target level for abiotic factor ai, and âi is the actual level of ai, in the normalized state vector for the A abiotic factors included in the model. Depending on the direction of artificial selection, the fitness of a flask ecosystem is based on maximizing or minimizing Φ. In each artificial selection experiment, three lines were selected based on the same initial random population: The “high” line was selected to maximize Φ, the “low” line was selected to minimize Φ, and the “random” line (where the source ecosystem used to create the batch of ecosystems for each iteration was chosen at random) acted as a control. All lines consisted of a number of iterations of directed selection followed by an equivalent number of iterations of random selection, to allow study of the relaxation of the selected response.
The different lines present different types of evolutionary “problem” for the artificial ecosystem selection process to “solve.” Because Φ measures the distance from a target, we may a priori say that the low line presents a more difficult problem than the high line, because converging on a target is more difficult than diverging from it; there are many ways to be far from a point in multidimensional space, but only one way to hit it. Furthermore, in a complex dynamic environment, holding an environmental variable close to a particular target level will often require correction in two directions. Because a single species can push any environmental variable in only one direction, at least two species may therefore be needed to provide the necessary opposing influences for the low line (target-seeking) problem. However, a variable can be moved away from a target by pushing in a single direction, so a single dominant species in the community may offer a good solution to the high line (target-avoiding) problem. Thus the high and low lines offer qualitatively different evolutionary problems that may demand qualitatively different ecosystem solutions.
Artificial ecosystem selection is an iterative process based on preferentially sampling from successive batches of flask ecosystems to create each succeeding batch. After a randomly seeded initial batch, at each iteration of the selection process, a new batch of flask ecosystems is created by inoculating sterile flasks with individuals from the fittest flasks of the previous iteration. A single inoculum of a fixed number of individuals is created by sampling at random from the source flasks, and identical copies of this inoculum are then used to seed the entire new batch of ecosystems. Two sampling methods are used: a propagule method, where the inoculum is drawn from a single source ecosystem, and a migrant pool method, where the inoculum takes individuals from several source ecosystems. The propagule method is analogous to asexual reproduction and should preserve ecological interactions among individuals. The migrant pool method is analogous to sexual reproduction and may better represent how new ecosystems form in nature. After inoculation, each ecosystem is propagated for a fixed period of Tprop timesteps before Φ is measured; the propagation time Tprop is experimentally varied in the range [2000,20000] and specifies the time between selection events. For more details see Methods.
Results
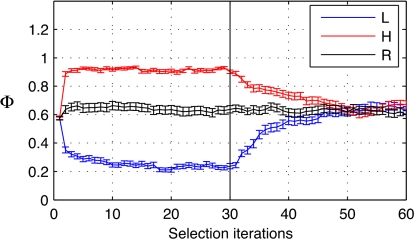
A robust response to artificial selection is seen in our model ecosystems (e.g., Fig. 1). For an arbitrarily chosen target vector, in both the high and low selected lines, the normalized abiotic environment state vector quickly diverges from the randomly selected control line. The high line is selected to maximize Φ (the distance of the actual abiotic environmental state from the target state), and there is a rapid initial increase in this distance followed by a leveling off. Similar behavior is displayed by the low line, except that the ecosystem-level selection in this case is for a decrease in Φ. When directed selection is removed (after 30 ecosystem selection iterations), the selected lines relax toward the nonselected condition (represented by the randomly selected control line ecosystems). The response to selection is very similar with both the propagule and the migrant pool sampling methods, and similar results are also achieved with different target vectors (SI Figs. 3 and 4).
Fig. 1.
Artificial ecosystem selection produces a strong adaptive response. Mean Φ ± 1 SE plotted. Here, 57 runs were performed by using propagule sampling with the default experimental settings (see Methods). Data are plotted for directed selection for either increase (high line, H) or decrease (low line, L) in distance of abiotic environment from target state, Φ, as well as for a random selection control line (R) that shows behavior in the absence of artificial ecosystem selection. Directed selection is stopped after iteration 30, at which point all ecosystem-level selection is random.
