Abstract
Oncocytic tumors are a distinctive class of proliferative lesions composed of cells with a striking degree of mitochondrial hyperplasia that are particularly frequent in the thyroid gland. To understand whether specific mitochondrial DNA (mtDNA) mutations are associated with the accumulation of mitochondria, we sequenced the entire mtDNA in 50 oncocytic lesions (45 thyroid tumors of epithelial cell derivation and 5 mitochondrion-rich breast tumors) and 52 control cases (21 nononcocytic thyroid tumors, 15 breast carcinomas, and 16 gliomas) by using recently developed technology that allows specific and reliable amplification of the whole mtDNA with quick mutation scanning. Thirteen oncocytic lesions (26%) presented disruptive mutations (nonsense or frameshift), whereas only two samples (3.8%) presented such mutations in the nononcocytic control group. In one case with multiple thyroid nodules analyzed separately, a disruptive mutation was found in the only nodule with oncocytic features. In one of the five mitochondrion-rich breast tumors, a disruptive mutation was identified. All disruptive mutations were found in complex I subunit genes, and the association between these mutations and the oncocytic phenotype was statistically significant (P = 0.001). To study the pathogenicity of these mitochondrial mutations, primary cultures from oncocytic tumors and corresponding normal tissues were established. Electron microscopy and biochemical and molecular analyses showed that primary cultures derived from tumors bearing disruptive mutations failed to maintain the mutations and the oncocytic phenotype. We conclude that disruptive mutations in complex I subunits are markers of thyroid oncocytic tumors.
Keywords: oncocytic tumors, heteroplasmy, homoplasmy, damaging mutation, microenvironment
Mutations in mitochondrial DNA (mtDNA) have been widely described in many types of tumors (1), and variant sequences have been reported in databases such as Mitomap (2) and HmtDB (3). Several groups have focused on the regulatory region of mtDNA, the displacement loop (D-loop) (4), whereas others investigated the D-loop in conjunction with various coding regions (5) or only a single coding region (6). Different approaches were undertaken to analyze the frequency of single polymorphisms in association with a specific tumor prevalence (7). The question regarding the pathogenic role for mtDNA mutations in cancer has already been debated (8, 9). However, the difficult task of attributing a causal role to mitochondrial variants in tumor development has not been accomplished so far. In fact, most variants found in tumors are also present as polymorphic variants in the control population, whereas for those inducing amino acid changes, it is difficult to prove a functional role relevant for tumor pathology (8).
Recently, we have attempted to clarify this point by clearly demonstrating the association between mtDNA mutations and defective oxidative phosphorylation in a cell line model of thyroid oncocytic tumor (10). The term “oncocytic” is used to designate lesions composed of cells with aberrant accumulation of mitochondria, resulting in a distinctive granular eosinophilic appearance on conventional histology. Tumors composed of oncocytic cells occur at various sites and are particularly common among thyroid neoplasms of follicular cell derivation (11). Oncocytic thyroid tumors have long been suspected to be more aggressive than their nononcocytic counterparts (11), and the presence of an oncocytic phenotype is now considered an adverse prognostic indicator for follicular thyroid carcinomas (12).
The exact relationship between mitochondrial accumulation in oncocytic cells and tumor development remains unknown. Several authors have performed gene expression and biochemical studies to investigate molecular aspects of this specific histological phenotype (13, 14). In particular, deficient complex I activity has been described in renal oncocytoma (15, 16), and a correlation between mitochondrial hyperplasia and tumorigenesis has been suggested (17). Most mtDNA changes reported in thyroid oncocytic tumors have been identified after partial sequencing of the mitochondrial genome, again without proven pathogenicity (18, 19). To the best of our knowledge, a systematic approach with complete sequencing of the whole mtDNA from tumor samples and strict mitochondrial genotype–phenotype correlation has not been carried out.
In the present study, we used a recently developed approach to sequence the whole mitochondrial genome from different types of tumors and control tissues. In silico analysis was performed on all amino acid changes with available software to predict their pathogenic potential (20). This process allowed a comprehensive investigation of all mtDNA variants in oncocytic lesions and control cases. Our data indicate that mtDNA disruptive complex I mutations are markers for the oncocytic phenotype.
Results
mtDNA Sequencing.
The entire mitochondrial genome was sequenced in 25 thyroid and 5 breast oncocytic lesions and 52 controls. More than 98% of mtDNA sequencing also was obtained from 20 additional formalin-fixed thyroid oncocytic samples. Sequencing results [deposited in the HmtDB database; see supporting information (SI) Table 3] are summarized in Tables 1 and 2 and SI Table 4. All mtDNA changes resulting in impaired protein synthesis (nonsense and frameshift alterations) were classified as “disruptive” mutations. Upon in silico prediction (see Materials and Methods), “probably damaging” or “possibly damaging” mutations were classified under the category of “potentially damaging” mutations.
