Abstract
Here, we show that an α-proteobacterium of the genus Asaia is stably associated with larvae and adults of Anopheles stephensi, an important mosquito vector of Plasmodium vivax, a main malaria agent in Asia. Asaia bacteria dominate mosquito-associated microbiota, as shown by 16S rRNA gene abundance, quantitative PCR, transmission electron microscopy and in situ-hybridization of 16S rRNA genes. In adult mosquitoes, Asaia sp. is present in high population density in the female gut and in the male reproductive tract. Asaia sp. from An. stephensi has been cultured in cell-free media and then transformed with foreign DNA. A green fluorescent protein-tagged Asaia sp. strain effectively lodged in the female gut and salivary glands, sites that are crucial for Plasmodium sp. development and transmission. The larval gut and the male reproductive system were also colonized by the transformed Asaia sp. strain. As an efficient inducible colonizer of mosquitoes that transmit Plasmodium sp., Asaia sp. may be a candidate for malaria control.
Keywords: malaria, symbiotic control, insect vector
One of the major objectives of malaria control programs is interference with parasite transmission by mosquito vectors (1). Mosquito genetic transformation has been developed for Anopheles gambiae (2) and Anopheles stephensi (3), the main malaria vectors in Africa and Asia, respectively, and transgenic mosquitoes have been developed that are impaired in parasite transmission (4). However, genetic manipulation tends to reduce mosquito fitness (5). There is current interest in the use of microorganisms as biological control agents of vector-borne diseases (6–8). Microorganisms associated with vectors could exert a direct pathogenic effect on the host (9), interfere with its reproduction (10, 11), or reduce vector competence (12, 13). Furthermore, the use of genetically modified bacteria to deliver antiparasite molecules has several advantages over the use of genetically modified vectors (14). Very little is currently known about mosquito-associated microbiota (14–16). Here, we show that bacteria of the genus Asaia are stably associated with An. stephensi, an Asian malarial mosquito vector. Asaia can be cultivated and genetically manipulated and can recolonize the insect host. As an efficient inducible colonizer of mosquitoes that transmit Plasmodium sp., Asaia sp. may be a candidate for malaria control.
Results and Discussion
16S rRNA Gene Libraries from Anopheles spp. and Asaia sp. PCR Screening.
Bacterial diversity associated with An. stephensi was initially explored by establishing three 16S rRNA gene libraries from total DNA of the abdomens of three laboratory-bred individuals. Analysis of the 16S rRNA gene sequence revealed that the libraries were dominated by sequences related to the genus Asaia [90% of the clones examined; see supporting information (SI) Table 2], an α-proteobacterium strictly related to acetic acid bacteria (17, 18). The obtained sequence showed >99% nucleotide identity with those of Asaia bogorensis and Asaia siamensis, two species previously isolated from tropical flowers (17, 18). Other clones were related to different α-proteobacteria (Acetobacter, Gluconobacter, and Sphingomonas; see SI Table 2). By using a 16S rRNA-based Asaia-specific PCR, amplicons were found in both males and females of all of the >300 individuals of An. stephensi tested derived from at least 10 different breeding batches (SI Table 3). The identity of the amplicons was confirmed by sequencing. PCR analysis showed that Asaia DNA is present in eggs, pupae, and different larval stages, as well as in various mosquito organs, including gut, salivary glands, ovaries, and testes. Asaia DNA was also found in all of the 60 field-collected individuals of Anopheles maculipennis captured during three different periods (June 2005, October 2005, and June 2006) and in their F1 obtained in the laboratory (SI Table 3). Asaia represented 20% of the clones in 16S rRNA gene libraries (SI Table 4). Asaia was sporadically (≈5% of the clones) found in 16S rRNA gene libraries from the total DNA of An. gambiae individuals collected in Burkina Faso (SI Tables 3 and 5). Interestingly, it has recently been reported that interference with the innate immune system of An. gambiae by transient silencing of AgDscam determines proliferation of A. bogorensis in mosquito hemolymph (19).
Diversity of bacteria within the 16S rRNA gene libraries was rather low, with relatively few phylotypes. Low bacterial diversity in Anopheles species by 16S rRNA gene sequencing has been reported, with six, two, and one bacterial species in An. arabiensis, An. gambiae sensu stricto, and An. funestus, respectively (16). We detected few operational taxonomic units within the γ-proteobacteria that were detected in other studies by 16S rRNA gene sequencing (16) and bacterial isolation (14, 16). This difference may be due to the different primer set used and emphasizes that the use of multiple primer sets in molecular microbial ecology widens the view of the actual diversity residing in a system (20).
Asaia Cultivation and Real-Time PCR Analysis.
