Abstract
The presence of repeated DNA sequences is a genomic liability, because interrepeat recombination can result in chromosomal rearrangements. The mismatch repair system prevents recombination between nonidentical repeats, but the mechanism of antirecombination has not been established. Although the MutS protein binds to base pair mismatches in heteroduplex DNA, the role of the MutL protein in preventing recombination is unknown. In a screen designed to identify new cellular functions that suppress deletion formation involving nonidentical DNA repeats, we isolated a mutL mutant having a separation-of-function phenotype. The mutant showed an increased frequency of deletions but not of mutations. The split phenotype is due to a decreased MutL level, indicating that recombination, but not replication editing, is highly sensitive to MutL level. By altering the MutL level, we found that the frequency of deletion-generating recombination is inversely related to the amount of cellular MutL. DNA sequence analysis of the recombined repeats shows that the tolerance of base pair mismatches in heteroduplex DNA is also inversely correlated with MutL level. Unlike recombination, correction of misincorporation errors by mismatch repair is insensitive to fluctuations in MutL level. Overproduction of MutS does not affect either of these phenotypes, suggesting that, unlike MutL, MutS is not limiting for mismatch repair activities. These results indicate that MutL (i) determines effective DNA homology in recombination processes and (ii) fine tunes the process of deletion formation involving repeated, diverged DNA sequences.
Keywords: deletions, DNA repeats, mismatch repair, recombination, replication
Prokaryotic genomes are riddled with DNA sequence repeats, which are found in intergenic regions, genes, and transposable elements (1). Repeats are even more abundant in eukaryotic genomes (2), and at least 34% of the human genome consists of such sequences (3). The presence of dispersed repeated DNA sequences is a genetic liability because interrepeat recombination can cause deletions, duplications, inversions or translocations, depending on the configuration and orientation of the repeat units. In humans, recombination between repeats is at the origin of disease-causing deletions, such as α-thalassemias, Duchenne muscular dystrophy, and familial hypercholesterolemia (4–7).
Recombination between repeats is constantly initiated by DNA damage and/or DNA replication blockage that results from exogenous and endogenous genomic insults. Therefore, natural selection for genome stability has resulted in the emergence of mechanisms that prevent interrepeat recombination. Newly arising repeats are identical at the DNA sequence level, but established repeats have usually diverged (8). The degree and distribution of sequence divergence are structural parameters that influence recombination between repeats because they affect the activity of recombination enzymes and determine whether the antirecombinogenic activity of mismatch repair (MMR) proteins is triggered (9). The Escherichia coli RecA protein is selective for sequence identity only at the initial stages of the recombination process. The minimum amount of sequence identity, called MEPS (minimal efficient processing segment) required for the efficient initiation of RecA-dependent strand exchange is 23–27 bb (10). Once initiated, strand exchange can be extended despite the presence of numerous mismatches and even large heterologies (11, 12). At this stage, recombination is controlled by the MMR system, which recognizes mismatches in heteroduplex regions and blocks in vitro RecA-catalyzed strand transfer and RuvAB-dependent branch migration (13, 14).
Therefore, inactivation of the MMR system in both prokaryotes and eukaryotes can lead to an increase in RecA-dependent recombination between nonidentical DNA sequences (15–17). Besides the hyperrecombination phenotype, MMR inactivation in prokaryotes and eukaryotes results in a mutator phenotype characterized by an increased frequency of base substitutions and frameshift mutations (18). MMR-deficient prokaryotic and eukaryotic mutants also show an increase in RecA-independent recombination events, such as replicative misalignment and single-strand annealing, which, along with RecA-dependent events, contribute to the generation of rearrangements mediated by sequence repeats (19, 20). These findings emphasize that molecular strategies for preventing interrepeat recombination are conserved throughout evolution.
The aim of this study was to identify not yet discovered cellular functions that suppress deletion formation mediated by nonidentical DNA repeat recombination. To this end, we devised a recombination assay that detects deletions between DNA repeats of 4% sequence divergence inserted on the E. coli chromosome. This extent of divergence is comparable with that observed among the ≈105 copies of LINEs (long interspersed elements) in the human genome (21). Using this experimental system, we screened a transposon-generated mutant library and found a mutL mutant that has a separation-of-function phenotype, i.e., it shows a hyperrecombination, but not mutator, phenotype. Because this split phenotype was due to a decreased MutL level, we concluded that a reduction in MutL specifically affects MMR-mediated recombination control. Analysis of the frequency and the structure of deletions from cells with different MutL and MutS levels suggest that MutL plays a pivotal role in controlling recombination efficacy.
Results
Construction and Characterization of the Recombination Assay.
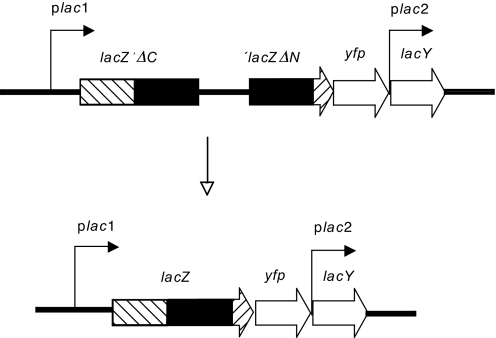
To score for deletions, we devised a recombination assay consisting of two truncated, nonfunctional chromosomal E. coli lacZ genes separated by a 0.5-kb spacer (Fig. 1). The lacZ fragments share a 1.3-kb long overlapping region that is either identical or 4% diverged because of nucleotide substitutions. The identical lacZ fragments are derived from the MG1655 lacZ gene, whereas the 4% diverged fragments are derived from the MG1655 and CFT073 lacZ alleles. A single recombination event between the two gene fragments reconstitutes a functional lacZ gene by deleting the intervening region (Fig. 1).