Table 1.
mtDNA mutations in oncocytic samples
| Sample | Diagnosis | Base change | Amino acid change | Gene | Het., % | % Sequenced | PSIC | Database |
|---|---|---|---|---|---|---|---|---|
| HCT26 | OCA | 3571insC | 101X | ND1 | − | 100 | − | Mitomap |
| HCT16 | OCAp | 3331del242bp | Frameshift | ND1 | 70 | 100 | − | Novel |
| HCT21 | OHTN | 3571insC | 101X | ND1 | 98 | 98.2 | − | Mitomap |
| HCT4 | OHTN | G5185A | W239X | ND2 | − | 98 | − | Novel |
| HCT33 | OCA | G4720A | W84X | ND2 | − | 100 | − | Novel |
| HCT27 | OCA | 11084delCA | 113X | ND4 | − | 100 | − | Novel |
| HCT28 | OCA | 11038delA | 99X | ND4 | − | 100 | − | Mitomap |
| HCT42 | OCA | 10885delT | 61X | ND4 | 35 | 100 | − | Novel |
| HCT38 | OFA | G11403A | W215X | ND4 | − | 100 | − | Novel |
| HCT1 | OHTN | G13414A | G360X | ND5 | 29 | 100 | − | Novel |
| HCT7 | OHTN | A13870T | K512X | ND5 | 25 | 100 | − | Novel |
| HCT29 | OCA | 13235insT | 311X | ND5 | − | 99 | − | Novel |
| HCT1 | OHTN | T13271C | L312P | ND5 | − | 100 | 2.637 | Novel |
| HCT40 | OFA | G13042A | A236T | ND5 | + | 100 | 2.111 | HmtDB |
| HCT23 | OHTN | G10537A | G35E | ND4L | − | 99.2 | 2.213 | Novel |
| HCT25 | OHTN | G12056A | E433K | ND4 | − | 99.5 | 2.101 | Novel |
| HCT36 | OCAp | G11475A | G239D | ND4 | − | 100 | 2.436 | Novel |
| HCT9 | OHTN | T11613C | L285P | ND4 | − | 100 | 2.568 | Novel |
| HCT18 | OCAp | G4975A | G169E | ND2 | − | 100 | 2.696 | Novel |
| HCT28 | OCA | G4831A | G121D | ND2 | − | 100 | 2.514 | Novel |
| HCT37 | OCAp | T3949C | Y215H | ND1 | − | 100 | 2.428 | Mitomap |
| HCT39 | OCA | G3392A | G29D | ND1 | − | 100 | 2.254 | Novel |
| HCT43 | OFA | T4222C | S306P | ND1 | − | 100 | 2.020 | Novel |
| HCT44 | OFA | T12797C | L154P | ND5 | − | 100 | 2.932 | Novel |
| A8836G | M104V | ATP6 | − | 2.376 | Mitomap | |||
| HCT30 | OFA | T15209C | Y155H | CYTB | − | 98.8 | 1.984 | Novel |
| HCT6 | OFA | T15674C | S310P | CYTB | − | 100 | 1.899 | HmtDB |
| HCT26 | OCA | A8836G | M104V | ATP6 | − | 100 | 2.376 | Mitomap |
| HCT5 | OHTN | G4148A | R281H | ND1 | − | 100 | 2.398 | Novel |
| HCT31 | OCAp | G8839A | A105T | ATP6 | − | 98.7 | 1.851 | HmtDB |
| BRCA13 | BRduct | 3331del242bp | Frameshift | ND1 | 20 | 100 | − | Novel |
| BRCA14 | BRduct | T15843C | M366T | CYTB | + | 100 | 2.458 | Novel |
| BRCA17 | BRduct | T15813G | V356G | CYTB | − | 100 | 1.806 | Novel |
Het., heteroplasmy (% of mutated); OCA, oncocytic thyroid carcinoma; OCAp, oncocytic thyroid carcinoma with papillary festures; OFA, onocytic follicular thyroid adenoma; OHTN, oncocytic hyperplastic thyroid module; BRduct, invasive ductal carcirnoma of the breast. Bold indicates disruptive mutations.