Asaia was isolated from An. stephensi according to the method reported for isolation from tropical flowers (17, 18). A preenrichment step in liquid medium at pH 3.5, followed by plating in carbonate-rich medium, resulted in the isolation of single, pink colonies capable of dissolving carbonate in the medium and generating dissolution haloes (Fig. 1 A and B). Colonies isolated from An. stephensi and An. maculipennis were confirmed to be Asaia sp. by 16S rRNA gene sequencing, with 99.6% and 99.8% nucleotide identity with A. bogorensis and A. siamensis, respectively (17, 18) (SI Fig. 4). Asaia sp. was abundant in the mosquito body, with bacterial counts of up to 9.8 × 105 colony forming units (CFU) per female and 9.8 × 104 CFU per male individuals. To evaluate the relative abundance of Asaia sp. in different organs of An. stephensi, we measured Asaia to total bacteria 16S rRNA gene Asaia to total bacteria copy ratio (ABR) with quantitative real-time PCR, by determining the Asaia sp. and total bacteria 16S rRNA gene copies in the gut, salivary glands, and female reproductive system (Table 1). Asaia sp. 16S rRNA gene copies constituted a mean of 41%, 25%, and 20% of the total 16S rRNA gene copies of the bacterial population in the gut, salivary glands, and female reproductive system, respectively, showing that this acetic bacterium represents the dominant bacterium in the population of An. stephensi examined in this study, particularly in the gut. Interestingly, high numbers of Asaia were also found in both the salivary glands, which have rarely been reported to be colonized by bacteria in other insects (21), and in the female reproductive system, where vertically transmitted endosymbionts are typically found.
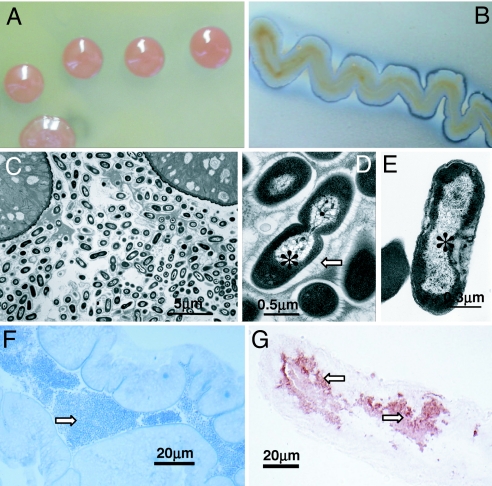
Fig. 1.
Images of Asaia sp. from An. stephensi. (A) Colonies of Asaia sp. on CaCO3-agarized medium, showing a characteristic pink color. (B) Haloes of carbonate solubilization due to acidification. (C) TEM micrograph of an An. stephensi adult midgut full of bacteria that probably belongs to genus Asaia. (D and E) Magnification of bacterial cells in the midgut (D), whose morphology resembles that of Asaia sp. in pure culture (E). Filamentous structures in the nucleoid region (asterisks), and an extracellular matrix with a fibrillar nature (arrow) can be noted. Cells of Asaia sp. in pure culture do not show the extracellular matrix observed in the mosquito midgut. (F) The midgut lumen of An. stephensi is full of bacterial cells (arrow). (G) ISH with an Asaia-specific probe showing a high density of the bacterium in the midgut lumen (arrows).
Table 1.
Asaia sp. and bacteria 16S rRNA gene copies and Asaia to bacteria 16S rRNA gene copy ratio (ABR) in different organs of An. stephensi
| Organ | No. of individuals* | Range of Asaia 16S rRNA gene copies† | Range of bacterial 16S rRNA gene copies† | ABR‡ | ABR range |
|---|---|---|---|---|---|
| Gut | 22 | 1.3 × 104 to 8.7 × 107 | 1.6 × 105 to 9.5 × 107 | 0.41 ± 0.28 | 0.03–0.91 |
| Salivary glands | 17 | 5.3 × 102 to 4.0 × 106 | 1.2 × 105 to 6.7 × 106 | 0.25 ± 0.25 | 0.004–0.62 |
| Female reproductive system | 17 | 7.5 × 101 to 3.4 × 106 | 4.0 × 104 to 4.9 × 106 | 0.20 ± 0.27 | 0.001–0.78 |
*Each individual was separately analyzed by triplicate real-time PCR.
†Minimum and maximum amounts of 16S rRNA copies detected.
‡ABR, Asaia to total bacterial 16S rRNA gene copy ratio. Mean ABR ± SD of the individuals reported in column 2 are given.
Transmission Electron Microscopy (TEM) and in Situ Hybridization (ISH) Analysis of Asaia in Mosquito.