Fig. 1.
Chromosomal construct used to measure deletion formation through recombination between identical or diverged repeats. Two nonfunctional lacZ genes are cloned in close proximity (0.5 kb) on the E. coli chromosome. The first lacZ gene contains a C-terminal deletion (lacZ′ΔC), whereas the second contains an N-terminal deletion (′lacZΔN). The two gene fragments share an overlapping region of 1.3 kb (filled boxes) that is 100% identical or 4% diverged at the DNA sequence level. Nonoverlapping lacZ gene C-terminal and N-terminal regions are presented as hatched boxes. A single recombination event between two gene fragments reconstitutes a functional lacZ gene and deletes the intervening region. plac, lactose promoter; lacZ, gene coding for β-galactosidase; lacY, gene coding for lactose permease; yfp, gene coding for yellow fluorescent protein.
To characterize this assay, we tested molecular mechanisms known to affect recombination between repeated sequences in E. coli. RecA-dependent recombination accounted for 88% and 95% of the deletions involving identical and diverged repeats, respectively (Table 1), whereas the remaining deletions were RecA-independent events, e.g., caused by replicative misalignment or single-strand annealing. DNA divergence between the two lacZ genes reduced the frequency of RecA-dependent deletions by 270-fold and that of RecA-independent deletions by 660-fold (Table 1). Inactivation of either the recF or recB gene had no or only a small effect on the deletion frequency, but the combined inactivation of both recF and recB almost completely abolished RecA-dependent recombination between diverged DNA repeats (Table 1). Substrates for the RecFOR pathway, single-strand breaks and gaps, can be converted to double-strand breaks that are processed by the RecBCD pathway, whereas the reverse is not possible (22). Therefore, it may be concluded that single-strand breaks and gaps are the dominant substrates for recombination initiation in the wild-type background, whereas a minority of recombination events is initiated by double-strand breaks.
Table 1.
Recombination phenotypes of strains with varying amounts of MutS and MutL proteins
| Genotype | Recombination frequency |
||
|---|---|---|---|
| Mean ± SEM | Fold difference | n | |
| 100% identical lacZ repeats | |||
| Wild-type | 6.8 × 10−4 ± 6.0 × 10−5 | 1.00 | 6 |
| recA | 7.9 × 10−5 ± 1.2 × 10−5 | 0.11* | 3 |
| mutS | 6.1 × 10−4 ± 5.5 × 10−5 | 0.89 | 3 |
| mutL | 6.5 × 10−4 ± 1.0 × 10−4 | 0.94 | 3 |
| mutLdown + tc | 6.7 × 10−4 ± 6.2 × 10−5 | 0.98 | 3 |
| Wild-type pVector | 1.8 × 10−3 ± 6.3 × 10−4 | 1.00 | 3 |
| Wild-type pMutL | 1.4 × 10−3 ± 2.8 × 10−4 | 0.79 | 3 |
| Wild-type pMutS | 2.2 × 10−3 ± 4.7 × 10−4 | 1.20 | 3 |
| 4% diverged lacZ repeats | |||
| Wild-type | 2.5 × 10−6 ± 2.0 × 10−7 | 1.00 | 6 |
| recA | 1.2 × 10−7 ± 1.0 × 10−8 | 0.05* | 4 |
| recF | 2.1 × 10−6 ± 6.4 × 10−7 | 0.83 | 3 |
| recB | 1.1 × 10−6 ± 1.1 × 10−7 | 0.45* | 5 |
| recF recB | 1.3 × 10−7 ± 1.6 × 10−8 | 0.05* | 6 |
| lexA1 | 2.3 × 10−6 ± 3.3 × 10−7 | 0.93 | 5 |
| mutS | 6.4 × 10−4 ± 7.9 × 10−5 | 251.14* | 4 |
| mutL | 5.1 × 10−4 ± 6.8 × 10−5 | 199.49* | 3 |
| mutH | 9.9 × 10−5 ± 2.9 × 10−5 | 38.90* | 3 |
| uvrD | 2.0 × 10−4 ± 7.5 × 10−5 | 81.30* | 4 |
| dam | 1.7 × 10−4 ± 1.2 × 10−5 | 65.99* | 5 |
| mutLdown+ tc | 1.1 × 10−5 ± 2.6 × 10−6 | 4.99* | 6 |
| mutLdown − tc | 3.1 × 10−4 ± 2.6 × 10−5 | 121.41* | 3 |
| Wild-type pVector | 5.2 × 10−6 ± 4.6 × 10−7 | 1.00 | 4 |
| Wild-type pMutL | 2.2 × 10−6 ± 3.5 × 10−7 | 0.42* | 6 |
| Wild-type pMutS | 8.1 × 10−6 ± 3.7 × 10−6 | 1.55 | 5 |
| recA | 1.2 × 10−7 ± 1.0 × 10−8 | 1.00 | 4 |
| recA mutS | 5.6 × 10−5 ± 5.7 × 10−6 | 440.37* | 3 |
| recA mutL | 7.0 × 10−5 ± 4.8 × 10−5 | 548.53* | 3 |
| recA mutH | 3.2 × 10−5 ± 1.7 × 10−5 | 254.27* | 3 |
| recA mutLdown + tc | 8.6 × 10−7 ± 9.9 × 10−8 | 6.80* | 3 |
| recA pVector | 5.8 × 10−7 ± 1.9 × 10−7 | 1.00 | 6 |
| recApMutL | 2.9 × 10−7 ± 5.7 × 10−8 | 0.49 | 6 |
| recApMutS | 5.9 × 10−7 ± 2.1 × 10−7 | 1.02 | 6 |
Genetic requirements for deletion formation between identical and diverged repeats are shown. Fold difference is the ratio of the value for a given strain to the value of the control strain, i.e., the first strain in every subdivision of the column. tc, tetracycline.