We explored the effects on the observed response of different sampling methods, varying the microbial mutation rate, and the ecosystem propagation time (Table 1). Selected ecosystem scores deviate significantly from control line scores, showing the effect of artificial ecosystem selection. There are inverse relationships between the size of the response to artificial ecosystem selection and mutation rate, and between the size of the response and propagation time (Table 1; SI Figs. 5–7). The rate of relaxation when directed selection is removed is directly proportional to the individual-level mutation rate (SI Fig. 8a). No significant relaxation occurs when mutation rate is zero, indicating that fit ecosystems in this scenario undergo a stable transition in ecological organization, i.e., a switch to a high-fitness ecological equilibrium. Relaxation rate is unrelated to the frequency of ecosystem selection events (SI Fig. 8 b and c). Results from the migrant pool and propagule sampling methods are similar for equivalent Pmut and Tprop. Perturbing the environmental fluxes (see SI Table 3) has a deleterious effect on performance, suggesting that both environment and community in general contribute significantly to the selected ecosystem function.
Table 1.
Mean performance (Φ) scores from artificial ecosystem selection experiments for control line, low-selected, and high-selected communities
| Sampling | Tprop | Pmut | Runs | Φ |
||
|---|---|---|---|---|---|---|
| Control | Low | High | ||||
| Propagule | 2000 | 0.01 | 46 | 0.67 | 0.19 | 0.94 |
| 5000 | 0.01 | 57 | 0.66 | 0.16 | 0.93 | |
| 10000 | 0.01 | 60 | 0.62 | 0.19 | 0.94 | |
| 20000 | 0.01 | 75 | 0.71 | 0.29 | 0.89 | |
| 5000 | 0 | 43 | 0.56 | 0.28 | 0.91 | |
| 5000 | 0.03 | 87 | 0.62 | 0.25 | 0.92 | |
| 5000 | 0.05 | 42 | 0.56 | 0.32 | 0.86 | |
| 5000 | 0.1 | 73 | 0.57 | 0.42 | 0.84 | |
| All | All | 483 | 0.63 | 0.27 | 0.90 | |
| Migrant | 5000 | 0.01 | 49 | 0.73 | 0.19 | 0.96 |
Results for the propagule sampling method, with varying propagation time Tprop and mutation rate Pmut, and for the migrant-pool sampling method at the default values.
Testing for Implicit Lower-Level Selection.
We tested (see Methods) whether the observed response to artificial ecosystem selection could be due to implicit selection at a lower level, by examining whether any species taken from an artificially selected community could achieve or exceed the performance of the intact selected community, either when allowed to develop in isolation as a clonal monoculture population or when placed in the context of a wild-type community (by adding individuals of the test species to the associated control line community). These are attempts to falsify the hypothesis:
H1: The adaptive response of the artificially selected ecosystems relies on the presence of multiple concurrently selected species.
With the observations:
O1: A species from within the community exists that in monoculture gives performance that equals or exceeds that of the selected community.
O2: A species from within the community exists that in the context of a wild-type community gives performance that equals or exceeds that of the selected community.
O3: A species from within the community exists that both as a monoculture population and in the context of a wild-type community gives performance that equals or exceeds that of the selected community.
We tested all species from each of the selected communities to see whether they satisfied observations O1, O2, and O3. For each community, each observation was satisfied if at least one species met the designated criterion. At first glance, it might appear that O3 is superfluous and corresponds to the intersection of O1 and O2. However, it is possible for O1 and O2 to be satisfied by different species from the same community; O3 adds a further distinction in recording the number of cases where the same species that satisfied O1 also satisfied O2.