Several body parts of female An. stephensi were examined by TEM, and the morphology of observed bacteria was compared with that of cultured Asaia. No bacteria were detected in the egg cell cytoplasm, whereas the midgut lumen contained large bacterial cell clusters with the typical Gram-negative architecture with a signature filamentous appearance of the nucleoid region similar to cultured Asaia (Fig. 1 C–E). The bacteria in the insect midgut were embedded within an extracellular matrix (Fig. 1D). The identity of the bacteria in An. stephensi midgut (Fig. 1F) was confirmed by ISH with Asaia-specific probes that indicated a high concentration of cells within the mucous substance lining the midgut epithelium (Fig. 1G). ISH signals with Asaia-specific probes were analogous to those observed by using bacterial probe EUB338, whereas no signals were observed in tissues treated with RNase or in the absence of the probe.
Mosquito Recolonization by Asaia Expressing Green Fluorescent Protein (Gfp).
The overall data indicated that Asaia bacteria are dominant in, and stably associated with, the midgut of An. stephensi and hence could be an interesting vector of antiparasite molecules in mosquitoes. The transformability of Asaia from An. stephensi was verified by electroporating Asaia sp. strain SF2.1 with plasmids pHM2 and pHM3, two replicative plasmids in the genera Acetobacter and Gluconobacter closely related to the genus Asaia (22). The plasmids transformed strain SF2.1 with an efficiency of 4.7 105 cells·μg−1 DNA. Plasmid pHM2 conferred both kanamycin resistance and the blue colony phenotype to the strain when the cells were grown in the presence of 5-bromo-4-chloro-3-indolyl-β-galactopyranoside, whereas plasmid pHM3 conferred resistance to the antibiotic but not blue colony phenotype. We cloned a Gfp cassette in plasmid pHM2 to label Asaia with an easy-to-follow optical marker for visually tracking the bacterium in the body of An. stephensi. The resulting plasmid pHM2-Gfp was used to transform strain SF2.1. We obtained strain SF2.1(Gfp) that was then used in recolonization experiments of An. stephensi adults and larvae.
The SF2.1(Gfp) strain recolonized An. stephensi adults fed with sucrose or blood-containing solutions in which the strain was resuspended. Fluorescent and laser-scanning confocal microscopy showed that the bacterium efficiently colonized the gut (Fig. 2), salivary glands (SI Fig. 5) and male reproductive system (Fig. 3 A–H). We examined a total of 140 insect adults from three cages fed with Asaia sp. strain SF2.1(Gfp). Most of the individuals (72%) showed fluorescent cells and microcolonies in the body. Because we do not know whether all of the individuals actually fed on the bacterial suspension, the lack of Asaia from some individuals, in the light of the high percentage of positives, could be explained with the lack of a meal. Fluorescent Asaia cells and microcolonies were observed in 65%, 32%, and 58% of the guts (91 individuals examined), salivary glands (59 females examined), and male reproductive systems (55 individuals examined). Colonization of the mosquito body by Asaia sp. strain SF2.1(Gfp) was stably maintained for the entire life span of the insect. Fluorescent cells and microcolonies were detected in all of the previously mentioned body parts (gut, salivary gland, and male reproductive system) of adult mosquitoes (10 individuals) up to 25 days, before insect death. Fluorescent cells and microcolonies were established in the gut after 24 h when mosquitoes were fed with blood and after 48 h when fed with sucrose-containing solutions. This difference in the timing of microcolony establishment could be due to the fact that sugar goes to the crop instead of mosquito midgut, determining a delay in dispersion to other portions of the gut. The consequent higher number of ingested bacterial cells likely allowed a more rapid colonization of the gut with the fluorescent Asaia. Strain SF2.1(Gfp) was capable of recolonizing salivary glands in a relatively short time (48 h) after ingestion of the sugar meal. Strain SF2.1(Gfp) was observed in the male reproductive system 48 h after the exposure of male mosquitoes to the sugar solution. Bacteria massively colonized testes, and, in particular, gonoducts (Fig. 3 A–H), demonstrating that the male reproductive system is a further subniche for Asaia in An. stephensi. Interestingly, large fluorescent microcolonies were observed within male gonoducts (Fig. 3H), indicating bacterial growth. Signals obtained by ISH with Asaia-specific probes also identified Asaia sp. in the spermatic bundles of An. stephensi males that were not exposed to strain SF2.1(Gfp) (Fig. 3I). A large number of bacterial cells resembling Asaia sp. was also observed by TEM in gonoducts of males not colonized by strain SF2.1(Gfp). These cells showed the typical morphology of Asaia sp. with signature filamentous structures in the nucleoid region and, like cultured Asaia cells (Fig. 1E), did not present the extracellular matrix that was observed in the mosquito midgut (Fig. 1D).