Table 2.
mtDNA mutations in nononcocytic samples
| Sample | Diagnosis | Base change | Amino acid change | Gene | Het., % | PSIC | Database |
|---|---|---|---|---|---|---|---|
| BRCA3 | BRduct | T12601C | F89L | ND5 | − | 2.086 | Novel |
| BRCA9 | BRduct | A13973T | Q546L | ND5 | − | 1.551 | HmtDB |
| BRCA10 | BRlob | T9903C | F233L | COXIII | − | 2.458 | HmtDB |
| BRCA5 | BRduct | T9119C | L198P | ATP6 | + | 2.429 | Novel |
| G5 | AST | T4016G | L237R | ND1 | + | 2.151 | Novel |
| G15 | AST | T11204C | F149L | ND4 | − | 1.513 | HmtDB |
| TC6 | FA | A12961G | S209G | ND5 | − | 1.521 | HmtDB |
| TC7 | HTN | T11204C | F149L | ND4 | − | 1.513 | HmtDB |
| TC12 | PTC | T11736C | L326P | ND4 | − | 2.552 | Novel |
| TC19 | PTC | 10116delAT | 31X | ND3 | − | − | Novel |
| TC8 | FTC | G3842A | W179X | ND1 | 38 | − | Novel |
| TC18 | PTC | C7441A | S513Y | COXI | − | 1.626 | Novel |
| TC16 | PTC | A8725G | T67A | ATP6 | − | 1.537 | Novel |
| TC4 | HTN | G8572A | G16S | ATP6 | − | 1.983 | HmtDB |
Het., heteroplasmy (% of mutated); AST, astrocytoma; BRduct, invasive ductal carcinoma of the breast; BRlob, invasive lobular carcinoma of the breast; FA, follicular thyroid adenoma; FTC, follicular thyroid carcinoma; HTN, hyperplastic thyroid module; PTC, papillary thyroid carcinoma. Bold indicates disruptive mutations.
Disruptive mutations were concentrated in a few mitochondrial genes coding for subunits of complex I of the respiratory chain (ND1, ND2, ND4, and ND5). Two samples (Table 1) harbored the same mutation (3571insC) in the ND1 gene that we recently reported in the XTC.UC1 oncocytic cell line, in which we demonstrated complete absence of the protein and defective activity of complex I (10).
Because of the physiological polyploidy of the mitochondrial genome, different variants of mtDNA can coexist in a single mitochondrion or in a single cell, which gives rise to the phenomena of homo- and heteroplasmy. Homoplasmy implies that all copies of the mitochondrial genome in cells and tissues harbor an identical sequence. Heteroplasmy denotes the coexistence of mtDNA copies carrying differences in their sequence. Thus, nonsynonymous mtDNA changes may lead to different protein variants, and the phenotypic effect of a mutation may become evident only if a certain threshold of heteroplasmy is reached. The heteroplasmic status of all mtDNA variants detected is reported in Tables 1 and 2. Heteroplasmy of all disruptive mutations was evaluated. As reported in Table 1, most disruptive mutations in the oncocytic samples were homoplasmic or with a high degree of heteroplasmy (>70%) and hence above the likely threshold for a damaging effect. For all disruptive mutations, the corresponding adjacent normal tissue was also analyzed, and in all cases mutations were somatic.
The presence of preferential combinations of mitochondrial polymorphisms (haplogroups) in our oncocytic samples versus nononcocytic tumor types was also investigated. Haplogroup definition did not reveal any statistically significant divergence from the previously reported haplogroup frequencies in European population (21).
Correlation of mtDNA Alterations with Clinico-Pathologic Features.
Immunohistochemistry and electron microscopy confirmed cytoplasmic accumulation of mitochondria in all oncocytic thyroid samples analyzed and an increased mitochondrial mass in five breast carcinomas. In the latter cases, the percentage of mitochondrion-rich cells was >80% in the tumor tissue, whereas for the thyroid oncocytic samples, the percentage was invariably >75%. No evidence of oncocytic differentiation was detected in the glioma group.
Twenty-six of the 45 oncocytic thyroid samples analyzed (57.8%) harbored 30 mtDNA mutations, 25 of which were in complex I genes. In 12/45 samples (26.7%), the mutations were disruptive and all occurred in complex I. In three samples, the disruptive mutations coexisted with potentially damaging missense mutations (Table 1). In 14/45 samples (31.1%), the mutations were potentially damaging missense changes, and in one case, two such mutations coexisted in the same lesion (Table 1). Fifteen of the 18 (83.3%) potentially damaging missense mutations identified in thyroid oncocytic samples (12 of which occurred in complex I) had a position-specific independent count (PSIC) > 2 [i.e., mutations were probably damaging (20)].
Eight of the 21 nononcocytic thyroid samples (38.1%) presented mtDNA mutations, 5 of which were in complex I. Only in 2/21 samples (9.5%) were the mutations, both occurring in complex I, disruptive, whereas in 6/21 samples (28.5%), the mutations were potentially damaging missense changes. Five of the six (83.3%) potentially damaging missense mutations identified in nononcocytic thyroid samples had a PSIC < 2 [i.e., mutations were possibly damaging (20)].