*Difference is significant according to Mann–Whitney test, P < 0.05.
Because double-strand breaks and single-strand gaps induce the SOS response, resulting in overexpression of the recombination genes recA, recN, and ruvAB (23), we also tested the involvement of this response in deletion formation between diverged DNA repeats by assessing the effect of the lexA1 mutation. The lexA1 allele encodes a noncleavable SOS repressor which prevents induction of the SOS response (24, 25). As shown in Table 1, this allele did not affect recombination efficiency, suggesting that the basal level of SOS recombination functions is not limiting for spontaneous deletion events.
Because MMR is known to be a potent inhibitor of recombination between nonidentical DNA sequences (9, 15), we tested the effect of the key E. coli MMR proteins MutS, MutL, and MutH on deletion events mediated by diverged repeats (26). MutS recognizes and binds to base pair mismatches, whereas MutL associates with MutS-mismatch complexes and activates the MutH protein. MutH is an endonuclease that recognizes and cuts unmethylated DNA strands 5′ to a GATC sequence. The GATC sites on the newly synthesized DNA strands are unmethylated because methylation of adenine by the Dam (DNA adenine methylase) protein lags behind replication by several minutes. Hence, MutH directs excision and resynthesis to the newly synthesized strands. Inactivation of MMR did not affect the frequency of deletions between identical sequences but specifically increased the frequency of both RecA-dependent and RecA-independent deletions between diverged sequences (Table 1). Remarkably, inactivation of the mutS or mutL gene was sufficient to restore the deletion frequency, despite a 4% sequence divergence, to the levels seen for the RecA-dependent and RecA-independent pathways of identical repeat recombination (Table 1). The effect of the mutH mutation on RecA-dependent deletions was smaller compared with the effects of the mutS and mutL mutations (38-, 251-, and 199-fold, respectively; Table 1), a difference that was observed in other recombination systems involving nonidentical DNA sequences (15, 27). These observations are consistent with a limited involvement of DNA synthesis generating unmethylated GATC sequences in the RecA-dependent deletion process.
In contrast, the RecA-independent deletion frequency involving diverged repeats increased to a similar extent upon inactivation of the mutS, mutL, and mutH genes (440, 548- and 254-fold, respectively; Table 1), comparable with the results obtained in a plasmid recombination assay (20). Because the mutS, mutL, and mutH mutants also show a similar mutator phenotype resulting from their defect in correcting misincorporation errors (28), we conclude that the mechanism of RecA-independent deletion formation in our system involves a newly synthesized unmethylated DNA strand that bypasses sequences between repeated homologies by misalignment. We presume that the misaligned hemimethylated heteroduplex is a substrate for MutSLH. An alternative RecA-independent deletion pathway, single-strand annealing, is not predominant because it does not involve the extensive synthesis of unmethylated GATC sequences required for MutH activity (29). Therefore, MMR activity is sufficient to prevent deletion formation by both RecA-dependent and RecA-independent recombination pathways that act on diverged sequence repeats.
Isolation and Characterization of Hyperrecombination Mutants.
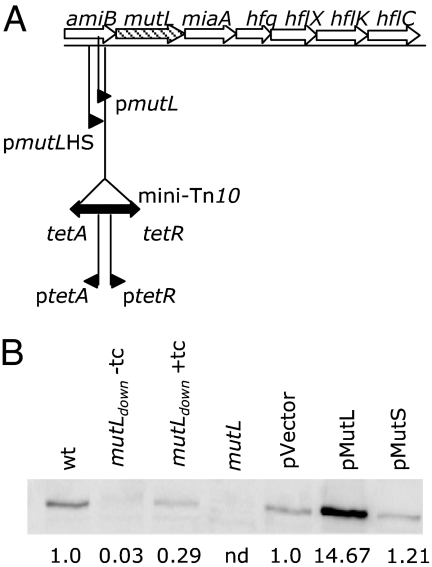
With this defined experimental system, we used a papillation assay to screen a mutant library generated by random insertions of the Tn10 transposon, which carries a tetracycline-resistant marker, to identify new mutants with an increased frequency of deletions mediated by nonidentical repeats [see supporting information (SI) Fig. 5 and SI Text for protocol]. Because all previously described mutants having such a phenotype are MMR-deficient mutators, we characterized only the nonmutator candidates. The mutator phenotype was revealed by an increased frequency of spontaneous mutations that confer resistance to rifampicin. Of 75,000 mutants, only 2 satisfied our criteria, i.e., they exhibited a hyperrecombination but not mutator phenotype. One of these, the rdgB mutation, had previously been found to stimulate recombination by generating more recombination substrates through the increased incorporation of hypoxanthine (30). Another mutation was localized to the amiB gene, which encodes a cell-wall amidase that is not known to be involved in recombination (31). Because the hyperrecombination phenotype of the amiB mutant was not reversed by complementation with a functional amiB gene (data not shown), we concluded that the inserted transposon most likely has a polar effect on transcription. The site of transposon insertion was found to reside between mutL gene promoters localized in the C-terminal region of the amiB gene and the mutL start codon (Fig. 2A). When this amiB mutant strain was grown in the presence of tetracycline, it contained ≈4-fold less MutL protein than the wild type (Fig. 2B). There was almost no detectable MutL protein in cells grown without tetracycline (Fig. 2B), suggesting that the transposon insertion prevents mutL gene expression from native promoters and that, in the presence of tetracycline, the low level of MutL results from induction of one of the transposon-borne promoters. Thus, we have isolated a mutL mutant strain with a 4-fold reduced level of MutL that shows a hyperrecombination but not mutator phenotype.
Fig. 2.