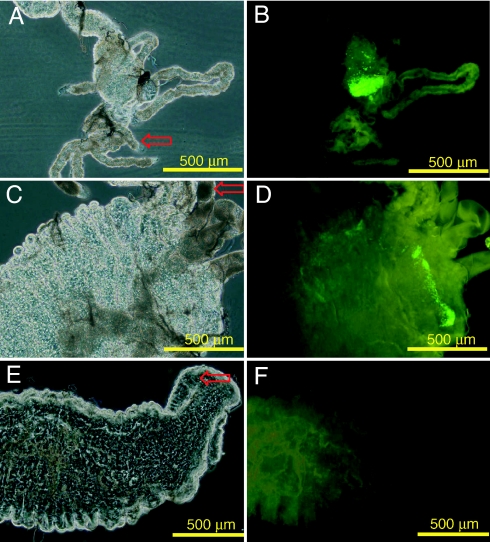
Fig. 2.
Recolonization of the adult An. stephensi gut by a Gfp-tagged Asaia. Phase contrast (A and C) and fluorescence (B and D) microscope images of Asaia sp. strain SF2.1(Gfp), recolonizing An. stephensi midgut. Male (A and B) and female (C and D) terminal portions of the midgut. Malpighian tubules are visible (red arrows). (E and F) Midgut terminal portion of a female fed with sucrose solution without Asaia sp. strain SF2.1(Gfp). (G and H) Laser scanning confocal microscope images of adult mosquito guts, showing high concentrations of strain SF2.1(Gfp) microcolonies.
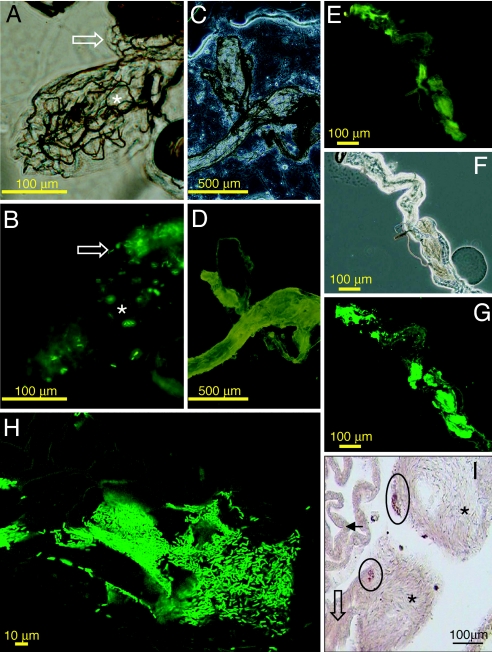
Fig. 3.
Colonization of male gonads of adult An. stephensi by Asaia sp. A large number of Asaia cells was found in the reproductive organs of adult, male An. stephensi. (A–D) Reproductive organs of an An. stephensi adult male fed (A and B) or not fed (C and D) with a sucrose-containing solution in which Asaia sp. SF2.1(Gfp) cells were suspended. A testis (asterisk) and initial portion of a gonoduct (arrow) are visible by phase contrast (A and C) and fluorescence (B and D) microscopy. Fluorescent Asaia sp. SF2.1(Gfp) cells are visible in the testis and in the gonoducts. (E–G) Male gonoduct, after colonization by Asaia sp. strain SF2.1(Gfp), visualized by fluorescent (E), phase contrast (F), and laser scanning confocal microscopy (G). The images indicate a high concentration of Asaia sp. is in the gonoduct. (H) Higher magnification of a microcolony of Asaia sp. strain SF2.1(Gfp) in the male gonoduct. (I) ISH showing cell clusters of Asaia sp. cells (circles) colonizing the male reproductive organs. Asterisks indicate spermatic bundles. A sperm duct (open black arrow) is visible. Intestine is indicated by a black arrow. The specimen pictured was taken from a male not exposed to strain SF2.1(Gfp).
Second and third instar larvae of An. stephensi were also fed with strain SF2.1(Gfp) that was resuspended in the larvae breeding water. Colonization of the larval gut was observed at 24 h after exposure. We examined a total of 24 larvae (12 each for L2 and L3 stages). Seven L2 and five L3 larvae showed massive colonization in the gut by large fluorescent microcolonies.
