Abstract
The checkpoint protein Rad9/Rad1/Hus1 heterotrimer (the 9-1-1 complex) is structurally similar to the proliferating cell nuclear antigen sliding clamp and has been proposed to sense DNA damage that leads to cell cycle arrest or apoptosis. Human (h) NEIL1 DNA glycosylase, an ortholog of bacterial Nei/Fpg, is involved in repairing oxidatively damaged DNA bases. In this study, we show that hNEIL1 interacts with hRad9, hRad1 and hHus1 as individual proteins and as a complex. Residues 290–350 of hNEIL1 are important for the 9-1-1 association. A significant fraction of the hNEIL1 nuclear foci co-localize with hRad9 foci in hydrogen peroxide treated cells. Human NEIL1 DNA glycosylase activity is significantly stimulated by hHus1, hRad1, hRad9 separately and the 9-1-1 complex. Thus, the 9-1-1 complex at the lesion sites serves as both a damage sensor to activate checkpoint control and a component of base excision repair.
INTRODUCTION
The eukaryotic genome is vulnerable to an array of DNA damaging agents of both endogenous and environmental origin. Reactive oxygen species (ROS) are commonly generated as by-products during mitochondrial oxidative phosphorylation, during inflammatory responses, and from exposure to ionizing radiation and various chemicals. Effects of ROS are believed to contribute to degenerative diseases such as aging, cancer and cardiovascular disease. The DNA damage induced by ROS includes strand breaks and oxidative base lesions that are typically repaired by the base excision repair (BER) pathway (1,2). The first step in BER is carried out by a lesion-specific DNA glycosylase, which cleaves the N-glycosidic bond between a base and deoxyribose. Using a base-flipping mechanism, glycosylases find lesions in the vast genomic DNA, and excise the damaged bases to generate toxic and mutagenic apurinic/apyrimidinic (AP) sites (3). Bifunctional glycosylases also possess an intrinsic AP-lyase (β- or β/δ-elimination) activity which cleaves 5′-phosphodiester bond (β lyase) or both 5′ and 3′ phosphodiester bonds (β/δ lyase) of the AP site. Escherichia coli Nei (Endo VIII) and Fpg (MutM), are prototypes of DNA glycosylases with β- and β/δ-elimination activities, respectively (4).
Mammalian homologs of E. coli Nei and Fpg have been identified and named Nei-like glycosylases (NEILs) (5–9). NEIL glycosylases containing β/δ-elimination activities excise a broad range of oxidatively damaged bases, including 5-hydroxyuracil (5-OHU), thymine glycol (Tg), uracil, 8-oxoguanine (8-oxoG) and ring-fragmented purines. The substrate specificities of NEIL1 and NEIL2 overlap with those of OGG1 (a functional homolog of E. coli Fpg) and NTH1 (a homolog of E. coli Endo III). However, NEILs have preference for single-stranded DNA, suggesting their roles in repairing transcribed or replicating DNA sequences (10). NEIL1-depleted cells show enhanced radiation sensitivity (11) in contrast to OGG1- and NTH1-null cells, which exhibit no increase in sensitivity to ROS and radiation (12,13). Inactivating mutations in the hNEIL1 gene is associated with gastric cancer (14). The neil1 knockout mice develop the metabolic syndrome with symptoms such as severe obesity, dyslipidemia, and fatty liver disease (15).
DNA repair is coordinated with cell-cycle progression and DNA-damage checkpoints (16,17). Checkpoints are activated upon DNA damage in order to arrest cell cycle progression and to enhance DNA repair or to induce apoptosis with excessive DNA damage. The loss of proper response to DNA damage can lead to genomic instability, and has been implicated in carcinogenesis. This activation requires the action of DNA-damage sensors and transducers, and effectors (18). Among these, Rad9, Rad1 and Hus1 form a heterotrimeric complex (the 9-1-1 complex) that exhibits structural similarity with the homotrimeric proliferating cell nuclear antigen (PCNA) sliding clamp (19–21). The 9-1-1 complex is loaded onto DNA by an alternative clamp loader Rad17/RFC2-5 (22–24). Moreover, the 9-1-1 complex, Rad17/RFC2-5 and PCNA co-localize in foci formed upon DNA damage (25,26). These data suggest a mechanism in which Rad17/RFC2-5 localizes to DNA lesions, allowing the recruitment of the 9-1-1 complex to these sites. Subsequently, the 9-1-1 complex serves as a recruitment platform for the checkpoint effector kinases such as Chk1 or Chk2, which are subsequently phosphorylated by ataxia telangiectasia mutated protein (ATM) or ATR (ATM- and Rad3-related protein). Additionally, a model has recently been proposed by two different groups, where the 9-1-1 complex and the Rad17/RFC2-5 clamp loader could stabilize stalled replication forks (23,27).
The link between checkpoint activation and recruitment of repair machineries to DNA lesions has been demonstrated through interaction and co-localization of checkpoint sensors with proteins involved in various DNA-repair processes upon DNA damage (26,28,29). However, the mechanism by which the sensor checkpoint proteins detect different types of DNA lesions remains elusive. It has been suggested that the checkpoint proteins may detect a common intermediate, such as single-stranded DNA coated by replication protein A (RPA) (30). RPA has been shown to directly interact with the 9-1-1 complex (31). Recently, several reports support a hypothesis that checkpoint proteins may require a series of ‘adaptors’ to recognize DNA damage (28,32–34). Such adaptor proteins may be DNA damage recognition proteins involved in mismatch repair, nucleotide excision repair, BER, and double-strand break repair.