Quantification of MutL protein level in different strains. (A) Position of the miniTn10 insertion within the amiB-mutL–miaA-hfq-hflX superoperon. pmutL and pmutLHS, mutL gene promoters; miniTn10 (hatched box), transposon; ptetA and ptetR, Tn10 tetracycline-inducible promoters. (B) Cellular level of MutL protein determined by immunoblotting. Values represent fold difference over the control calculated from the average of three independent experiments. wt, wild-type strain; mutLdown, mutant strain with miniTn10 insertion in the amiB gene; pMutL and pMutS, wild-type strains carrying plasmids that overexpress MutL or MutS proteins, respectively; pVector, wild-type strain carrying control plasmid; tc, tetracycline; nd, not detected.
Recombination and Mutation Phenotypes as a Function of MutL Concentration.
To examine the effects of altering the cellular levels of MutL protein on recombination and mutagenesis, we assessed strains having 15-fold more, 4-fold less, and 33-fold less MutL than the control (Fig. 2B). Mutagenesis was measured by determining the frequency of substitution mutations conferring resistance to rifampicin and nalidixic acid. As previously observed for frameshifts (32), the frequency of substitution mutations was unaffected by 15-fold MutL overexpression, suggesting that the cellular level of MutL protein is not limiting for replication editing (Table 2). The mutation frequency was increased in the strain with 33-fold less MutL, but it was unaffected in the strain with 4-fold less MutL (Table 2). The mutation spectrum in the rpoB gene in rifampicin-resistant mutants that arose in the strain with 4-fold less MutL was identical to that in the wild-type strain but very different from that in the mutL-deficient strain (Table 3). Therefore, neither the nature nor the frequency of substitution mutations is affected by decreasing the level of MutL by a factor of four. To determine whether the absence of a mutagenesis effect in the strain with 4-fold less MutL is due to a low number of mismatches generated per replication cycle that could be easily repaired by this level of MutL, we increased the number of replication errors by treating cells with the base analogue 2-aminopurine. However, the frequency of substitution mutations increased in a similar dose-dependent manner in both the wild-type strain and in the mutant with 4-fold less MutL, after 2-aminopurine treatment (SI Fig. 6). In conclusion, a 4-fold reduction in the MutL level does not affect the correction of misincorporation errors.
Table 2.
Frequency of spontaneous mutations conferring resistance to rifampicin and nalidixic acid
| Genotype | Mutation frequency per 109 cells |
||
|---|---|---|---|
| Mean ± SEM | Fold difference | n | |
| RifampicinR | |||
| Wild type | 19.3 ± 3.7 | 1.0 | 39 |
| mutLdown + tc | 22.4 ± 4.3 | 1.1 | 39 |
| mutLdown − tc | 2,468 ± 286 | 127.8* | 20 |
| mutL | 6,504 ± 394 | 336.6* | 32 |
| Wild-type pVector | 37.2 ± 14.2 | 1.0 | 19 |
| Wild-type pMutL | 51.8 ± 20.4 | 1.3 | 20 |
| Wild-type pMutS | 23.8 ± 3.5 | 0.6 | 10 |
| Nalidixic acidR | |||
| Wild type | 2.7 ± 0.6 | 1.0 | 17 |
| mutLdown + tc | 2.4 ± 0.6 | 0.9 | 16 |
| mutLdown − tc | 990 ± 116 | 361.3* | 20 |
| mutL | 1,450 ± 144 | 531.2* | 35 |
Fold difference is the ratio of the value for a given strain to the value of the control strain, i.e., the first strain in every subdivision of the column. tc, tetracycline.
*Difference is significant according to Mann–Whitney test, P < 0.05.
Table 3.
Percentage of different mutation types in rpoB
| Genotype | Transitions |
Transversions |
||||
|---|---|---|---|---|---|---|
| AT→GC | GC→AT | AT→TA | AT→CG | GC→TA | GC→CG | |
| Wild type | 28 | 49 | 15 | 4 | 2 | 2 |
| mutLdown + tc | 27 | 43 | 20 | 2 | 9 | 0 |
| mutL | 88 | 11 | 0 | 0 | 0 | 0 |
Fifty-three, 56, and 44 rpoB genes of wild-type, mutLdown + tc, and mutL rifampicinR mutants were analysed by sequencing, respectively; For sequencing protocol see SI Text. tc, tetracycline.
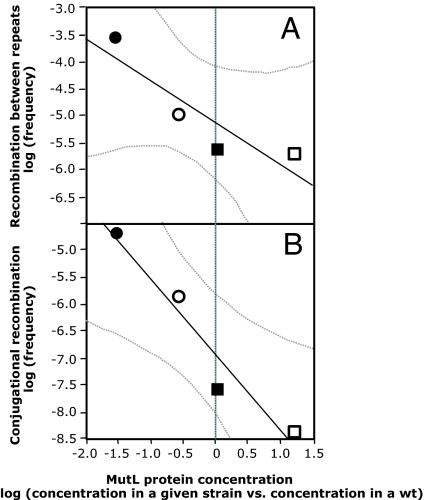
We then measured the effects of altering cellular MutL levels on RecA-dependent recombination. We observed an inverse relationship between the MutL level and the frequency of recombination between nonidentical repeats (Fig. 3A), whereas altering MutL levels had no detectable effect on recombination between identical sequences (Table 1). Very similar results were obtained for recombination in interspecies conjugational crosses involving bacterial partners diverged by 20% (Fig. 3B; SI Table 5). Thus, the efficacy of RecA-dependent recombination between nonidentical DNAs is very sensitive to fluctuations in MutL levels.
Fig. 3.