The observation of Asaia in the male gonoduct suggested that transmission of Asaia from male to female may occur during mating. We thus caged 16 virgin females with 24 males previously fed with a cell suspension of strain SF2.1(Gfp). Eight of the 16 females showed fluorescent Asaia cells and microcolonies in the spermatheca and the gut. It is possible that the percentage of Asaia-positive females could be biased by a low mating rate due to a nonoptimal male-to-female ratio in the cage. Finally, 16 females of An. stephensi previously fed with strain SF2.1(Gfp) were allowed to have a blood meal and lay eggs. Thirty-two of the hatched larvae were then examined (eight individuals for each larval stage from L1 to L4) and 19 of these showed a massive colonization by fluorescent cells and microcolonies in the gut, indicating that the bacterium is transmitted to the offspring. This result is congruent with PCR data showing the presence of Asaia sp. DNA in preadult stages of An. stephensi. There is thus an overall consistency of results indicating that colonization of mosquitoes occurs early during their development. It is reasonable to assume that infection of mosquito larvae occurs by acquisition of Asaia sp. from the environment, if we consider, for example, the results reported above on the colonization of larvae by Asaia strain SF2.1(Gfp) released in the breeding water. However, the fluorescent Asaia cells that were detected in larvae generated by females fed on SF2.1(Gfp) strain indicate that Asaia is also transmitted from mother to offspring. Understanding whether this transmission is direct (e.g., based on some forms of transovarial transmission or egg smearing) or indirect (e.g., adult mosquitoes contaminate the environment during egg laying, and bacteria then infect the progeny) still remain to be determined.
Perspectives.
Flowers and plant-derived materials have been reported as the natural habitats of Asaia spp. (17, 18, 23). Strains of the genus Asaia have also been isolated in bottled fruit-flavored drinks (24), and A. bogorensis was recently isolated from the blood of an i.v. drug user in Finland (25). Based on the results reported here, mosquitoes of the genus Anopheles could represent another environmental niche for Asaia where it lives at a particularly high density in the gut. This acetic acid bacterium could be taken up by mosquitoes from their environment, i.e., from water during the larval stages or from flowers during the first sugar meals as an adult. Thereafter, particular physiological and metabolic requirements would allow infection of the insect body. It would be interesting to study the environmental sources of infection of the mosquito body. Bacteria of the genus Asaia have not been previously reported as associated with any other entomological system, except with Scaphoideus titanus (Hemiptera: Cicadellidae), the vector of Flavescence Dorée in grapevines (8). The identification of bacteria of the genus Asaia in insect species of different orders like Diptera and Hemiptera suggests that they may be widespread symbionts of insects. The presence of Asaia in the three Anopheles species that we have examined and its very high prevalence in different developmental stages of An. stephensi and An. maculipennis indicate that Asaia may play an important role in the biology of the host that will be investigated in future studies. Here, we have shown that Asaia is capable of efficiently crossing body barriers and colonizing different organs in a short time, like guts and salivary glands, both important sites of Plasmodium parasites' life cycle. The ability of Asaia sp. to recolonize the mosquitoes was also confirmed by the capacity of the bacterium to grow quickly in several different body parts, as shown by the formation of microcolonies (Fig. 3H) and the identification of cells undergoing division (see for example Fig. 1D). The high level of mosquito colonization reached by Asaia sp. after feeding indicates that horizontal transmission by the oral route can efficiently lead to the infection of the mosquito body and could possibly be exploited in the field for insect colonization.
Vertical transmission to the progeny is commonly observed in insects for intracellular bacterial symbionts (26), whereas extracellular midgut bacteria are generally regarded as opportunistic microorganisms acquired from the environment (14). However, a clear-cut distinction between environmental acquisition and vertical transmission of symbionts is not always easy to establish, such as in the case of microorganisms living in the gut of wood-feeding cockroaches and termites (27). Our results show that the Anopheles–Asaia symbiotic system might represents such a borderline situation, where acquisition from the environment is likely the most common source of infection for both preadult and adult stages, but where transmission from mother to offspring might also occur, leading to an efficient exploitation of the insect niche by this environmental bacterium. The efficiency by which Asaia might exploit the insect niche is also highlighted by the capacity demonstrated by strain SF2.1(Gfp) of colonizing different body parts and by its transmission from male to female during mating, as it has recently been shown for beneficial secondary symbionts in aphids (28).
Easy acquisition by both mosquito adults and larvae, culturability, cryogenic preservability, and the easy transformability of Asaia spp. make it a candidate for paratransgenic control (29) of malaria (14), through the production of anti-Plasmodium molecules (30–34) or anti-mosquito factors (15) by engineered bacterial strains (30).
Materials and Methods
Mosquitoes.
An. stephensi samples came from a colony reared since 1988 in the insectary at the University of Camerino. An. maculipennis specimens were sampled in June and October 2005 and June 2006, inside a cowshed of a small farm close to Orte, in central Italy. An. gambiae specimens were field-collected in various villages of Burkina Faso (West Africa) in the period 2002–2004.
DNA-Based Analysis of the Mosquitoes' Microflora.