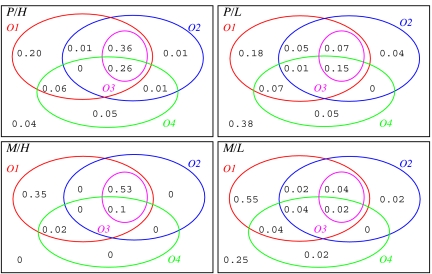
We tested every ecosystem that was artificially selected by using the propagule sampling method (483 in each line, giving 966 in total) (Fig. 2). These results ignore differences in propagation time and mutation rate between runs (for more detailed results, see SI Tables 4 and 5). For observations O1 and O2, there is a marked difference between the high- and low-selected ecosystems. In ecosystems selected for high Φ, a species giving better performance than the intact selected community when grown as a monoculture population was found (O1) in 89% of cases, and a species that induced better performance in the control line community was found (O2) in 65% of cases. The overlap is large; in 63% of cases, both O1 and O2 were satisfied, with a single species that satisfied both O1 and O2 simultaneously being found (O3) in almost all of these (62% of cases). This leaves 9% of high-selected ecosystems where no species can match the performance of the selected community either in monoculture or in the context of the wild-type community. In ecosystems selected using the propagule method for low Φ, the observations are satisfied less often: O1 in 53% of cases, O2 in 32% of cases, with an overlap of 28% of cases, of which a single species that satisfied O1 and O2 simultaneously was found (O3) in 22% of cases. This leaves 43% of low-selected ecosystems for which no species could match the performance of the intact selected community, either in monoculture or in the wild-type community. Results for the migrant pool sampling method (Fig. 2) show that O1 and O2 are satisfied in an even greater fraction of cases; of the 49 cases tested in each line, all of the high line cases satisfied O1, and 63% of cases satisfied O2. All of the cases that satisfied O2 also satisfied O3. Low-selected ecosystems with migrant pool sampling were again less likely to satisfy O1 or O2; 71% of cases satisfied O1, 14% satisfied O2, and 27% cases satisfied neither.
Fig. 2.
Fractions of the 483 ecosystems artificially selected using propagule sampling (P) and the 49 ecosystems selected using migrant pool sampling (M), in both high (H) and low (L) lines, that satisfy observations O1–O4. O1 indicates cases where at least one species from the selected community gives performance equal to or better than the intact community when grown in isolation as a monoculture. O2 indicates cases where at least one species from the selected community gives equal or better performance in the context of the wild-type community. O3 indicates cases where the same species gives equal or better performance both as a monoculture and in a wild-type community, i.e., it represents a subset of the intersection between O1 and O2. O4 indicates cases where the sum of contributions of individual species (excluding interactions) equals or exceeds the performance of the selected community (with interactions).
These results show that the majority of ecosystems (91% cases with the propagule sampling method, 100% with the migrant pool method) artificially selected for high Φ contain a single species that can outperform the intact community in some context. However, in ecosystems selected for low Φ, no single better species could be found in a significant proportion of cases (43% with propagule sampling, 27% with migrant pool sampling), showing that multiple species were involved in performing the selected function in these cases.
Testing for Selection Acting on Interactions.
We sought to establish whether the response to artificial ecosystem selection depended on interactions among different species by measuring the fitness score (Φ) for each species in each selected community when grown as a monoculture, then taking the sum of the contributions of all member species, weighted by the fraction of the community made up of that type (see Methods). This gives the expected score for the community in the absence of interactions between species (ΦE), which can be compared with the actual observed score (ΦO). Any significant difference between these values implies the presence of interactions among species in determining community performance. This method is similar to tests for biodiversity effects in determining the overall yield of plant communities (e.g., ref. 17). Formally, we attempted to falsify the hypothesis:
H2: The observed response to artificial ecosystem selection is due to selection acting on interactions among species.
With the observation:
O4: The expected performance in the absence of interactions among species is not significantly different from, or is better than, the observed performance of the intact community.
For ecosystems selected with propagule sampling, O4 is satisfied (hence interactions between species are insignificant or deleterious) in the minority of cases for both high-selected (38%) and low-selected (28%) ecosystems. For ecosystems selected with the migrant pool method, O4 is satisfied even less often (12% cases in both high and low lines). Hence beneficial interactions play a significant role in the function of the majority of artificially selected ecosystems.