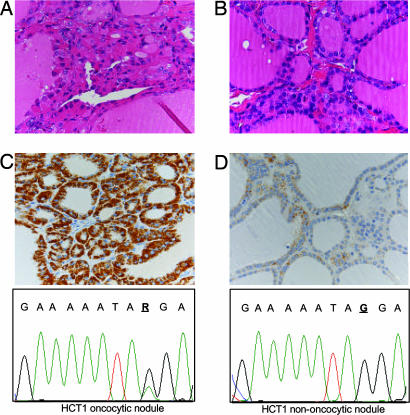
In one case, three separate hyperplastic nodules from the same thyroid gland were analyzed. Only one nodule presented oncocytic change, and only in this nodule was a disruptive heteroplasmic nonsense mutation in ND5 found (Table 1 and Fig. 1). Heteroplasmy evaluation showed that 29% of ND5 copies were mutated in the nodule.
Fig. 1.
Analysis of separate hyperplastic nodules with and without oncocytic phenotype from the same thyroid (case HTC1, Table 1). (Upper) Histologic appearance of the hyperplastic oncocytic nodule (A) and of one hyperplastic nodule without oncocytic change (B). Immunohistochemistry with antibodies specific for human mitochondria confirms the increased mitochondrial mass in the oncocytic nodule (C) compared with the nononcocytic one (D). (Lower) The heteroplasmic nonsense mutation of the ND5 (G13414A) gene in the oncocytic and nononcocytic nodule. (Magnification: ×400.)
Among the 15 breast carcinoma samples examined, there were 4 potentially damaging missense variants (26.7%) and no disruptive mutations (Table 2). Among the five mitochondrion-rich breast tumors, one harbored a heteroplasmic disruptive mtDNA mutation (Table 1 and Fig. 2), whereas two cases had potentially damaging missense mutations. Two potentially damaging missense variants (12.5%) and no disruptive mutations were detected in the glioma group. Overall, the oncocytic phenotype was associated with mtDNA mutations. The association was significant in both cases: when considering only disruptive mutations (P = 0.001) and when all mutations were taken into account (P = 0.0013). Patient age, sex, and size of the lesion were not associated with mtDNA mutations (SI Table 5).
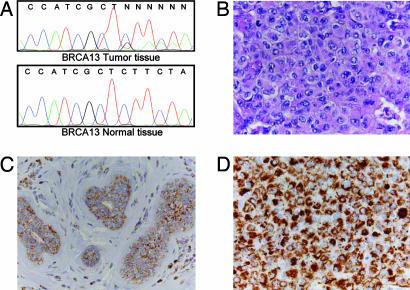
Fig. 2.
Disruptive mtDNA mutation in a mitochondrion-rich breast carcinoma (case BRCA13, Table 1). (A) Electropherograms showing the heteroplasmic deletion in the ND1 gene in tumor (Upper) and perilesional normal (Lower) tissue. (B) Histologic appearance of the breast carcinoma showing neoplastic cells with abundant eosinophilic cytoplasm and oncocytic features. (C and D) Immunohistochemistry with antibodies specific for human mitochondria confirms the increased mitochondrial mass in the tumor (D) compared with the nonneoplastic perilesional breast parenchyma (C). (Magnification: ×400.)
Primary Cell Cultures.
To address further the correlation between mtDNA mutations and biochemical phenotype of oncocytic tumors, primary cultures were established from thyroid samples according to availability of material. We focused on the two cultures derived from oncocytic tumors and bearing a disruptive mutation. Although no unequivocal tumor markers are available to ascertain the transformed phenotype of cultured cells, higher production of IL-6 has been shown to occur in thyroid carcinoma (22). Accordingly, IL-6 secretion was increased significantly in all tumor-derived primary cultures compared with corresponding normal cultures from the same patient (SI Fig. 4 and SI Materials and Methods).
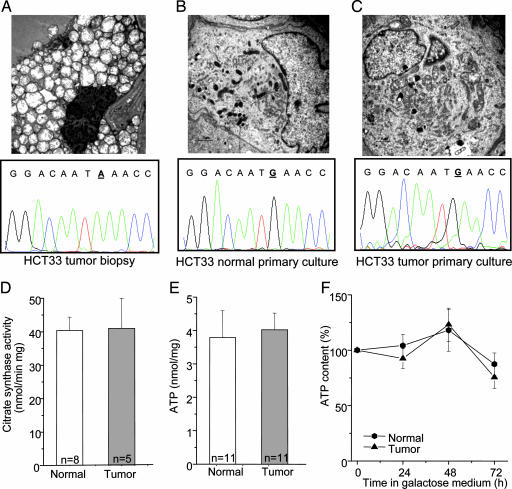
Interestingly, no mutations were detected in any of the three primary tumor cultures originating from tumors presenting a disruptive mutation (data not shown). Accordingly, electron microscopy showed that both fresh and paraffin-retrieved (data not shown) biopsies from oncocytic tumors were composed of large cells rich in closely packed, swollen mitochondria (Fig. 3A). In contrast, cultured cells from oncocytic lesions retained only a few large mitochondria with frequently observed secondary lysosomal structures (Fig. 3B) and showed a filamentous mitochondrial network similar to that of their normal cultured cell counterpart (Fig. 3C and SI Fig. 4). Furthermore, no difference was detected in the citrate synthase activity, a widely accepted biochemical indicator for mitochondrial mass (Fig. 3D). The ATP content of normal and tumor thyroid primary cultures was similar in glucose medium (Fig. 3E) and also during incubation in galactose-containing medium [i.e., under conditions leading to a dramatic reduction of glycolytic rate and forced use of oxidative phosphorylation for ATP production (Fig. 3F)]. Given that cells with defects in oxidative phosphorylation are unable to maintain their ATP content in galactose-containing medium (23), these results indicate that oxidative phosphorylation was not impaired in tumor cells.