DNA extraction from whole insects and organs/tissues was performed as described (35). Extracted DNA was used as template in PCRs with universal 16S rRNA bacterial primers 27F 5′-TCGACATCGTTTACGGCGTG-3′ and 805R 5′-AGAGTTTGATCCTGGCTCAG-3′ and conditions reported in SI Materials and Methods. For Asaia-specific PCR, primers sets 20F–1500R, 520F–520R, and 920F–920R (17) and primers Asafor (5′-GCGCGTAGGCGGTTTACAC-3′) and Asarev (5′-AGCGTCAGTAATGAGCCAGGTT-3′) have been used following conditions described elsewhere (17) (see also SI Materials and Methods). Sequences of the purified amplicons were analyzed by using BLASTn (www.ncbi.nlm.nih.gov/blast/) and RDPII (http://rdp.cme.msu.edu), and comparative analysis was performed by ClustalX v.1.83 (36) and Treecon v.1.3b software (Van de Peer, University of Konstanz, Konstanz, Germany). Bacteria 16S rRNA gene copies in the total DNA extracted from insect organs were determined by quantitative real-time-PCR in a I-cycler thermal cycler (Bio-Rad, Hercules, CA) by using primers 357F (5′-CTACGGGAGGCAGCAG-3′) and 907R (5′-CCGTCAATTCCTTTGAGTTT-3′) (37). For Asaia, primers Asafor and Asarev were used. The reactions were performed with Brilliant Sybr green qPCR Master Mix (Stratagene, La Jolla, CA).
TEM and ISH.
Adults of An. stephensi were dissected in saline. Semithin sections for light microscopy and thin sections for TEM were prepared and examined as described (8). ISH was performed on paraffin-embedded sections of mosquito organs as described (38). Formamide concentration was adjusted at 30%, and hybridization temperature was set at 46°C. Probe Eub338 was used as a bacterial positive control, and probes Asaia1 (5′-AGCACCAGTTTCCCGATGTTAT-3′) and Asaia2 (5′-GAAATACCCATCTCTGGATA-3′), designed to target the 16S rRNA were used for specific detection of Asaia cells. All of the probes were 5′-labeled with digoxigenin, and anti-digoxigenin/horseradish peroxidase antibodies were used as secondary antibodies. Staining was performed with 3-amino-9-ethylcarbazole and observation was with a light microscope (see also SI Materials and Methods).
Asaia Isolation, Identification, Count in Mosquito Individuals, and Transformation.
Asaia was isolated by a preenrichment step in liquid medium (pH 3.5) and hence plated in agarized medium as described (17, 18). Asaia colonies were tentatively identified by colony morphology and the formation of carbonate dissolution haloes in agar plates. Identification was confirmed by 16S rRNA gene sequencing. Asaia cells in the whole insect were counted in 96-well microtiter plates by the most-probable number method in liquid medium, followed by plating on calcium carbonate-containing medium (17, 18) from the growth-positive wells. Colony identity was confirmed by 16S rRNA gene sequencing of randomly picked colonies. Asaia sp. cells were electroporated with plasmids pHM2 and pHM3 (21) as reported in SI Materials and Methods. A gfp gene cassette was cloned in pHM2 plasmid as reported in SI Materials and Methods and electroporated in Asaia sp. strain SF2.1 isolated from an An. stephensi female. Successful transformants were confirmed by fluorescence microscopy, plasmid analysis, and PCR of the gfp gene cassette as reported in SI Materials and Methods. A Gfp-tagged transformant, designated as strain SF2.1(Gfp), was hence selected for mosquito recolonization experiments.
Mosquito Recolonization by Asaia sp. SF2.1(Gfp).
Asaia sp. SF2.1(Gfp) was grown 24 h at 30°C in GLY medium (SI Materials and Methods). Cells were harvested by centrifugation, washed three times in 0.9% (wt/vol) NaCl and adjusted to 103 or 108 cells per ml−1 in 30 ml of H2O/5% (wt/vol) sucrose solution or in 1 ml of human blood to feed adult mosquitoes. For monitoring long-term colonization of An. stephensi, the suspension containing 103 cells per ml−1 was supplemented with 50 μg·ml−1 of kanamycin to avoid the loss of plasmid from bacterial cells. Cages were prepared to feed An. stephensi adult with 2 × 108 or 4 × 108 cells per ml−1 of Asaia sp. strain SF2.1(Gfp), respectively. After 24, 48, 72, and 96 h of exposure to the feeding solutions containing strain SF2.1(Gfp), mosquitoes were dissected in PBS. One additional cage was fed with 4 × 103 cells per ml−1 of Asaia sp. strain SF2.1(Gfp), resuspended in 5% (wt/vol) sucrose solution. After 2 days feeding, the cotton swab containing strain SF2.1(Gfp) was removed and replaced with a new sterile 5% (wt/vol) sucrose solution supplemented with kanamycin. Mosquitoes fed with 4 × 103 cells per ml−1 of Asaia sp. strain SF2.1(Gfp) were sampled every 2–3 days up to 25 days after initial exposure to the bacterium. To feed 2nd and 3rd instar larvae, 108 cells per ml−1 were released in 40-ml batches of breeding water (each containing 60 larvae). The mosquitoes and the larvae were maintained with their usual diet. Guts, salivary glands, and reproductive organs were examined with a IX71 fluorescent microscope (Olympus, Melville, NY), and a MRC600 laser scanning confocal microscope (Bio-Rad). All tissues were fixed with 4% paraformaldeyde for 10 min at 4°C, with the exception of salivary glands. The slides were then mounted in glycerol–PBS for analysis. Whole larvae were directly observed without dissection after several washings in sterile distilled water.