Combined Tests.
All combinations of O1–O4 are summarized in Fig. 2. If we look for cases where multiple species and beneficial interactions between them are an essential part of the selected community function (i.e., we reject any cases where any of O1, O2, or O4 is satisfied, noting that O3 is subsumed into the overlap between O1 and O2), with propagule sampling, we are left with 4% of high-selected and 38% of low-selected cases. With migrant pool sampling, we are left with no high-selected cases and 25% of low-selected cases.
For both high and low lines, the likelihood of H1 and H2 being satisfied varies inversely with the mutation rate Pmut (SI Table 4). When Pmut = 0, even the high-selected ecosystems satisfy H1 and H2 in 30% of cases; this is because without mutation, a single species that is alone capable of performing the selected function cannot evolve, so solutions based on interactions among multiple species are more likely. No obvious relation exists between satisfaction of H1 and H2 and the time between selection events.
Discussion
We have demonstrated a robust response to artificial selection in our model ecosystems. Our results suggest that individual-level selection pressure has a degrading effect on the response to artificial ecosystem selection, as expected from evolutionary theory. Between artificial ecosystem selection events, individual-level selection pressures on metabolic requirements and environmental preferences cause the species composition of the population to change. Although at the individual level this change is adaptive, at the higher level it amounts to drift, phenotypic variation without selection pressure. The genetic composition of different communities moves in different directions due to ecological interactions and the random individual-level mutations occurring within them. Artificial ecosystem selection prunes away those communities that move in the wrong direction and creates replicant variations of those that move in the right direction. Thus artificial ecosystem selection can be viewed as an external steering of the ongoing ecological and evolutionary processes within the microbial community that moves it along a different trajectory from that which it would have followed under the influence of individual-level selection pressure alone.
If a species from within a selected community is found that as a monoculture matches or exceeds the performance of the selected community, then it could be argued that ecosystem-level selection was implicitly selecting this species, and that the rest of the community is irrelevant to the observed response. Considering each species in the context of a wild-type community (supplied here by the control line community from the same selection run), we can identify cases where a single species may be solely responsible for the observed response to selection but requires the presence of a non-specific background community for the desired property to be expressed, as was observed in artificial selection experiments performed on beetle communities (6, 7). In this scenario, the background community acts as a non-evolving part of the environment that could have been incorporated into selection experiments at a lower level, hence community-level or ecosystem-level selection is not required. However, if the expression of the desired property requires a particular community, then selection above the level of the species or clonal group is required, because multiple species are concurrently selected. If multiple species are required and beneficial interactions are contributing significantly to community performance, then it suggests that selection has acted on those interactions, i.e., selection has acted on higher-level traits.
Our two sets of tests establish that a significant fraction of artificially selected ecosystem responses (especially on the low line) cannot be accounted for by a single type of organism within the community, and that interactions among different types of organism contribute significantly to the response in the majority of cases. The tests performed may be combined in an attempt to falsify (or not) the proposition that the ecosystem selection process acts on traits above the level of individuals or species. What qualifies as a falsification depends on one's view as to the reasonableness of the tests, but if we adopt the harsh criterion that any one of our observations O1–O4 amounts to a falsification, we are still left with a significant fraction of ecosystems on the low line where higher-level selection has acted on higher-level traits.
Almost all of the high-selected ecosystems (91% with propagule sampling, 100% with migrant pool sampling) appear to have a selected function that is based on the strong contribution of a single species. A further 5% with propagule sampling show no contribution of interactions, i.e., a response that could be due to the individual contributions of two or more species without interaction. Such solutions could be found by lower-level selection methods, leaving only 4% of cases with propagule sampling where higher-level traits were selected. However, with the low-selected ecosystems, a significant number of cases (38% with propagule sampling, 25% with migrant pool sampling) have a selected function based on beneficial interactions among multiple species. Higher-level selection is necessary more often in the low than in the high line, because selecting for low Φ (being close to the target environmental state) is a priori a harder task, requiring more than one type of microbe to be present in the community, whereas a good high-Φ solution may be found with a single microbe type. Single-species solutions are found more often in ecosystems selected with migrant pool sampling (which mixes individuals from several source communities and is thus analogous to sexual reproduction), because this method tends to break associations among species and thereby decrease the chance that particular species interactions will be stably transmitted.