Fig. 3.
Characterization of normal and tumor primary cell cultures. (A–C) Ultrastructure of thyroid tumor biopsy shows mitochondrial hyperplasia (A Upper) lacking in normal and tumor primary cultures (B Upper and C Upper). Electropherograms showing the mutated base (bold and underlined) in the tumor biopsy and the wild-type base at the same position in both normal and tumor primary culture (A Lower, B Lower, and C Lower). (D–F) Citrate synthase activity (D), total ATP levels in glucose (E), and activity during incubation in galactose-containing medium (F) are shown. Data points F are means ± SD of at least five different experiments.
Discussion
In this study, we have demonstrated that the oncocytic phenotype is associated with disruptive mutations in complex I subunits genes.
The role of mitochondria in the process of tumorigenesis has been debated widely, and the literature has flourished with studies investigating the association between mtDNA variants and tumors (1). However, careful survey of all mitochondrial variants reported in association with cancer makes it difficult to accept that silent and even missense mutations may be only causally related or predisposing to tumorigenesis. Nevertheless, controversial technical aspects have warranted skepticism (24). Given the very complex nature of tumorigenesis, it is difficult to attribute a causal role to single mitochondrial mutations, and a more-than-one-hit hypothesis is more plausible. In this context, mitochondrial mutations may play their part as one of the strikes leading to tumor development.
In the oncocytic samples, we found a larger prevalence of nonsense and frameshift mutations caused by insertions or deletions in coding regions of mtDNA, in most cases (8 of 12) occurring early in sequence, so that dramatic disruption of the protein was easily predictable. Only two such mutations were found in the control group. Statistical analysis showed a clear correlation between the presence of such disruptive mutations and the oncocytic phenotype. All of the disruptive mutations were concentrated in complex I subunits, whereas analysis of the corresponding samples from the normal adjacent tissue showed the somatic origin of the mutations in all cases. This finding further supports the hypothesis that dysfunction of complex I may play a role in tumor development, as previously proposed for thyroid (10) and renal oncocytoma (15, 16).
A higher prevalence of missense mutations was also found in oncocytic compared with nononcocytic samples (Tables 1 and 2). Additional functional studies will be needed to substantiate their pathogenicity.
One case (HCT1), from which three different nodules were analyzed, presented a heteroplasmic nonsense mutation in the ND5 gene in only one of the nodules. Double-blinded histopathological examination confirmed that only the nodule with the mutation presented oncocytic changes, suggesting that this specific mutational event may be responsible for the oncocytic phenotype.
Interestingly, the only case of breast carcinoma harboring a disruptive mutation was a mitochondrion-rich tumor, and potentially damaging mutations were present in two additional mitochondrion-rich breast carcinomas. Oncocytic carcinomas of the breast are rare tumors, although the prevalence of mitochondrion-rich breast carcinomas with oncocytic features, such as the cases presented in this study, might be underestimated (25). mtDNA mutations have been described in breast carcinomas (1, 5) without being correlated to a mitochondrion-rich phenotype. Our findings in breast carcinoma strengthen the link between pathogenic mtDNA changes and mitochondrial hyperplasia.
A link between mtDNA disruptive mutations and the oncocytic phenotype also was provided by the thyroid primary culture experiments. None of the primary tumor cultures showed evidence of the disruptive mutations found in the original biopsies. Accordingly, the oncocytic phenotype was lost during culture, and no differences between tumor and the corresponding normal tissue cultures were found. These data indicate that, under the culture conditions used in this study, cells bearing the mutations are selected against.