Correlation between varying the level of MutL protein and the efficacy of recombination between diverged DNA sequences. (A) RecA-dependent recombination between 4% diverged lacZ repeats (R2 = 0.79). (B) RecA-dependent conjugational recombination between Salmonella enterica serovar Typhimurium Hfr and Escherichia coli F− recipients, which are 20% diverged between homologous genes (R2 = 0.94). Wild-type strain (filled squares), strain with 33-fold less MutL (filled circles), strain with 4-fold less MutL (open circles), and wild-type strain overexpressing MutL (open squares) are shown. The dotted curves show the 95% confidence interval around the regression. For MutL protein quantification see Fig. 2B.
Simultaneous overexpression of the mutS and mutL genes was shown to decrease the frequency of conjugational recombination between weakly diverged bacterial genomes by several orders of magnitude, but which of the two proteins is limiting was not determined (33). Overexpression of mutS (18-fold, data not shown) did not affect either deletion formation involving diverged repeats or interspecies recombination (Table 1 and SI Table 5). Therefore, the control of RecA-dependent recombination cannot be enhanced by increasing the concentration of the MutS mismatch detector protein, but it can be improved by overproducing MutL, which acts at a later stage of MMR (Fig. 3 A and B). In contrast, RecA-independent recombination, although very sensitive to decreases in the MutL level and to mutL inactivation, was not significantly affected by MutL or MutS overexpression (Table 1).
The Relationship Between MutL Protein Concentration and the Structure of Recombinants.
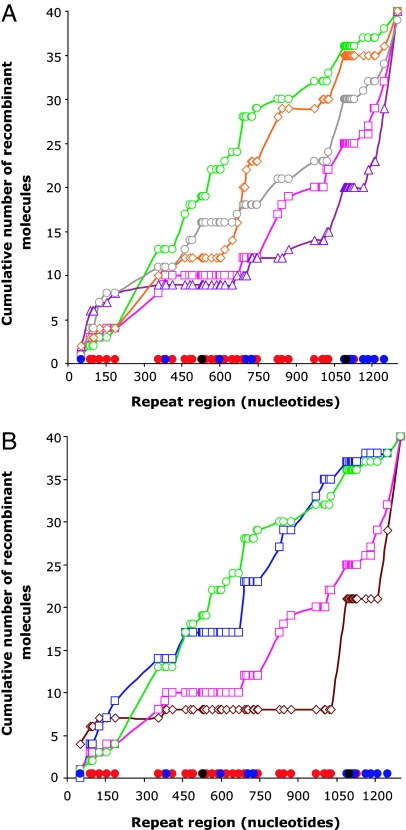
To explore how the recombination is regulated by MMR at the molecular level, we investigated the structure of deletion products isolated from strains with different concentrations of MutL or MutS proteins (Fig. 4A and SI Fig. 7A). The two diverged truncated lacZ genes differ by 62 base substitutions, which can be used as markers to delimit regions in which strand exchange took place. In the RecA-proficient strain, the position of the junction between the two parental sequences corresponds to the strand exchange initiation site and/or to the DNA heteroduplex resolution site, in 95% of the deletion products (Table 1). In the mutL− strain, the positions of junctions were evenly distributed across the entire length of the aligned lacZ repeats, whereas in the MutL-overproducing strain, they almost exclusively localized to the two borders of the lacZ repeats (Fig. 4A and SI Fig. 7A). The distribution of junctions in the wild-type strain and in the mutant with 4-fold less MutL was intermediate between these two extreme cases, indicative of a MutL concentration effect. The sequence analysis of recombinant lacZ genes showed that, with increasing cellular MutL concentration, recombinants with junctions in the middle of the lacZ repeats became progressively underrepresented; this region contains the highest density of mismatches well recognized by MMR (Fig. 4A and SI Fig. 7A). This observation can be accounted for by blockage of strand exchange initiation and/or by blockage of DNA heteroduplex extension with increasing MutL concentration.
Fig. 4.
Structure of lacZ+ recombinants as a function of cellular MutL and MutS level. For each recombinant, the junction is assigned to a region delimited by mismatches in aligned lacZ repeats. The cumulative distribution of junction positions for the strains examined is shown. Mismatches that are well (red filled circles; G–T and A–C), less efficiently (blue filled circles; A–G and C–T), or not at all (black filled circles; C–C) recognized by the MMR system are shown on the x axes. (A) RecA-dependent lacZ+ recombinants. Wild-type strain (pink open squares and lines), mutL strain (green open circles and lines), strain with 4-fold less MutL (red open diamonds and lines), wild-type strains overexpressing MutL (purple open triangles and lines), and MutS proteins (gray open circles and lines) are shown. (B) RecA-independent lacZ+ recombinants. recA strain (brown open diamonds and lines) and recA mutL strain (blue open squares and lines) are shown. For comparison wild-type strain (pink open squares and lines) and mutL strain (green open circles and lines) are given. For sequencing protocol see SI Text.
Contrary to the overproduction of MutL, MutS overproduction did not decrease the deletion frequency (Table 1), but it modified the distribution of junction points (Fig. 4A and SI Fig. 7A). Junctions were distributed even more randomly than in the wild-type strain, which could be explained by the stabilization of mismatched heteroduplexes by an excess of MutS.
MMR deficiency significantly changed the distribution of resolution points, not only in the RecA-proficient strain, but also in the recA− background (Fig. 4B and SI Fig. 7B). The extent to which the junction distribution was modified by MMR in RecA-deficient and RecA-proficient strains suggests that RecA-independent deletion events are more efficiently prevented by MMR than are RecA-dependent events (Fig. 4 and SI Fig. 7). This interpretation is in agreement with our genetic data showing that MMR more significantly inhibits RecA-independent deletion events than RecA-dependent events (Table 1).