The flask ecosystems used in the experiments reported here meet the three criteria for units of selection (18). Phenotypic variation among units occurs by sampling error when inoculating a new batch of flasks from the previous batch and by mutation during ecosystem development. Differential fitness based on this variation is externally imposed by the nature of the artificial selection process. Heritability can be inferred from the observed response to selection [although no direct measurements were made of the similarity of offspring ecosystems to their parent ecosystem(s)]. If there were no heritability, no sustained deviation from the control line would have been observed. The heritable information is encoded in the genotypes of the different microbe species and their relative frequencies in the flask communities, and transmitted by the inoculation method, leading to the formation of new ecosystems similar to the source ecosystem that provided their inoculum.
Although the simulated ecosystems used here meet the criteria for units of selection within the artificial ecosystem selection scenario, caution must be exercised in drawing any inferences concerning ecosystem-level selection in nature. The artificial ecosystem selection experiments described here [and elsewhere (refs. 1, 2, 11–13)] impose highly specialized conditions: the ecosystems are isolated by hosting each microbial population in a separate container; the transmission mechanism by which individuals are sampled and used to inoculate new flasks is externally provided; and differential fitness at the ecosystem level results from an arbitrary measure of the abiotic environment and does not take into account differences in viability or proliferation. Hence, although we note a strong response to artificial ecosystem selection using the more naturally plausible migrant pool sampling method, all we can say regarding the operation of higher-level selection in nature is that if the same experimental conditions (that were externally imposed here) occur in a natural setting, it is theoretically possible for selection at the level of the ecosystem to shape ecosystem properties and the underlying community. It has been argued elsewhere that spatial separation of semi-isolated populations may provide similar conditions and allow some forms of higher-level selection to operate (19). A spatial version of this model is currently in preparation that will be used in part to determine the scope (if any) for natural ecosystem selection in a more realistic setting.
Methods
Each run in an artificial ecosystem selection experiment involves a batch of 20 flasks, subjected to the same randomly generated set of flux parameters for nutrients and abiotic factors throughout, giving identical environmental conditions in the absence of microbial activity. At the start of each run, the liquid medium in each flask is allowed to reach equilibrium before being seeded with a microbial inoculum. An iteration of ecosystem selection is defined as the creation of a new batch of flask ecosystems (using microbes sampled from the fittest ecosystems of the previous iteration), the propagation of these ecosystems for a fixed period, and the subsequent assignation of a fitness score to each flask. For the first iteration, the initial batch of flask ecosystems is seeded with inocula made up of microbes with randomly generated genotypes. In subsequent iterations, the inoculum for each flask is made up of individuals sampled (with replacement) from the fittest flasks from the previous iteration. For propagule sampling, 100 individuals were randomly sampled from the fittest ecosystem and used to seed all 20 ecosystems in the next batch. For migrant pool sampling, 25 individuals were randomly sampled from each of the four fittest flask ecosystems and the resulting 100 individuals used to seed the next batch of 20 ecosystems.
Flask ecosystem fitness was based on the distance (Φ) of the abiotic environment from an arbitrary target state (see Eq. 1). To measure Φ, the target and actual state vectors of the abiotic environment are normalized, so that the vector of relative proportions of all abiotic factors is the ecosystem property on which artificial selection is based. Φ is measured on the final state of each flask after each iteration. This method should reduce noise, because the state of the abiotic environment is in part a cumulative function of the environment-altering activity of the population over time. The high line was selected to maximize Φ. The low line was selected to minimize Φ. For the control line, source ecosystems were selected at random from the previous batch. The randomly selected control line was used, because an abiotic line would not control for any inherent tendencies of the flask ecology to alter their environment in a particular way irrespective of higher-level selection. All lines also had to meet a criterion of viability, that is, only flask ecosystems with a living microbial population could be selected to provide inocula for the next iteration.