Recent investigations have established a link between the human 9-1-1 complex and the BER pathway. We have shown that the 9-1-1 complex physically and functionally interacts with the MutY homologs (MYHs) of Schizosaccharomyces pombe and human (35,36). In addition, Hus1 phosphorylation is dependent on MYH expression in S. pombe (35). The 9-1-1 complex has been shown to interact with and stimulate other BER enzymes, which include polymerase β (Polβ) (37), flap endonuclease 1 (FEN1) (38,39), and DNA ligase l (40,41). In the present report, we demonstrate that hNEIL1 DNA glycosylase physically and functionally interacts with Rad9, Rad1 and Hus1 as individual proteins and as a complex. The interacting site of the 9-1-1 complex is localized to the C-terminal domain of hNEIL1. Our results strongly suggest that the 9-1-1 complex not only serves as a damage sensor to activate checkpoint control, but it is also a component of the BER pathway and may provide a platform for different factors involved in BER.
MATERIALS AND METHODS
Human cell culture
Human HeLa S3 and 293 cell lines were purchased from American Type Cell Culture (ATCC). HeLa cells were cultured in modified Ham's F-12 (Mediatech, Herndon, VA, USA) supplemented with 10% fetal bovine serum (FBS). The 293 cells were cultured in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% FBS. At 90% confluence, cells were transfected with pFLAG-NEIL1 (see below) using Fugene6 (Roche, Nutley, NJ, USA). The cells were replanted at 24 h after the transfection. Cell extracts were prepared as described (42,43). The protein concentration was determined by Bio-Rad's protein assay (Bio-Rad, Hercules, CA, USA).
Construction of expression plasmids pFLAG-NEIL1
To generate the FLAG-tagged NEIL1 expression construct, hNEIL1 cDNA encoded residues 1-390 was polymerase chain reaction (PCR) amplified with primers hNEIL1-5′-EcoRI and hNEIL1-3′-XbaI (listed in Table S1 in the supplementary data) using template pRESETB-hNEIL1 (8). The PCR products were digested with EcoRI and XbaI, then ligated into EcoRI and XbaI digested pFLAG-CMV 5.1 (Sigma, St. Louis, MO, USA) vector for expression of C-terminal FLAG-tagged NEIL1. The cloned NEIL1 gene sequence was confirmed by DNA sequencing using CMV30 and CMV24 (Sigma) sequencing primers.
Construction of expression plasmids for glutathione-S-transferase (GST)-fusions and His-hNEIL1 protein in E. coli
The full-length cDNA encoded residues 1–390 of hNEIL1 was amplified by PCR method by Pfu DNA polymerase using the primers hNEIL1-F and hNEIL1-R (listed in Table S1 in the supplementary data), and template pFLAG-NEIL1. The PCR products were digested with BglII and XhoI and ligated into the BamHI-XhoI-digested pGEX-4T-2 vector (GE Health, Princeton NJ, USA) and pET-21a vector (EMD Biosciences, San Diego, CA, USA) to yield plasmids pGEX-NEIL1 and pET-NEIL1, respectively. The sequences of the cloned genes were confirmed by DNA sequencing. The plasmids pGEX-3X-hHus1 (hHus1 coding region inserted at BamHI and XhoI sites) and pGEX-4T3-hRad9 (hRad9 coding region inserted at BamHI and EcoRI sites), which contained GST-tagged hHus1 and hRad9, respectively, were obtained from Dr A. E. Tomkinson (University of Maryland, Baltimore, MD, USA). The pGEX-3X-hRad1 plasmid (Rad1 coding region inserted at BamHI and EcoRI sites) was obtained from Dr E. Y. Lee (University of California, Irvine, CA, USA) through Dr A. E. Tomkinson.
Construction and expression of hRad9, hRad1 and hHus1 in E. coli
The cDNA of hHus1, hRad1 and hRad9 were amplified by PCR from GST-Rad9, GST-Rad1 and GST-Hus1 plasmids. The sequences of forward and reverse primers for these PCR reactions are given in Table S1 in the supplementary data. The hHus1 gene was cloned between the BamH1 and Not1 sites of pET-21a (EMD Biosciences) to obtain the clone pET21a-hHus1 as described (36). The hRad9 gene was cloned between the BglII and XhoI sites of pACYCDuet-1 (EMD Biosciences) to obtain the clone pACYCD-hRad9. The hRad1 gene was cloned between the BamH1 and SalI sites of pACYCD-hRad9 to obtain pACYCD-hRad1-hRad9. The hHus1, hRad1, and hRad9 proteins were tagged with a C-terminal His, N-terminal His, and C-terminal S-tag, respectively.
The BL21 Star cells (Stratagene, La Jolla, CA, USA) harbouring the expression plasmids, pET21a-hHus1 and pACYCD-hRad1-hRad9, were cultured in LB broth containing 100 μg/ml of ampicillin and 50 μg/ml of chloramphenical at 37°C. Protein expression was induced at an A590 of 0.6 by the addition of iso-propylthiogalactoside (IPTG) to a final concentration of 0.2 mM and the cells were grown at 20°C and then harvested 16 h later.