Discussion
Studies of the effects of DNA sequence divergence on the efficiency of homologous recombination show that there is an exponential decrease in recombination frequency with a linear increase in DNA divergence (33, 34). This relationship results from an exponential decrease in the number of available MEPSs, i.e., minimal effective DNA homology during recombination process (33). A change in the concentration of MMR proteins changes the slope of this relationship by altering MEPS length, e.g., an increase in the concentration of MMR proteins increases the size of MEPS and thus reduces the efficiency of recombination. In light of our data, we propose that the MutL protein plays a key role in determining MEPS size. MutL moderates the ATP-dependent dissociation of MutS from DNA, thus considerably increasing the longevity of the MutS-mismatch complex (35). More cellular MutL protein probably results in a longer sequestration of MutS protein on mismatches and therefore more effectively blocks the strand exchange process. It was indeed observed that the inhibition of in vitro strand exchange between 3% diverged DNA substrates increases with rising concentration of MutL protein at a constant level of MutS protein (L. Worth, Jr., personal communication). Similarly, the probability at which MutS binds to heteroduplex DNA containing a single mismatch in vitro increases with rises in MutL (36).
Three processes that are regulated by MMR are differentially affected by MutL overproduction. Although the MMR-mediated control of RecA-dependent recombination is improved by overproducing MutL (Table 1 and SI Table 5), MutL overproduction has no effect on the correction of misincorporation errors (Table 2) or on the control of RecA-independent recombination (Table 1). The mechanism of RecA-independent deletion formation in our assay involves a newly synthesized DNA strand that skips sequences between the repeats by misalignment. This is suggested by the major role of MutH, which cleaves unmymethylated GATC sequences on newly synthesized DNA strands, in the suppression of RecA-independent deletions (Table 1). Therefore, like the correction of misincorporation errors, RecA-independent recombination between repeats involves newly replicated DNA.
How can we explain the differential effect of MutL overexpression on replication and recombination? A recent study showed that overproduction of MutSΔ800, a mutant protein with a C-terminal 53-aa deletion, restores proficiency for mutation avoidance but does not affect the control of recombination between nonidentical sequences in a mutS background (37). This finding suggests that MMR-mediated control of replication and recombination may not involve the same mechanism. The processing of mismatches generated during replication and recombination may require different proteins, or the same proteins could be present at different levels during recombination and replication. For example, the β-clamp (the processivity factor for DNA polymerases) is known to interact with MutS and MutL, and this binding is essential for the correction of replication misincorporation errors (38). The β-clamp is expected to be found around replication forks, where it could target mismatch repair to replication forks and stabilize MutL–MutS-mismatch complexes. Although RecA-dependent recombination is frequently associated with localized DNA synthesis (39), the β-clamp is probably present at higher concentrations at DNA replication forks than associated with intermediates in homologous recombination. Consequently, a higher MutL concentration may facilitate the formation of mismatch-repair complexes on homologous recombination intermediates and/or increase their stability, whereas it would not change the efficiency of misincorporation error correction and of RecA-independent recombination control. The stabilization of MutL–MutS-mismatch complexes may facilitate the recruitment of factors that disrupt recombination intermediates, such as UvrD. Indeed, in vitro studies showed that UvrD helicase activity rises with increasing MutL concentration (40).
Curiously, decreasing MutL levels by 4-fold does not affect the correction of misincorporation errors (Table 2), but it does attenuate RecA-independent recombination (Table 1). The difference between these two processes is that a heteroduplex resulting from misincorporation errors contains a single mismatch, whereas a heteroduplex resulting from replication misalignment is long and contains many mismatches (up to 62 in our case). The disruption of both types of heteroduplex requires the MutS, MutL, and MutH proteins (Table 1 and ref. 26). However, whereas a single MutL–MutS-mismatch complex may be sufficient for processing a single mismatch, several complexes may be required for disruption of long heteroduplex molecules containing multiple mismatches.
In conclusion, our data show that the MutL protein plays a pivotal role in fine-tuning the efficacy of MMR-mediated editing of rearrangements resulting from recombination between repeated diverged DNA sequences. The remarkable conservation of the MMR MutS and MutL components throughout evolution has directed and accelerated research in mammals by fast piloting exploration in bacteria and yeast. Hence, the present work has relevance to a number of topics in cancer research. It is intriguing that the only MMR gene found to be epigenetically down-regulated in human mutator tumors is the mutL ortholog hMLH1 (41). In mammals, loss of heterozygosity caused by mitotic crossovers, resulting in the expression of recessive mutations, can be suppressed by parental sequence polymorphism and depends on MLH-1 activity (42). Furthermore, hMLH1-fluorescent foci are used to detect chiasmata, because hMLH1 localizes to meiotic cross-over points (43), perhaps as an obligate element of the fidelity of recombination?
Materials and Methods
Bacterial Strains, Plasmids, and Media.
Strains, plasmids, and media are described in SI Text and in SI Table 4.
Immunoblotting.
Protein samples were prepared from bacterial cultures inoculated with <100 cells and grown until exponential phase (OD600 = 0.5) in LB at 37°C. Tetracycline was added or omitted from media as indicated. Cells from 25-ml cultures were harvested by centrifugation at 4°C, and pellets were lysed by resuspending in Bugbuster (Novagene, Madison, WI) according to the manufacturer's protocol. The total protein concentration was determined with a Bradford assay kit (Sigma–Aldrich, Saint Quentin Fallavier, France). Total protein (25–50 μg) was loaded onto 10% or 7.5% SDS polyacrylamide gels for detection of MutL (70 kDa) or MutS (97 kDa), respectively. Proteins were electrotransferred to nitrocellulose membranes that were incubated with MutL or MutS antisera overnight. The intensities of MutL and MutS bands were measured with a PhosphorImager (Molecular Dynamics, Sunnyvale, CA).
Determination of Deletion Frequencies.