Supplementary Material
Acknowledgments
We thank K. J. Heller (Institute for Microbiology, Federal Dairy Research Center for Nutrition and Food, Kiel, Germany) for providing us plasmids pHM2 and pHM3 for transformation experiments of Asaia sp., Dr. L. J. Halverson (Iowa State University, Ames, IA) for the gfp gene cassette, P. Ballarini for the help with the fluorescent and confocal microscopy analysis, K. Bourtzis for critically reading the manuscript, and L. Margulis for encouragement, discussion, and suggestion.
Abbreviations
- Gfp
green fluorescent protein
- ISH
in situ hybridization
- TEM
transmission electron microscopy.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The 16S rRNA gene sequences of isolates reported in this paper have been deposited in the DNA Data Bank of Japan/European Molecular Biology Laboratory/GeneBank databases (accession nos. AM404260, AM404261, and AM404262).
This article contains supporting information online at www.pnas.org/cgi/content/full/0610451104/DC1.
References
- 1.Atkinson PW, Michel K. Genesis. 2002;32:42–48. doi: 10.1002/gene.10026. [DOI] [PubMed] [Google Scholar]
- 2.Grossman GL, Rafferty CS, Clayton JR, Stevens TK, Mukabayire O, Benedict MQ. Insect Mol Biol. 2001;10:597–604. doi: 10.1046/j.0962-1075.2001.00299.x. [DOI] [PubMed] [Google Scholar]
- 3.Catteruccia F, Nolan T, Loukeris TG, Blass C, Savakis C, Kafatos FC, Crisanti A. Nature. 2000;405:959–962. doi: 10.1038/35016096. [DOI] [PubMed] [Google Scholar]
- 4.Ito J, Ghosh A, Moreira AL, Wimmer EA, Jacobs-Lorena M. Nature. 2002;417:452–455. doi: 10.1038/417452a. [DOI] [PubMed] [Google Scholar]
- 5.Catteruccia F, Godfray HC, Crisanti A. Science. 2003;299:1225–1227. doi: 10.1126/science.1081453. [DOI] [PubMed] [Google Scholar]
- 6.Beard CB, Durvasula RV, Richards FF. Emerg Infect Dis. 1998;4:581–591. doi: 10.3201/eid0404.980408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beard CB, Cordon-Rosales C, Durvasula RV. Annu Rev Entomol. 2002;47:123–141. doi: 10.1146/annurev.ento.47.091201.145144. [DOI] [PubMed] [Google Scholar]
- 8.Marzorati M, Alma A, Sacchi L, Pajoro M, Palermo S, Brusetti L, Raddadi N, Balloi A, Tedeschi R, Clementi E, et al. Appl Environ Microbiol. 2006;72:1467–1475. doi: 10.1128/AEM.72.2.1467-1475.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schnepf E, Crickmore N, Van Rie J, Lereclus D, Baum J, Feitelson J, Zeigler DR, Dean DH. Microbiol Mol Biol Rev. 1998;62:775–806. doi: 10.1128/mmbr.62.3.775-806.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zabalou S, Riegler M, Theodorakopoulou M, Stauffer C, Savakis C, Bourtzis K. Proc Natl Acad Sci USA. 2004;101:15042–15045. doi: 10.1073/pnas.0403853101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zchori-Fein E, Gottlieb Y, Kelly SE, Brown JK, Wilson JM, Karr TL, Hunter MS. Proc Natl Acad Sci USA. 2001;98:12555–12560. doi: 10.1073/pnas.221467498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beard CB, Dotson EM, Pennington PM, Eichler S, Cordon-Rosales C, Durvasula RV. Int J Parasitol. 2001;31:621–627. doi: 10.1016/s0020-7519(01)00165-5. [DOI] [PubMed] [Google Scholar]
- 13.Baldridge GD, Burkhardt NY, Simser JA, Kurtti TJ, Munderloh UG. Appl Environ Microbiol. 2004;70:6628–6636. doi: 10.1128/AEM.70.11.6628-6636.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Riehle MA, Jacobs-Lorena M. Insect Biochem Mol Biol. 2005;35:699–707. doi: 10.1016/j.ibmb.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 15.Khampang P, Chungjatupornchai W, Luxananil P, Panyim S. Appl Microbiol Biotechnol. 1999;51:79–84. doi: 10.1007/s002530051366. [DOI] [PubMed] [Google Scholar]