The first set of experiments was designed to look at the effects on the response to artificial ecosystem selection of different sampling methods, varying mutation rate, and varying the propagation time (the time period for which each ecosystem was allowed to develop before fitness testing). The default settings are a target vector (ā1,ā2,ā3) = (0.2, 0.3, 0.5), propagation time, Tprop = 5,000 timesteps, and individual mutation rate, Pmut = 0.01, i.e., a 1% chance of a new allele value at each offspring locus during each reproduction event. First, the effect of different sampling methods is considered for the default settings. Second, the effect of varying the mutation rate (Pmut) during microbe reproduction is examined for the same target vector and propagation time, with the propagule sampling method. Values for Pmut are taken from the set {0,0.01,0.03,0.05,0.1}. Third, the effect of varying the propagation time (Tprop) is examined for the default target vector and individual-level mutation rate, again with the propagule method. Values for Tprop (measured in simulation time steps) are taken from the set (2,000, 5,000, 10,000, 20,000). Each run has a population of 20 flask ecosystems, undergoing 60 ecosystem selection iterations (e.g., 300,000 time steps when Tprop = 5,000), with 30 iterations of directed selection followed by a further 30 iterations of random selection. In each run, the initial seed population and flux parameters were held fixed for all flask ecosystems and treatments (high, low, and random), thus giving identical initial conditions. However, in each experiment, a number of runs are undertaken with different initial seed populations and flux parameters; this repetition allows reliable results to be generated, despite the high level of stochasticity in the system. Φ values are recorded for each ecosystem at the end of each iteration. For each run, Φ is then averaged over all flasks in the population. Finally, Φ is averaged over all runs in each experiment to give the values used in the results.
The evolved ecosystems were tested to determine their performance in perturbed conditions and to provide data for the second group of experiments. For these tests, a baseline value for Φ is required for each artificially selected ecosystem. This is obtained by running a mock iteration of ecosystem selection: 20 identical propagules of 100 individuals sampled from the fittest ecosystem in the final (30th) iteration of directed ecosystem selection, were run forward with all parameters duplicated from the selection experiments but with mutation disabled (Pmut = 0) to prevent it changing the species composition of the community. Short runs were used (Tprop = 2,000), because in the absence of mutation, this is sufficient for the ecosystem to reach a reasonably steady state. The mean final value of Φ measured across all 20 replicated ecosystems was taken to be the baseline score for comparison. The effect of perturbations to the material fluxes through the flask environment was found by running a similar test on the selected community with randomly chosen flux parameters that were different from its normal conditions. For each selected ecosystem, a propagule of 100 individuals was used to inoculate 20 flasks, in each of which the flux parameters were randomly generated (all other parameters were kept the same). After a mock iteration, Φ was measured for each of the 20 variant sets of flux parameters.
The second group of experiments was designed to establish the basis of the response to artificial ecosystem selection and whether it was created by selection acting above the level of the clonal group. This was done by looking for species in each selected community that were capable of inducing the observed functionality on their own. For every species in each selected ecosystem, a propagule of 100 individuals was allowed to develop as a monoculture population. After a mock iteration, Φ was measured for each clonal variant. In another test, for every species from each selected ecosystem, 25 individuals were combined with 75 randomly sampled individuals from the associated control line community (because these control line communities are adapted to the same environmental conditions as the artificially selected community, the control line community acts as a non-selected wild-type community). After a mock iteration, Φ was measured for each composite community. Twenty repetitions were performed and mean Φ scores taken to account for variation between runs.