Bacterial cultures were inoculated with <100 cells to ensure that no preexisting lacZ+ cells were present in the starting inoculum. Cells were grown in LB supplemented with antibiotics when needed, with shaking overnight at 37°C. Recombinants were selected by plating on M63 medium with lactose as sole carbon source (0.2%), supplemented with antibiotics when needed, as follows. (i) Plates were first overlaid with top agar containing scavenger E. coli cells (NEC222) with a nonrevertible deletion of the lacZ gene, which were used to remove any contaminating nonlactose carbon sources in the medium. Scavengers were used in a 10-fold excess relative to tester strains. (ii) On top, a second layer of top agar containing an appropriate dilution of a tester strain was poured. Plates were incubated at 37°C for 48 h. X-Gal and IPTG were added to plates to facilitate the visualization of lacZ+ recombinants. Total cell number was determined by plating appropriate dilutions of the tester strain on M63 medium with glucose as carbon source (0.2%). When MutS and MutL overexpression from multicopy plasmids was assessed, scavenger cells containing control plasmid were used.
Conjugational Crosses.
Interspecies crosses involved S. enterica serovar Typhimurium Hfr SA955 TetR donor and E. coli F− recipients MG1655 wild type, mutL, mutL mutant with 4-fold less MutL, wild-type pMutL, or wild-type pMutS. In intraspecies crosses, E. coli Hfr P4X TetR was used as a donor instead of S. typhimurium Hfr. For conjugation, overnight cultures were diluted 50-fold in LB (supplemented with tetracycline when needed) and grown to ≈108 cells per ml. Donor and recipient were mixed at a 1:1 ratio and immediately deposited on a sterile 0.45-μm pore size filter, which was then incubated on prewarmed rich-medium agar (supplemented with tetracycline when needed). After 1 h at 37°C, cells were resuspended in 10 mM MgS04 and separated by vortexing for 2 min. The exconjugants were plated on minimal M63 medium lacking leucine to select for leu+ recombinants and containing nalidixic acid to counterselect the Hfr donor cells (and supplemented with tetracycline when needed). Recombinants were scored after 48-h incubation at 37°C.
Determination of Mutation Frequencies.
Bacterial cultures were inoculated with <100 cells to ensure that no preexisting mutants were present in the starting inoculum. Cells were grown in LB supplemented with antibiotics when needed, with shaking overnight at 37°C. Appropriate dilutions of cells were plated on selective media (LB containing 100 μg/ml rifampicin or 40 μg/ml nalidixic acid) to detect rifampicin-resistant (RifR) or nalidixic acid-resistant (NalR) colonies and on LB to determine the total number of colony-forming units. Colonies were scored after 24 h of incubation at 37°C.
Supplementary Material
Acknowledgments
We thank A. Lindner for help with the construction of the recombination assay, M. Vulic for advice; the M. Pende laboratory for help with immunoblotting experiments; P. Modrich (Duke University, Durham, NC) and M. Marinus (University of Massachusetts Medical School, Worcester, MA) for their generous gifts of MutL and MutS polyclonal antibodies; S. Rosenberg (Baylor College of Medicine, Houston, TX) and T. Palmer (John Innes Centre, Norwich, U.K.) for providing mutL and amiB plasmids; and M. Pende, M. A. Petit, M. Vulic, S. Delmas, and L. Worth for comments on the manuscript. This work was supported by a grant from the Agence Nationale de la Recherche. M.E. was supported by grants from the Association pour la Recherche Contre le Cancer and the Ligue Nationale Française Contre le Cancer.
Abbreviations
- MEPS
minimal efficient processing segment
- MMR
mismatch repair.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0610149104/DC1.
References
- 1.Achaz G, Rocha EP, Netter P, Coissac E. Nucleic Acids Res. 2002;30:2987–2994. doi: 10.1093/nar/gkf391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Achaz G, Netter P, Coissac E. Mol Biol Evol. 2001;18:2280–2288. doi: 10.1093/oxfordjournals.molbev.a003774. [DOI] [PubMed] [Google Scholar]
- 3.Weiner AM. Curr Opin Cell Biol. 2002;14:343–350. doi: 10.1016/s0955-0674(02)00338-1. [DOI] [PubMed] [Google Scholar]
- 4.Lehrman MA, Schneider WJ, Sudhof TC, Brown MS, Goldstein JL, Russell DW. Science. 1985;227:140–146. doi: 10.1126/science.3155573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harteveld KL, Losekoot M, Fodde R, Giordano PC, Bernini LF. Hum Genet. 1997;99:528–534. doi: 10.1007/s004390050401. [DOI] [PubMed] [Google Scholar]
- 6.Suminaga R, Takeshima Y, Yasuda K, Shiga N, Nakamura H, Matsuo M. J Hum Genet. 2000;45:331–336. doi: 10.1007/s100380070003. [DOI] [PubMed] [Google Scholar]
- 7.Burwinkel B, Kilimann MW. J Mol Biol. 1998;277:513–517. doi: 10.1006/jmbi.1998.1641. [DOI] [PubMed] [Google Scholar]
- 8.Radman M. Biochimie. 1991;73:357–361. doi: 10.1016/0300-9084(91)90101-6. [DOI] [PubMed] [Google Scholar]
- 9.Matic I, Rayssiguier C, Radman M. Cell. 1995;80:507–515. doi: 10.1016/0092-8674(95)90501-4. [DOI] [PubMed] [Google Scholar]