- 16.Lindh JM, Terenius O, Faye I. Appl Environ Microbiol. 2005;71:7217–7223. doi: 10.1128/AEM.71.11.7217-7223.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamada Y, Katsura K, Kawasaki H, Widyastuti Y, Saono S, Seki T, Uchimura T, Komagata K. Int J Syst Evol Microbiol. 2000;50:823–829. doi: 10.1099/00207713-50-2-823. [DOI] [PubMed] [Google Scholar]
- 18.Katsura K, Kawasaki H, Potacharoen W, Saono S, Seki T, Yamada Y, Uchimura T, Komagata K. Int J Syst Evol Microbiol. 2001;51:559–563. doi: 10.1099/00207713-51-2-559. [DOI] [PubMed] [Google Scholar]
- 19.Dong Y, Taylor HE, Dimopoulos G. PLoS Biol. 2006;4:e229. doi: 10.1371/journal.pbio.0040229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ben-Dov E, Shapiro OH, Siboni N, Kushmaro A. Appl Environ Microbiol. 2006;72:6902–6906. doi: 10.1128/AEM.00849-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng Q, Aksoy S. Insect Mol Biol. 1999;8:125–132. doi: 10.1046/j.1365-2583.1999.810125.x. [DOI] [PubMed] [Google Scholar]
- 22.Mostafa HE, Heller KJ, Geis A. Appl Environ Microbiol. 2002;68:2619–2623. doi: 10.1128/AEM.68.5.2619-2623.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bae S, Fleet GH, Heard GM. J Appl Microbiol. 2006;100:712–727. doi: 10.1111/j.1365-2672.2006.02890.x. [DOI] [PubMed] [Google Scholar]
- 24.Moore JA, McCalmont M, Xu J, Millar BC, Heaney N. Appl Environ Microbiol. 2002;68:4130–4131. doi: 10.1128/AEM.68.8.4130-4131.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tuuminen T, Heinäsmäki T, Kerttula T. J Clin Microbiol. 2006;44:3048–3050. doi: 10.1128/JCM.00521-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moran NA. Curr Biol. 2006;16:R866–871. doi: 10.1016/j.cub.2006.09.019. [DOI] [PubMed] [Google Scholar]
- 27.Nalepa CA, Bignell DE, Bandi C. Insect Socieaux. 2001;48:194–201. [Google Scholar]
- 28.Moran NA, Dunbar HE. Proc Natl Acad Sci USA. 2006;103:12803–12806. doi: 10.1073/pnas.0605772103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bextine B, Lauzon C, Potter S, Lampe D, Miller TA. Curr Microbiol. 2004;48:327–331. doi: 10.1007/s00284-003-4178-2. [DOI] [PubMed] [Google Scholar]
- 30.Ro DK, Paradise EM, Ouellet M, Fisher KJ, Newman KL, Ndungu JM, Ho KA, Eachus RA, Ham TS, Kirby J, et al. Nature. 2006;440:940–943. doi: 10.1038/nature04640. [DOI] [PubMed] [Google Scholar]
- 31.Pidiyar VJ, Jangid K, Patole MS, Shouche YS. Am J Trop Med Hyg. 2004;70:597–603. [PubMed] [Google Scholar]
- 32.Aguilar R, Jedlicka AE, Mintz M, Mahairaki V, Scott AL, Dimopoulos G. Insect Biochem Mol Biol. 2005;35:709–719. doi: 10.1016/j.ibmb.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 33.Mourya DT, Gokhale MD, Pidiyar V, Barde PV, Patole M, Mishra AC, Shouche Y. Acta Virol. 2002;46:257–260. [PubMed] [Google Scholar]
- 34.Pumpuni CB, Beier MS, Nataro JP, Guers LD, Davis JR. Exp Parasitol. 1993;77:195–199. doi: 10.1006/expr.1993.1076. [DOI] [PubMed] [Google Scholar]
- 35.Favia G, Dimopoulos G, della Torre A, Toure YT, Coluzzi M, Louis C. Proc Natl Acad Sci USA. 1994;91:10315–10319. doi: 10.1073/pnas.91.22.10315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. Nucleic Acids Res. 1997;25:4876–4878. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muyzer G, de Waal EC, Uitterlinden AG. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beninati T, Lo N, Sacchi L, Genchi C, Noda H, Bandi C. Appl Environ Microbiol. 2004;70:2596–2602. doi: 10.1128/AEM.70.5.2596-2602.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.