Expression and purification of hRad9, hRad1, hHus1 and the 9-1-1 complex in baculovirus system
The Sf9 insect cells (Invitrogen, Princeton, NJ, USA) were grown in 80 ml of Sf-900 II SFM complete medium (GIBCO/BRL) in suspension to 3 × 106 cells/ml and then infected with baculovirus vectors containing cDNA encoding FLAG-hRad9 (obtained from Dr Alan Tomkinson), FLAG-hRad1(obtained from Dr Aziz Sancar), or FLAG-hHus1 (obtained from Dr Aziz Sancar) and supplemented with 0.2% FBS. For expression of the 9-1-1 complex, 500 ml culture of the Sf9 insect cells were coinfected with a mixture of three viruses carrying cDNA encoding hRad9, hRad1 and hHus1. About 48 h after infection, the cells were harvested by centrifuge at 1500 × g for 2 min. The cell pellets were lysed in 10 packed cell volumes of lysis buffer [(50 mM Tris-HCl, pH 7.5, 0.5% Nonidet P-40, 0.3 M NaCl, and 0.1 mM phenylmethylsulfonyl fluoride (PMSF)] and incubated on ice for 15 min before centrifuged for 30 min at 32 000 × g. The supernatants were incubated with 1 ml (for the 9-1-1 complex) or 0.5 ml (for each subunit) of 50% slurry of Anti-FLAG M2 Affinity Gel (Sigma) at 4°C overnight. The resin was then washed four times with lysis buffer and the proteins were eluted with 1 ml (for h9-1-1 complex) or 0.45 ml (for each subunit) elution buffer [50 mM Tris–HCl, pH 7.5, 0.05% Nonidet P-40, 0.3 M NaCl, 0.1 mM PMSF and 200 μg/ml FLAG peptide (Sigma)]. FLAG-hRad9, FLAG-hRad1 and FLAG-hHus1 were dialysed against lysis buffer without Nonidet P-40 and stored at −80°C. The 9-1-1 complex was further purified by a Sepharose-12 gel filtration column (GE Health) with a buffer containing 20 mM KPO4, pH 7.4, 0.1 mM EDTA, 0.2 M KCl, 10% glycerol, 0.5 mM dithiothreitol (DTT), 0.1 mM PMSF. The column was run at 0.25 ml/min and 0.25 ml fractions were collected. By comparison to the size markers (bovine thyroglobin, apoferritin, β-amylase, and bovine serum albumin), the 9-1-1 complex eluted in a position corresponding to a mass of about 120 kDa, which is in line with the theoretical value (110 kDa) of a trimeric complex. The samples were divided into small aliquots and stored at −80°C. The protein concentration was determined by the Bradford method (44).
The human 9-1-1 complex expressed in the baculovirus system used in the initial experiments was kindly provided by Dr Howard Lieberman (Columbia University).
Other proteins used
Untagged hNEIL1 and deletion constructs were purified as described (8,45). His-tagged hNEIL1 was purified from E. coli BL21 Star cells (Stratagene) that harbour the expression plasmid pET-NEIL1 by Ni-NTA resin (QIAGEN, Valencia, CA, USA) and then by 1 ml Hi-Trap Q and SP columns (GE Health) as described (8,45). Human His-tagged hHus1 expressed in E. coli BL21 Star cells (Stratagene) was purified by Ni-NTA resin (QIAGEN) and 1 ml Heparin column (GE Health) as described (36). The human 9-1-1 complex expressed in E. coli BL21 Star cells was partially purified by Ni-NTA resin (QIAGEN) harboring the expression plasmids, pET21a-hHus1 and pACYCD-hRad1-hRad9.
GST pull-down assay
The BL21 Star cells (Stratagene) harbouring the GST expression plasmids were cultured in LB broth containing 100 μg/ml of ampicillin. Protein expression was induced as described above. The cell paste, from a 500 ml culture, was resuspended in 9 ml of buffer G (50 mM Tris–HCl, pH 7.4, 150 mM NaCl and 2 mM EDTA) containing 0.5 mM DTT and 0.1 mM PMSF and treated with lysozyme (1 mg/ml) for 30 min at 4°C. After sonication, the solution was centrifuged at 10 000 × g for 20 min and the supernatant was saved. The GST-tagged proteins were immobilized on glutathione-sepharose 4B (GE Health) as described (43). GST fusion proteins (500 ng) were incubated with purified protein or cell extracts in 0.2 ml volume at 4°C with shaking overnight. After centrifugation at 1000 × g for 2 min, the pellets were washed five times with 1 ml of buffer G containing Nonidel P-40. Bound proteins were eluted by boiling in SDS loading buffer [30 mM Tris-HCl, pH 6.8, 5% (v/v) glycerol, 1% SDS, 0.5 mg/ml bromophenol blue and 1% β-mercapoethanol] and resolved on a 12% SDS polyacrylamide gel. The proteins were subsequently analysed by western blot using the corresponding antibodies according to established methods.
Ni-affinity binding
His-Select (Sigma) magnetic beads (20 μl suspension) were washed once with water, equilibrated with binding buffer (50 mM Tris–HCl, pH 7.5, 200 mM NaCl, 10 mM imidazole), and then incubated with His-tagged hHus1 (500 ng) in binding buffer for 1 h at 4°C with gentle rocking. The beads were then pelleted using a magnetic separator and washed three times with binding buffer then equilibrated with interaction buffer (50 mM Tris–HCl, pH 7.5, 400 mM NaCl, 10 mM imidazole). Wild-type and mutant hNEIL1 (250 ng) in interaction buffer were incubated with the beads for 2 h at 4°C with gently rocking. Again the beads were pelleted on a magnetic separator and washed three times with final wash buffer (50 mM Tris–HCl, pH 7.5, 500 mM NaCl, 10 mM imidazole). The NEIL1/Hus1 complex was eluted with 20 μl of SDS loading buffer and resolved on a 12% SDS polyacrylamide gel. The presence of NEIL1 was examined by western analysis with anti-NEIL1 antibody.
Far-western analysis
Wild-type and mutant hNEIL1 were separated by 10% SDS-PAGE and transferred to nitrocellulose membrane. The membrane was washed with 1× PBS and treated with 6 M guanidine-HCL in PBS for 10 min at 4°C. The proteins were then renatured with serial dilutions of guanidine-HCL in PBS, diluted by 1 mM DTT in PBS at 4°C to a final concentration of 0.09 M guanidine-HCL. After blocking with 5% non-fat dry milk and Tween-20 in PBS for 45 min at 4°C, the membrane was incubated with 10 pmol/ml of his-tagged hHus1 in blocking solution containing 1 mM DTT and 100 mM trimethylamine-N-oxide dihydrate (TMAO) for 3 h at 4°C. Subsequent western blotting was performed using anti-His-tag antibody followed by anti-Rabbit IgG HRP-conjugated secondary antibody (Amersham Biosciences, Piscataway, NJ, USA).