The glycolytic shift in cancer cells observed initially by Warburg (26), a phenomenon currently exploited for diagnostic purposes by positron emission tomography, led to the idea that mitochondrial damage, forcing cells to rely on glycolysis for ATP production, may confer a selective advantage in the hypoxic environment surrounding the tumor (27). Nonetheless, it has been suggested that severe mutations impairing oxidative phosphorylation may be lost once the tumor cells return to a high-oxygen environment as during cell culture (8). Hence, the in vivo microenvironment may have a fundamental influence on conditions that allow the mutation to arise, be propagated, and eventually shift to homoplasmy. It is worth noting that different mechanisms may be involved for the maintenance of the oncocytic phenotype. For example, the XTC.UC1 cell line is the only existing cellular model of thyroid oncocytoma (28). XTC.UC1 is an immortalized cell line, derived from a metastasis, and possibly the accumulation of genetic damage may have contributed to the positive selection of oncocytic cells.
Mitochondrial dysfunction might also arise from mutations in nuclear genes encoding for mitochondrial proteins (29, 30). A mutation screening of nuclear-coded oxidative phosphorylation subunits was not performed in this study. Mutations in the nuclear-encoded complex I subunits may account for the percentage of oncocytic cases in which mitochondrially encoded complex I subunits were not mutated; hence, the percentage of complex I mutations in oncocytic tumors could be underestimated. In fact, it has been shown that some types of hereditary tumors are characterized by mitochondrial defects (31). The presence of germ-line changes in mitochondria-related genes and their potential involvement in oncocytic tumor development further suggests a complex interplay between nuclear and mitochondrially encoded genes in promoting the oncocytic phenotype.
In conclusion, this study shows a statistically significant prevalence of disruptive mutations in genes coding for complex I of the electron transport chain in thyroid oncocytic tumors. It is likely that these mutations may arise as a secondary hit in tumor development and that the oncocytic phenotype, characterized by mitochondrial hyperplasia, may be strictly correlated with these mutations. We therefore propose this type of mutation as a molecular marker of oncocytic phenotype in thyroid tumors.
Materials and Methods
Tissue Samples, Clinico-Pathologic Features, and Immunohistochemistry.
Samples were obtained from the pathology units of Bologna University Medical School at Bellaria and S. Orsola-Malpighi Hospitals. From 11 thyroid oncocytic samples and 48 controls, excess lesional and/or perilesional tissue was obtained fresh and stored frozen at −80°C before analysis. Samples were diagnosed according to established criteria (12). Twenty-two were hyperplastic thyroid nodules (16 of them oncocytic), 10 were follicular thyroid adenomas (7 of them oncocytic), 22 were oncocytic thyroid carcinomas, 12 were thyroid carcinomas without oncocytic features (11 papillary, 1 follicular), 20 were breast carcinomas (5 of which had oncocytic features), and 16 were gliomas. The cytoplasmic content of mitochondria was visualized on tissue sections by using routine immunohistochemical methods (25). At the time of diagnosis, the average patient age was 53 for patients with oncocytic lesions compared with 58 for controls. Average lesional size was 2.6 cm for the oncocytic lesions versus 3 cm for thyroid controls. All tumors considered for the study were sporadic. Clinical information was obtained by chart review. Handling of samples and clinical data proceeded in accordance with internal review-board-approved protocols.
DNA Extraction and mtDNA Sequencing.
DNA was extracted with the Qiagen kit (Qiagen, Valencia, CA) according to the manufacturer's protocols. mtDNA was sequenced with the recently developed MitoAll resequencing kit (Applera, Foster City, CA) and analyzed as described in ref. 10. Haplogroup and subhaplogroup affiliations of all samples investigated were assigned as described in ref. 21, and whenever possible, heteroplasmy was confirmed by cloning as described in ref. 10.
Prediction Analysis of Amino Acid Substitutions.
PolyPhen (www.tux.embl-heidelberg.de/ramensky/polyphen.cgi) was used to predict the possible impact of amino acid substitutions on the protein. The program is based on sequence comparison with homologous proteins; profile scores (PSIC) are generated for the allelic variants and represent the logarithmic ratio of the likelihood of a given amino acid occurring at a particular site relative to the likelihood of this amino acid occurring at any site (background frequency). PSIC score differences >2 indicate a damaging effect, scores between 1.5 and 2 suggest that the variant is possibly damaging, and scores <1.5 indicate that the variant is benign (20).
Electron Microscopy.
For electron microscopy, small fresh-tissue biopsies or cell pellets obtained from primary cultures of both lesional and perilesional thyroid tissue were processed according to previously published protocols (32).
Primary Cultures.
Of 66 thyroid samples collected, 29 primary cultures were established from both the tumor and the normal tissue. Twelve of these were from oncocytic samples, and two of these originated from biopsies in which a disruptive mutation was present (HCT33 and HCT38, Table 1). One culture was derived from a nononcocytic tumor (TC8, Table 2) bearing a disruptive mutation.
H1/10P Culture Growth Medium.
H1/10P culture growth medium, a basic culture proliferation medium with composition similar to that previously used for human thyroid cells, was used (33, 34) with and without 50 μg/ml uridine to allow growth of cells with impaired mitochondrial function.