To verify O4, the expected score (ΦE) for each intact community was compared with the observed community score (ΦO). ΦE was found as the weighted sum of the monoculture Φ values for each species in the selected community:
 |
where S is the number of species, pi is the fraction of the total community made up of species i, and Φi is the monoculture score of species i. In the comparisons of ΦO and ΦE used in verifying O4, mean values from all 20 repetitions of the baseline test of the selected community were used and a significant difference among them was said to exist only if the absolute difference between the mean values for ΦO and ΦE was greater than the sum of their standard deviations.
Further details of the model are in SI Text. Model parameter values are given in SI Table 2, and any deviations from these values are noted in the text.
Supplementary Material
Acknowledgments
This work was supported by the Leverhulme Trust.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0610038104/DC1.
References
- 1.Swenson W, Wilson DS, Elias R. Proc Natl Acad Sci USA. 2000;97:9110–9114. doi: 10.1073/pnas.150237597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Swenson W, Arendt J, Wilson DS. Environ Microbiol. 2000;2:564–571. doi: 10.1046/j.1462-2920.2000.00140.x. [DOI] [PubMed] [Google Scholar]
- 3.Sober E, Wilson DS. Unto Others: The Evolution and Psychology of Unselfish Behavior. Cambridge, MA: Harvard Univ Press; 1997. [Google Scholar]
- 4.Goodnight CJ. Popul Ecol. 2005;47:3–12. [Google Scholar]
- 5.Wade MJ. Evolution (Lawrence, Kans) 1977;31:134–153. doi: 10.1111/j.1558-5646.1977.tb00991.x. [DOI] [PubMed] [Google Scholar]
- 6.Goodnight CJ. Evolution (Lawrence, Kans) 1990;44:1614–1624. doi: 10.1111/j.1558-5646.1990.tb03850.x. [DOI] [PubMed] [Google Scholar]
- 7.Goodnight CJ. Evolution (Lawrence, Kans) 1990;44:1625–1636. doi: 10.1111/j.1558-5646.1990.tb03851.x. [DOI] [PubMed] [Google Scholar]
- 8.Goodnight CJ, Stevens L. Am Nat. 1997;150:S59–S79. doi: 10.1086/286050. [DOI] [PubMed] [Google Scholar]
- 9.Traulsen A, Nowak MA. Proc Natl Acad Sci USA. 2006;103:10952–10955. doi: 10.1073/pnas.0602530103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilson DS. Ecology. 1992;73:1984–2000. [Google Scholar]
- 11.Penn A. In: Banzhaf W, Christaller T, Dittrich P, Kim JT, Ziegler J, editors. Advances in Artificial Life: Proceedings of the 7th European Conference on Artificial Life (ECAL 2003); Heidelberg, Germany: Springer; 2003. pp. 659–666. [Google Scholar]
- 12.Penn A, Harvey I. In: Pollack J, Bedau M, Husbands P, Ikegami T, Watson RA, editors. Proceedings of the Ninth International Conference on the Simulation and Synthesis of Living Systems (ALIFE 9); Cambridge, MA: MIT Press; 2004. pp. 352–357. [Google Scholar]
- 13.Penn A. Brighton, UK: University of Sussex; 2005. PhD thesis. [Google Scholar]
- 14.Chow SS, Wilke CO, Ofria C, Lenski RE, Adami C. Science. 2004;305:84–86. doi: 10.1126/science.1096307. [DOI] [PubMed] [Google Scholar]
- 15.Williams HTP. Leeds, UK: University of Leeds; 2006. PhD thesis. [Google Scholar]
- 16.Williams HTP, Lenton TM. Oikos. 2007 in press. [Google Scholar]
- 17.Loreau M. Oikos. 1998;82:600–602. [Google Scholar]
- 18.Lewontin RC. Annu Rev Ecol Syst. 1970;1:1–18. [Google Scholar]
- 19.Johnson CR, Boerlijst MC. Trends Ecol Evol. 2002;17:83–90. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.