Co-immunoprecipitation
Extracts (1 mg) derived from HeLa cells expressing FLAG-NEIL1 were precleared by adding 30 μl Protein G agarose (Invitrogen) for 1–4 h at 4°C. After centrifugation at 1000 × g, the supernatant was incubated with 4 μg of polyclonal anti-FLAG or anti-His (HA) overnight at 4°C. Protein G agarose (30 μl) was added and incubated for 4–12 h at 4°C. After centrifugation at 1000 × g, the supernatant was saved and the pellet was washed. Both the supernatant (∼10% of total volume) and pellet fractions were resolved on a 12% SDS–PAGE and western blot analysis for hRad9 was performed.
Western blotting and antibodies
Proteins were separated on SDS-polyacrylamide gels and transferred to nitrocellulose membrane. The membranes were blocked with PBS with 0.1% Tween-20 and 10% nonfat dry milk, reacted with primary antibodies, and then incubated with horseradish peroxidase-linked second antibodies with wash between each step (46). Western blotting was detected by the Enhanced Chemiluminescence (ECL) analysis system (GE Health) according to the manufacturer's protocol. Human NEIL1 polyclonal antibody was from Alpha Diagnostics. Polyclonal and monoclonal antibodies of Rad9 are from Stratagene and Imegenex, respectively. His-tag antibody is from BD Bioscience while anti-FLAG and HA are from Sigma.
Immunofluorescence staining
Human HeLa cells were transiently transfected with pFLAG-NEIL1, replanted at 24 h after the transfection in Lab-Tek chamber slides (NUNC) overnight, treated with 5 mM H2O2 for 40 min, and then recovered in serum-free media for 6 h. The cells were fixed with 4% formaldehyde for 15 min at room temperature, and permeabilized at room temperature in PBS-0.1% Triton X-100 for 10 min. After being blocked in PBS containing 15% FBS for 15 min at 37°C, the cells were reacted with FLAG monoclonal antibody (Sigma/Aldrich) and hRad9 polyclonal antibody (Stratagene) at 37°C for 30 min. Next, the cells were washed three times for 15 min each in PBS and incubated with Alexa Fluor 594 goat anti-rabbit and Alexa Fluro 488 goat anti-mouse antibodies (Invitrogen) at a 1:250 dilution in PBS for 30 min at 37°C. The cells were then washed three times in PBS. Nuclear DNA was counterstained with 4′,6′-diamidineno-a-phenylindole (DAPI) (Vector Laboratories, Burlingame, CA, USA). Images were captured with a Nikon E400 fluorescent microscope with an attached CCD camera.
NEIL1 glycosylase activity assay
The 54-mer duplex DNA substrate containing Tg (listed in Table S1 in the supplementary data) for hNEIL1 was a gift from Dr Susan Wallace at the University of Vermont. The strand containing Tg was labeled at the 5′ end with [γ-32P]ATP by polynucleotide kinase and then was annealed with the other strand as described by Lu et al. (47). The hNEIL1 reaction (10 μl) contained 2.5 mM HEPES (pH 7.8), 1 mM DTT, 2.5% Glycerol, 50 mM KCl, 50 μg/ml bovine serum albumin, 0.5 mM EDTA and 1.8 fmol of DNA substrate. The hHus1 or 9-1-1 complex was added immediately after hNEIL1 and reactions proceeded at 37°C for 30 min. About 5 μl of formamide dye (90% formamide, 10 mM MEDTA, 0.1% xylene cyanol, and 0.1% bromophenol blue) was added to the sample, which was heated at 90°C for 3 min and 7 μl of the mixture was loaded onto a 14% polyacrylamide sequencing gel containing 7 M urea. The gel images were viewed on a PhosphorImager and quantified using the ImageQuant software (Molecular Dynamics). The area at the product position in the no protein control lane was used to subtract out the background signal. The hNEIL1 cleavage activity was calculated by the percentage of product over total DNA (product plus intact bands).
RESULTS
The human 9-1-1 complex interacts with hNEIL1
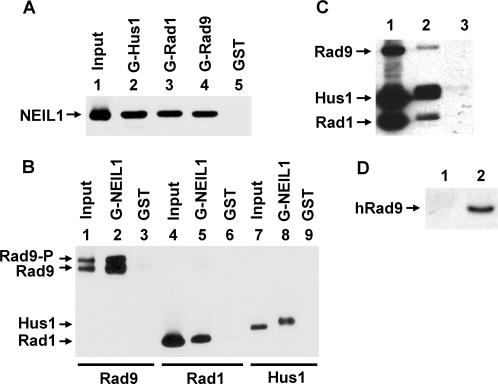
We have shown that the Rad9/Rad1/Hus1heterotrimer interacts with MYH in S. pombe and human cells (35,36). To determine whether the 9-1-1 complex interacts with other DNA glycosylases, we tested hNEIL1 because it is also involved in repairing DNA lesions derived from oxidative damage. We used the GST pull-down assay to show the physical interactions of hNEIL1 with hRad9, hHus1 and hRad1. GST-hHus1, GST-hRad1, or GST-hRad9 fusion protein bound to glutathione-Sepharose was incubated with purified hNEIL1 protein (residues 2–390). As shown in Figure 1A, hNEIL1 could be pulled down by GST-hHus1, GST-hRad1 and GST-hRad9. As a negative control, hNEIL1 did not bind to GST alone (lane 5). In reciprocal experiments, GST-hNEIL1 fusion protein bound to glutathione-Sepharose could pull down hRad9, hRad1 and hHus1 expressed in the baculovirus-transfected insect cells (Figure 1B). Both phosphorylated and un-phosphorylated hRad9 interact with hNEIL1 (Figure 1B, lane 2). Thus, hNEIL1 binds to all three subunits of the 9-1-1 complex. The individual proteins used in Figure 1A, B were expressed separately in E. coli or insect cells, thus hNEIL1 can interact with hHus1, hRad1 and hRad9 when they are not in a complex. In addition, we also incubated immobilized GST-hNEIL1 fusion protein with the partially purified 9-1-1 complex expressed in E. coli. All three subunits of the 9-1-1 complex could be pulled down by GST-hNEIL1 (Figure 1C, lane 2). In addition, the physical interaction between hNEIL1 and hHus1 was demonstrated by Ni-affinity binding. His-tagged hHus1 bound to Ni-beads could pull down hNEIL1 (Figure 2B, lane 1). Finally, Far-western analysis also confirmed the interaction of hNEIL1 and hHus1 (Figure 2C, lane 1).