Reverse Transcription.
Reverse transcription was performed for the following markers to confirm selection of thyroid cells: thyroid transcription factor 1, thyroid-stimulating hormone receptor, and thyroid peroxidase (data not shown).
ATP Assay and Citrate Synthase Activity.
Cells from primary culture (3 × 105) were seeded into six-well plates and incubated in H1/10P medium or in glucose-free H1/10P medium supplemented with 5 mM galactose, 5 mM Na-pyruvate, and 10% FBS. ATP was determined with the luciferin/luciferase assay (10). Citrate synthase activity was measured as described in ref. 10.
Statistical Analysis.
Statistical analysis was performed with the Fisher's exact test. A P value <0.05 was considered to be statistically significant.
Supplementary Material
Acknowledgments
We thank Dr. Luca Morandi (Bellaria Hospital, Bologna) for help in sample collection and L'Oreal Italia “Per le Donne e la Scienza” for fellowship support (to A.M.P.). This work was supported by Associazione Italiana Ricerca sul Cancro (AIRC) Grant 1145 (to G.T.) and partially supported by grants from Fondo Italiano Ricerca di Base, Rome (FIRB), and European Commission Project SH-2005-2.2.0-2 “HERMIOINE” (to G.R.).
Abbreviation
- PSIC
position-specific independent count.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the HmtDB database (accession nos. listed in SI Table 3).
This article contains supporting information online at www.pnas.org/cgi/content/full/0703056104/DC1.
References
- 1.Penta J-S, Johnson F-M, Wachsman J-T, Copeland W-C. Mut Res. 2001;488:119–133. doi: 10.1016/s1383-5742(01)00053-9. [DOI] [PubMed] [Google Scholar]
- 2.Brandon M-C, Lott M-T, Nguyen K-C, Spolim S, Navathe S-B, Baldi P, Wallace D-C. Nucleic Acids Res. 2005;33:D611–D613. doi: 10.1093/nar/gki079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Attimonelli M, Accetturo M, Santamaria M, Lascaro D, Scioscia G, Pappada G, Russo L, Zanchetta L, Tommaseo-Ponzetta M. BMC Bioinformatics. 2005;6(Suppl 4):S4. doi: 10.1186/1471-2105-6-S4-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lievre A, Chapusot C, Bouvier A-M, Zinzindohoue F, Piard F, Roignot P, Arnould L, Beaune P, Faivre J, Laurent-Puig P. J Clin Oncol. 2005;23:3517–3525. doi: 10.1200/JCO.2005.07.044. [DOI] [PubMed] [Google Scholar]
- 5.Parrella P, Xiao Y, Fliss M, Sanchez-Cespedes M, Mazzarelli P, Rinaldi M, Nicol T, Gabrielson E, Cuomo C, Cohen D, et al. Cancer Res. 2001;61:7623–7626. [PubMed] [Google Scholar]
- 6.Petros J-A, Baumann A-K, Ruiz-Pesini E, Amin M-B, Sun C-Q, Hall J, Lim S, Issa M-M, Flanders W-D, Hosseini S-H, et al. Proc Natl Acad Sci USA. 2005;102:719–724. doi: 10.1073/pnas.0408894102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Canter J-A, Kallianpur A-R, Parl F-F, Millikan R-C. Cancer Res. 2005;65:8028–8033. doi: 10.1158/0008-5472.CAN-05-1428. [DOI] [PubMed] [Google Scholar]
- 8.Brandon M, Baldi P, Wallace D-C. Oncogene. 2006;25:4647–4662. doi: 10.1038/sj.onc.1209607. [DOI] [PubMed] [Google Scholar]
- 9.Chatterjee A, Mambo E, Sidransky D. Oncogene. 2006;25:4663–4674. doi: 10.1038/sj.onc.1209604. [DOI] [PubMed] [Google Scholar]
- 10.Bonora E, Porcelli A-M, Gasparre G, Biondi A, Ghelli A, Carelli V, Baracca A, Tallini G, Martinuzzi A, Lenaz G, et al. Cancer Res. 2006;66:6087–6096. doi: 10.1158/0008-5472.CAN-06-0171. [DOI] [PubMed] [Google Scholar]
- 11.Tallini G. Virchows Arch. 1998;433:5–12. doi: 10.1007/s004280050209. [DOI] [PubMed] [Google Scholar]
- 12.DeLellis R-A, Lloyd R-V, Heitz P-U, Eng C, editors. World Health Organization Classification of Tumors: Pathology and Genetics of Tumours of the Endocrine Organs. Lyon, France: IARC Press; 2005. pp. 57–72. [Google Scholar]