- 10.Shen P, Huang HV. Genetics. 1986;112:441–457. doi: 10.1093/genetics/112.3.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bianchi ME, Radding CM. Cell. 1983;35:511–520. doi: 10.1016/0092-8674(83)90185-x. [DOI] [PubMed] [Google Scholar]
- 12.Lichten M, Fox MS. Proc Natl Acad Sci USA. 1984;81:7180–7184. doi: 10.1073/pnas.81.22.7180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Worth L, Jr, Clark S, Radman M, Modrich P. Proc Natl Acad Sci USA. 1994;91:3238–3241. doi: 10.1073/pnas.91.8.3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fabisiewicz A, Worth L., Jr J Biol Chem. 2001;276:9413–9420. doi: 10.1074/jbc.M005176200. [DOI] [PubMed] [Google Scholar]
- 15.Rayssiguier C, Thaler DS, Radman M. Nature. 1989;342:396–401. doi: 10.1038/342396a0. [DOI] [PubMed] [Google Scholar]
- 16.Datta A, Adjiri A, New L, Crouse GF, Jinks Robertson S. Mol Cell Biol. 1996;16:1085–1093. doi: 10.1128/mcb.16.3.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Wind N, Dekker M, Berns A, Radman M, te Riele H. Cell. 1995;82:321–330. doi: 10.1016/0092-8674(95)90319-4. [DOI] [PubMed] [Google Scholar]
- 18.Schofield MJ, Hsieh P. Annu Rev Microbiol. 2003;57:579–608. doi: 10.1146/annurev.micro.57.030502.090847. [DOI] [PubMed] [Google Scholar]
- 19.Sugawara N, Paques F, Colaiacovo M, Haber JE. Proc Natl Acad Sci USA. 1997;94:9214–9219. doi: 10.1073/pnas.94.17.9214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lovett ST, Feschenko VV. Proc Natl Acad Sci USA. 1996;93:7120–7124. doi: 10.1073/pnas.93.14.7120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin SL, Voliva CF, Hardies SC, Edgell MH, Hutchison CA., III Mol Biol Evol. 1985;2:127–140. doi: 10.1093/oxfordjournals.molbev.a040340. [DOI] [PubMed] [Google Scholar]
- 22.Kowalczykowski SC, Dixon DA, Eggleston AK, Lauder SD, Rehrauer WM. Microbiol Rev. 1994;58:401–465. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Courcelle J, Khodursky A, Peter B, Brown PO, Hanawalt PC. Genetics. 2001;158:41–64. doi: 10.1093/genetics/158.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mount DW, Low KB, Edmiston SJ. J Bacteriol. 1972;112:886–893. doi: 10.1128/jb.112.2.886-893.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walker GC. Microbiol Rev. 1984;48:60–93. doi: 10.1128/mr.48.1.60-93.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Modrich P, Lahue R. Annu Rev Biochem. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- 27.Abdulkarim F, Hughes D. J Mol Biol. 1996;260:506–522. doi: 10.1006/jmbi.1996.0418. [DOI] [PubMed] [Google Scholar]
- 28.Radman M, Wagner R. Annu Rev Genet. 1986;20:523–538. doi: 10.1146/annurev.ge.20.120186.002515. [DOI] [PubMed] [Google Scholar]
- 29.Paques F, Haber JE. Microbiol Mol Biol Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bradshaw JS, Kuzminov A. Mol Microbiol. 2003;48:1711–1725. doi: 10.1046/j.1365-2958.2003.03540.x. [DOI] [PubMed] [Google Scholar]
- 31.Tsui HC, Zhao G, Feng G, Leung HC, Winkler ME. Mol Microbiol. 1994;11:189–202. doi: 10.1111/j.1365-2958.1994.tb00300.x. [DOI] [PubMed] [Google Scholar]
- 32.Harris RS, Feng G, Ross KJ, Sidhu R, Thulin C, Longerich S, Szigety SK, Winkler ME, Rosenberg SM. Genes Dev. 1997;11:2426–2437. doi: 10.1101/gad.11.18.2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vulic M, Dionisio F, Taddei F, Radman M. Proc Natl Acad Sci USA. 1997;94:9763–9767. doi: 10.1073/pnas.94.18.9763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roberts MS, Cohan FM. Genetics. 1993;134:401–408. doi: 10.1093/genetics/134.2.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schofield MJ, Nayak S, Scott TH, Du C, Hsieh P. J Biol Chem. 2001;276:28291–28299. doi: 10.1074/jbc.M103148200. [DOI] [PubMed] [Google Scholar]
- 36.Drotschmann K, Aronshtam A, Fritz HJ, Marinus MG. Nucleic Acids Res. 1998;26:948–953. doi: 10.1093/nar/26.4.948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Calmann MA, Nowosielska A, Marinus MG. Nucleic Acids Res. 2005;33:1193–1200. doi: 10.1093/nar/gki263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lopez de Saro FJ, Marinus MG, Modrich P, O'Donnell M. J Biol Chem. 2006;281:14340–14349. doi: 10.1074/jbc.M601264200. [DOI] [PubMed] [Google Scholar]
- 39.Cox MM. Proc Natl Acad Sci USA. 2001;98:8173–8180. doi: 10.1073/pnas.131004998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guarne A, Ramon-Maiques S, Wolff EM, Ghirlando R, Hu X, Miller JH, Yang W. EMBO J. 2004;23:4134–4145. doi: 10.1038/sj.emboj.7600412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heinen CD, Schmutte C, Fishel R. Cancer Biol Ther. 2002;1:477–485. doi: 10.4161/cbt.1.5.160. [DOI] [PubMed] [Google Scholar]
- 42.Shao C, Deng L, Chen Y, Kucherlapati R, Stambrook PJ, Tischfield JA. Oncogene. 2004;23:9017–9024. doi: 10.1038/sj.onc.1208148. [DOI] [PubMed] [Google Scholar]
- 43.Baker SM, Plug AW, Prolla TA, Bronner CE, Harris AC, Yao X, Christie DM, Monell C, Arnheim N, Bradley A, et al. Nat Genet. 1996;13:336–342. doi: 10.1038/ng0796-336. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.