Figure 1.
Physical interaction between hNEIL1 and the 9-1-1 complex. (A) hNEIL1 binds to all subunits of the 9-1-1 complex. GST-hHus1 (lane 2), GST-hRad1 (lane 3), GST-hRad9 (lane 4), and GST alone (lane 5) were immobilized on glutathione-sepharose and incubated with purified hNEIL1 (residues 2–390, 100 ng). The pellets were fractionated by a 12% SDS-PAGE followed by western blot analysis with the hNEIL1 antibody. Lane 1 contains 30 ng (30% of the total input) of hNEIL1. (B) Binding of hRad9, hRad1 and hHus1 to GST-hNEIL1. GST-tagged NEIL1 containing residues 1–390 (lanes 2, 5 and 8) or GST beads (lanes 3, 6 and 9) were incubated with FLAG-tagged hRad9, hRad1, or hHus1 (∼300 ng) expressed in insect cells. The pellets were fractionated by a 12% SDS-PAGE followed by western blot analysis with the FLAG antibody. Lanes 1, 4 and 7 contain 10 ng (10% of the total input) of hRad9, hRad1 and hHus1, respectively. The slow-migrating band of Rad9 is phosphorylated. (C) Binding of the 9-1-1 complex to GST-hNEIL1. GST-tagged NEIL1 containing residues 1–390 (lane 2) or GST beads (lane 3) were incubated with the partially purified 9-1-1 complex (300 ng) expressed in E. coli. The hHus1, hRad1, and hRad9 proteins were tagged with a C-terminal His, N-terminal His, and C-terminal S-tag, respectively. Lane 1 contains 90 ng (30% of the total input) of the partially purified 9-1-1 complex. The western blot was detected by a mixture of the antibodies against His-tag and S-tag. (D) Coimmunoprecipitation of hRad9 with FLAG-tagged hNEIL1 containing residues 1–390. HeLa cells were transiently transfected with pFLAG-NEIL1. Immunoprecipation was performed with antibody against FLAG and the western blot was detected by the antibody against hRad9 (Imegenex) (lane 2). Lane 1 is a negative control in which the immunoprecipation was performed with antibody against His-tag.
Figure 2.
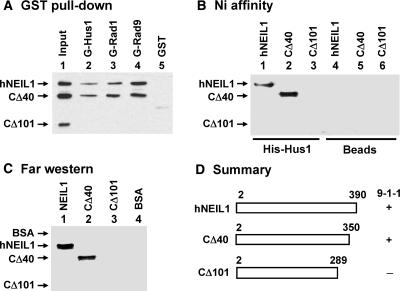
Determination of regions within hNEIL1 involved in binding to the 9-1-1 complex. (A) Binding of hNEIL1 deletion mutants to GST-hRad9, GST-hRad1 and GST-Hus1. Immobilized GST-hHus1 (lane 2), GST-hRad1 (lane 3), GST-hRad9 (lane 4), and GST alone (lane 5) were incubated with a mixture of 100 ng each of purified processed intact hNEIL1 (residues 2–390), hNEIL1-CΔ40 (residues 2–350) and hNEIL1-CΔ101 (residues 2–289). The pellets were fractionated on a 10% SDS–PAGE followed by western blot analysis with the hNEIL1 antibody. Lane 1 contains 30 ng each of hNEIL1, hNEIL1-CΔ40, and hNEIL1-CΔ101 (30% of the total input). (B) Binding of hNEIL1 deletion mutants to His-tagged hHus1. His-hHus1 bound to His-Select (Sigma) magnetic beads (lanes 1–3) or Beads alone (lanes 4–6) were incubated with wild-type or mutant hNEIL1. The presence of hNEIL1 in the pellet was examined by western analysis with anti-NEIL1 antibody. (C) Far-western analysis. Processed intact and mutant hNEIL1 were separated by 10% SDS–PAGE and transferred onto nitrocellulose membrane. Lane 4 contains BSA. The proteins on the membrane were renatured and incubated with 10 pmol/ml of His-tagged hHus1. Subsequent western blotting was performed using His-tag antibody. (D) Graphic depiction of hNEIL1 constructs and the summary of binding results of these constructs with the 9-1-1 complex from Figure 2A–C. The intact hNEIL1 contains 390 amino acid residues, however, processed intact hNEIL1 contains residues 2–390. hNEIL1-CΔ40 and hNEIL1-CΔ101 contain residues 2–350 and 2–289, respectively. The ‘+’ and ‘−’ listed on the right of each construct indicate positive and negative interactions with the 9-1-1 complex, respectively.
The interaction between hNEIL1 and the 9-1-1 complex was also demonstrated by co-immunoprecipitation. We used FLAG antibody to co-immunoprecipitate hRad9 from extracts derived from HeLa S3 cells being transfected with pFLAG-hNEIL1 plasmid. As shown in Figure 1D (lane 2), hRad9 could be immunoprecipitated by FLAG antibody using extracts derived from HeLa cells expressing FLAG-tagged hNEIL1.