- 13.Baris O, Savagner F, Nasser V, Loriod B, Granjeaud S, Guyetant S, Franc B, Rodien P, Rohmer V, Bertucci F, et al. J Clin Endocrinol Metab. 2004;89:994–1005. doi: 10.1210/jc.2003-031238. [DOI] [PubMed] [Google Scholar]
- 14.Hervouet E, Godinot C. Mitochondrion. 2006;6:105–117. doi: 10.1016/j.mito.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 15.Simonnet H, Alazard N, Pfeiffer K, Gallou C, Beroud C, Demont J, Bouvier R, Schagger H, Godinot C. Carcinogenesis. 2002;23:759–768. doi: 10.1093/carcin/23.5.759. [DOI] [PubMed] [Google Scholar]
- 16.Simonnet H, Demont J, Pfeiffer K, Guenaneche L, Bouvier R, Brandt U, Schagger H, Godinot C. Carcinogenesis. 2003;24:1461–1466. doi: 10.1093/carcin/bgg109. [DOI] [PubMed] [Google Scholar]
- 17.Yeh J-J, Lunetta K-L, van Orsouw N-J, Moore F-D, Jr, Mutter G-L, Vijg J, Dahia P-L, Eng C. Oncogene. 2000;19:2060–2066. doi: 10.1038/sj.onc.1203537. [DOI] [PubMed] [Google Scholar]
- 18.Maximo V, Soares P, Lima J, Cameselle-Teijeiro J, Sobrinho-Simoes M. Am J Pathol. 2002;160:1857–1865. doi: 10.1016/S0002-9440(10)61132-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rogounovitch T, Saenko V, Yamashita S. Endocr J. 2004;51:265–277. doi: 10.1507/endocrj.51.265. [DOI] [PubMed] [Google Scholar]
- 20.Ramensky V, Bork P, Sunyaev S. Nucleic Acids Res. 2002;30:3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carelli V, Achilli A, Valentino M-L, Rengo C, Semino O, Pala M, Olivieri A, Mattiazzi M, Pallotti F, Carrara F, et al. Am J Hum Genet. 2006;78:564–574. doi: 10.1086/501236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Russell J-P, Engiles J-B, Rothstein J-L. J Immunol. 2004;172:4059–4067. doi: 10.4049/jimmunol.172.7.4059. [DOI] [PubMed] [Google Scholar]
- 23.Robinson B-H, Petrova-Benedict R, Buncic J-R, Wallace D-C. Biochem Med Metab Biol. 1992;48:122–126. doi: 10.1016/0885-4505(92)90056-5. [DOI] [PubMed] [Google Scholar]
- 24.Salas A, Yao Y-G, Macaulay V, Vega A, Carracedo A, Bandelt H-J. PLoS Med. 2005;2:e296. doi: 10.1371/journal.pmed.0020296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Damiani S, Eusebi V, Losi L, D'Adda T, Rosai J. Am J Surg Pathol. 1998;22:221–230. doi: 10.1097/00000478-199802000-00011. [DOI] [PubMed] [Google Scholar]
- 26.Warburg O. The Metabolism of Tumors. London: Constable; 1930. [Google Scholar]
- 27.Gatenby R-A, Gillies R-J. Nat Rev Cancer. 2004;4:891–899. doi: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- 28.Zielke A, Tezelman S, Jossart G-H, Wong M, Siperstein A-E, Duh Q-Y, Clark O-H. Thyroid. 1998;8:475–483. doi: 10.1089/thy.1998.8.475. [DOI] [PubMed] [Google Scholar]
- 29.Zeviani M, Spinazzola A, Carelli V. Curr Opin Genet Dev. 2003;13:262–270. doi: 10.1016/s0959-437x(03)00052-2. [DOI] [PubMed] [Google Scholar]
- 30.DiMauro S. Biochem Biophys Acta. 2004;1658:80–88. doi: 10.1016/j.bbabio.2004.03.014. [DOI] [PubMed] [Google Scholar]
- 31.Eng C, Kiuru M, Fernandez M-J, Aaltonen L-A. Nat Rev Cancer. 2003;3:193–202. doi: 10.1038/nrc1013. [DOI] [PubMed] [Google Scholar]
- 32.Ambrosini-Spaltro A, Salvi F, Betts C-M, Frezza G-P, Piemontese A, Del Prete P, Baldoni C, Foschini M-P, Viale G. Virchows Arch. 2006;448:442–448. doi: 10.1007/s00428-005-0137-6. [DOI] [PubMed] [Google Scholar]
- 33.Curcio F, Ambesi-Impiombato F-S, Perrella G, Coon H-G. Proc Natl Acad Sci USA. 1994;91:9004–9008. doi: 10.1073/pnas.91.19.9004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perrella G, Fabbro D, Damante G, Di Loreto C, Beltrami C-A, Curcio F, De Filippi R, Ambesi-Impiombato F-S. Adv Clin Path. 1997;1:191–197. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.