Mapping the 9-1-1 interacting domain within hNEIL1
By using truncated hNEIL1 proteins, we determined the region of hNEIL1 engaged in the physical interaction with the 9-1-1 complex. The results are shown in Figure 2A–C and summarized in Figure 2D. In Figure 2A, immobilized GST-hHus1, GST-hRad1 and GST-hRad9 proteins were incubated with 100 ng each of processed intact hNEIL1 (residues 2–390), CΔ40 (residues 2–350), and CΔ101 (residues 2–289). The hNEIL-CΔ40 construct retained interactions with hHus1, hRad1 and hRad9; however, the CΔ101 construct exhibited no interaction with the 9-1-1 complex. Ni affinity binding (Figure 2B) and Far-western analysis (Figure 2C) also confirmed that any subunits of the 9-1-1 complex could interact with CΔ40 but not with the CΔ101 construct. Thus, residues 290–350 of hNEIL1 are essential for the interaction of hNEIL1 with the 9-1-1 complex.
Co-localization between hNEIL1 and 9-1-1
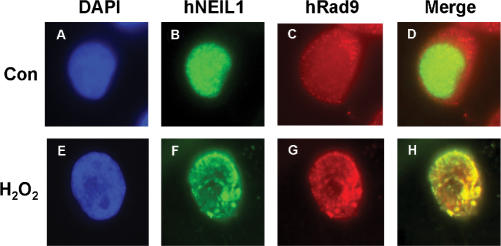
Next, we tested whether hNEIL1 and hRad9 translocate to the same nuclear foci following H2O2 treatment, by immunofluorescent staining analyses. FLAG-hNEIL1 appeared granulated in faint spots throughout the nucleus of untreated HeLa cells expressing FLAG-hNEIL1 (Figure 3B). Human Rad9 protein molecules, as detected by polyclonal antibodies of Rad9 from Stratagene, were distributed to both nucleus and cytoplasm in untreated cells (Figure 3C). Inside the nucleus, a few sites of co-localization of hRad9 and hNEIL1 were observed in control cells (Figure 3D). In H2O2-treated cells, hNEIL1 and hRad9 formed discrete nuclear foci (Figure 3F and G). The majority of the hNEIL1 nuclear foci were found to co-localize with hRad9 foci in H2O2-treated cells (Figure 3H). This data indicates that hNEIL1 and the 9-1-1 complex translocate to repair foci following DNA damage.
Figure 3.
Co-localization of hNEIL1 with hRad9 following oxidative stress. HeLa cells were transiently transfected with pFLAG-hNEIL1. About 24 h after transfection, cells were treated with 5 mM H2O2 for 40 min and then allowed to recover for 6 h (E–H). Control cells were not treated with H2O2 (A–D). The cells were stained with antibody against FLAG (Sigma) (green, B and F) and anti-hRad9 antibody (Stratagene) (red, C and G). (A) and (E) are the DAPI-stained nuclei. (D) is the merged images of (B) and (C). (H) is the merged images of (F) and (G). Co-localization of hNEIL1 (green) and hRad9 (red) is visualized as yellow.
The hNEIL1 activity can be enhanced by hHus1, hRad1, hRad9 and the 9-1-1 complex
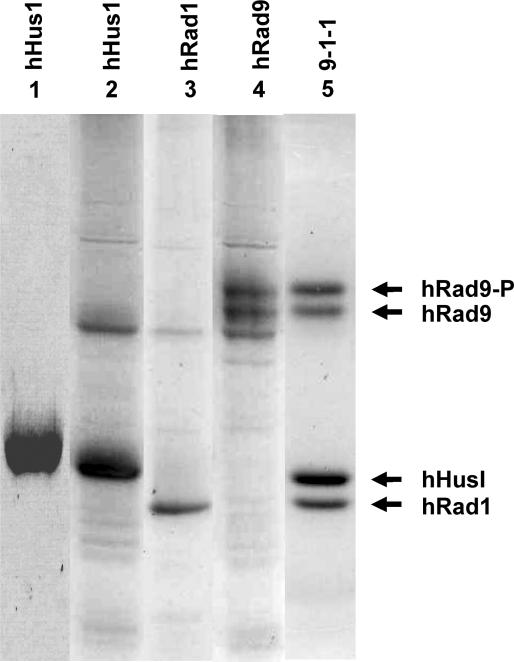
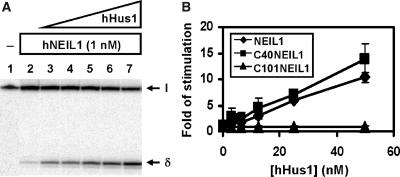
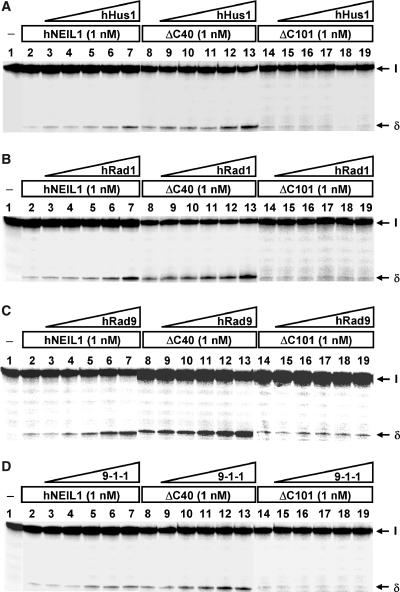
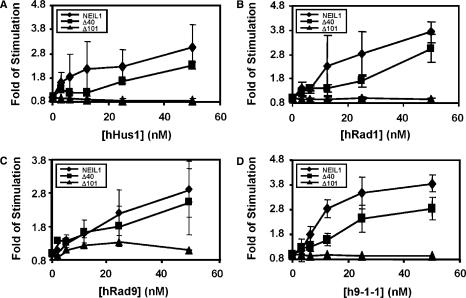
The above results show that hNEIL1 physically interacts with hHus1, hRad1, and hRad9 as individual proteins and as a complex. We then tested whether the glycosylase and β/δ-elimination activities of hNEIL1 can be enhanced by hHus1, hRad1, hRad9, or the 9-1-1 complex. We purified hHus1 expressed in E. coli as well as purified hHus1, hRad1, hRad9 and the 9-1-1 complex expressed in the baculovirus-transfected insect cells (Figure 4). We added increasing amounts of purified hHus1, hRad1, hRad9 and the 9-1-1 complex to the hNEIL1 glycosylase reactions containing the Tg substrate. As shown in Figure 5A (lanes 3–7), the strand cleavage activity of hNEIL1 by its combined glycosylase and β/δ-elimination actions was enhanced significantly by hHus1 protein expressed in bacteria. The difference between hNEIL1 (1 nM) alone and hNEIL1 with 20 nM of hHus1 was ∼5-fold (Figure 5B, diamons). Human Hus1 alone at 50 nM did not have glycosylase activity on the substrate containing Tg (data not shown). A similar stimulation effect on the hNEIL1 strand cleavage activity was observed separately with hHus1, hRad1, hRad9, and the 9-1-1 complex expressed in the baculovirus system (Figure 6A–D, lanes 1–6). Interestingly, hHus1, hRad1, hRad9, separately or in the 9-1-1 complex stimulated hNEIL1 activity to a similar extent (Figure 7A–D, diamonds).
Figure 4.
SDS–PAGE of purified human Hus1, Rad9, Rad1 and the 9-1-1 complex. Lane 1, purified His-tagged hHus1 expressed in E. coli. Lanes 2–4, FLAG-tagged hHus1, hRad1, and hRad9 were individually expressed in insect cells and purified by FLAG-antibody (M2) affinity column. Lane 5, the 9-1-1 complex was expressed in insect cells with three types of baculovirueses; and purified by M2 affinity column and a gel filtration column (Superose 12). The slow-migrating band of Rad9 is phosphorylated (hRad9-P). Proteins were stained by Coomassie Blue.
Figure 5.
hNEIL1 activity was stimulated by hHus1 expressed in bacteria. (A) Human Hus1 enhances the activities of processed full-length hNEIL1 containing residues 2-390. Lane 1, thymine glycol (Tg)/A-containing DNA substrate. Lane 2, 1.8 fmol (90 pM) of DNA substrate was incubated with hNEIL1 (1 nM). Lanes 3–7 are similar to lane 2 but with added 3.125, 6.25, 12.5, 25 and 50 nM hHus1, respectively. The products were separated on a 14% DNA sequencing gel. Arrows mark the intact DNA substrate (I) and the βδ-elimination product (δ). (B) Quantitative analyses of fold stimulation of hHus1 on processed full-length hNEIL1 (diamonds), NEIL1-CΔ40 (squares), and NEIL1-CΔ101 (triangles) activities from three experiments. The error bars reported are the standard deviations of the averages.
Figure 6.
hNEIL1 activity was stimulated by hHus1, hRad1, hRad9 and the 9-1-1 complex expressed in the baculovirus-transfected insect cells. Human Hus1 (A), hRad1 (B), hRad9 (C) and the 9-1-1 complex (D) enhance the activities of processed full-length hNEIL1 and NEIL1-CΔ40, but not NEIL1-CΔ101. (A)–(D), Lane 1, thymine glycol (Tg)/A-containing DNA substrate and lane 2, 1.8 fmol (90 pM) of DNA substrate was incubated with hNEIL1 (1 nM). (A)–(C), Lanes 3–7 are similar to lane 2 but with added 3.125, 6.25, 12.5, 25 and 50 nM hHus1, hRad1, or hRad9, respectively. (D), Lanes 3–7 are similar to lane 2 but with added 4, 8, 16, 32 and 64 nM of the 9-1-1 complex, respectively. Lanes 8–13 are similar to lanes 2–7 except using 1 nM hNEIL1-CΔ40. Lanes 14–19 are similar to lanes 2–7 except using 1 nM hNEIL1-CΔ101. The products were separated on a 14% DNA sequencing gel. Arrows mark the intact DNA substrate (I) and the βδ-elimination product (δ).
Figure 7.
Quantitative analyses of fold stimulation of hHus1, hRad1, hRad9 and the 9-1-1 complex expressed in the baculovirus system on processed full-length hNEIL1 (diamonds), NEIL1-CΔ40 (squares), and NEIL1-CΔ101 (triangles) activities from three experiments. The representative gels are shown in Figure 6. The error bars reported are the standard deviations of the averages.
Both C-terminal truncated NEIL1 proteins, CΔ40 and CΔ101, retained the strand cleavage activity on DNA substrates containing Tg as the intact protein (Figure 6A–D, lanes 2, 8, 14). Quantization of several assays indicated ∼8% of the DNA substrate were cleaved by intact, CΔ40 and CΔ101 hNEIL1 proteins. Thus, the hHus1 interaction region (residues 290–350) of hNEIL1 (Figure 2) is not essential for the hNEIL1 activity. The strand cleavage activity of CΔ40 could be enhanced by hHus1, hRad1, hRad9 and the 9-1-1 complex (Figure 6A–D, lanes 8–13; and Figures 5B, 7A–D, squares). However, hHus1, hRad1, hRad9 and the 9-1-1 complex could not enhance the activity of CΔ101 (Figure 6A–D, lanes 14–19; and Figures 5B, 7A–D, triangles). Thus, the 9-1-1 complex enhances the hNEIL1 activity through direct physical interaction.
DISCUSSION
Oxidative DNA base damage is repaired mainly by the BER pathway. It has been suggested that the BER pathway involves highly coordinated processes governed by protein–protein and protein–DNA interactions (48–51). In this study, we show that all three subunits of the 9-1-1 complex individually interact with hNEIL1. The strand cleavage activity of hNEIL1 is stimulated by hHus1, hRad1, hRad9, separately and the 9-1-1 complex with similar degrees (Figure 7). The 9-1-1 complex is likely not dissociated in the hNEIL1 reaction because it eluted as one single peak from the gel filtration column and it migrated as one single band on the native gel. Thus, the formation of the 9-1-1 complex is not a prerequisite for hNEIL1 stimulation. Because the functional interaction is parallel with the physical interaction between hNEIL1 and the 9-1-1 complex, the 9-1-1 complex stimulates hNEIL1 by direct contact with hNEIL1. Recently, the 9-1-1 complex has been shown to interact with and stimulate the enzymes involved in BER including MYH (35,36), Polβ (37), FEN1 (38,39), and DNA ligase l (40,41). Thus, the 9-1-1 complex is not only a DNA damage sensor (18) but is also involved in the hNEIL1-dependent BER pathway.
In eukaryotes, there are two major subpathways for BER: a single-nucleotide short patch and a 2-10 nucleotide long patch pathway (2,52–54). The short patch BER pathway is usually independent of PCNA and requires five proteins: a glycosylase, APE1, Polβ and DNA ligase III/XRCC1 heterodimer (55). The long patch BER pathway requires a glycosylase, APE1, replication factor C (RFC), FEN1, DNA polymerases δ/ɛ (Polδ/ɛ) (or Polβ), RPA, and DNA ligase I (54,56). Because the 9-1-1 complex can interact with DNA ligase I but not DNA ligase III (40), it has been suggested that it may mainly be involved in the long-patch BER. Earlier studies suggested that the hNEIL1-dependent BER is of the short-patch type (57). The 9-1-1 complex may also be a component of the short-patch BER such as the hNEIL1-dependent pathway because of its interaction with DNA polymerase β and DNA ligase III/XRCC1 heterodimer (57). The hNEIL1-dependent BER pathway is unique for its independence of APE1 and interactions with polynucleotide kinase and DNA ligase III/XRCC1 (57).
We have shown that the 9-1-1 complex physically and functionally interacts with MYH in S. pombe and human cells (35,36). Human NEIL1 is the second glycosylase to be identified to interact with the 9-1-1 complex. Both glycosylases reduce mutagenesis induced by oxidative damage. NEIL1 mainly acts on oxidized pyrimidines while MYH removes adenines or 2-hydroxyadenine misincorporated opposite G or 7,8-dihydro-8-oxo-guanine (8-oxoG) (58–60). The reaction mechanism of NEIL1 is distinct from MYH. MYH is a monofunctional glycosylase, whereas NEIL1 is a bifunctional glycosylase with β/δ-elimination activity (57). Both NEIL1 and MYH activities can be stimulated by Hus1 and the 9-1-1 complex. Unlike MYH, which interacts with the 9-1-1 complex mainly via the Hus1 subunit (35,36), hNEIL1 interacts with hHus1, hRad1, and hRad9 equally. In addition, both hMYH and hNEIL1 co-localize with hRad9 following H2O2 treatment. NEIL1 expression has been shown to be induced by ROS (61), however, MYH expression level remains almost the same after ionizing radiation and hydrogen peroxide treatments (35,36).
Our mapping analyses indicate that the 9-1-1 complex interacting domain is localized to residues 290–350 of hNEIL1. A consensus PCNA binding motif [QXX(L/V)XXF(F/Y)] is found in many proteins involved in DNA replication, DNA repair, DNA methylation, and chromatin assembly (62,63). Recently the list of proteins that interact with 9-1-1 has been expanding. The interacting regions of hMYH and S. pombe MYH have been mapped to specific motifs (36). An alignment of residues 290–350 of hNEIL1 with residues 295–350 of hMYH (Figure 8) reveals some conserved features. Particularly, V311 of hNEIL1 is conserved to V315 of hMYH that has been shown to be important for the interaction between hMYH and hHus1 (36). The crystal structure of hNEIL1 containing residues 2–342 has been determined (64). However, there is no identifiable density beyond residues 290. Thus, the region containing residues 290–350 of hNEIL1 is likely flexible. It is possible that this region becomes structured in the presence of the 9-1-1 complex and then this conformational change promotes the catalytic activity of hNEIL1.
Figure 8.
Alignment of the hHus1 binding motifs of hNEIL1 and hMYH. Sequences are: Homo sapiens NEIL1 (hNEIL1, accession No. AAO74826) and Homo sapiens MYH (hMYH, accession No. U63329). V315 of hMYH has been shown to be important for the interaction between hMYH and hHus1 (36). Identical amino acid residues are shaded in black and conserved residues are boxed in gray. V311 of hNEIL1 and V315 of hMYH are marked by a star.
It is interesting to note that residues 290–350 of hNEIL1 are also important for its interactions with Polβ (57), DNA ligase III (57), XRCC1 (57), and PCNA (Hong Dou, Corey Theriot and Sankar Mitra, unpublished data). How so many partner proteins interact with hNEIL1 within this common 61-residue region is unclear. Our preliminary data suggest that PCNA at equimolar levels slightly reduces the physical interaction between hNEIL1 and hHus1. Because the structure of the 9-1-1 complex resembles that of PCNA sliding clamp (19–21), it has been proposed that the 9-1-1 complex acts as a damage-specific substitute for PCNA (38,39). The 9-1-1 complex may replace PCNA when active PCNA is depleted by p21 during cell cycle arrest in response to DNA damage (65). It will be interesting to find the mechanism by which hNEIL1 switches its partners from PCNA to the 9-1-1 complex.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Drs Evan Y. Lee (University of California, Irvine, CA, USA) and Alan E. Tomkinson (University of Maryland, Baltimore, MD, USA) for kindly providing the plasmids. We thank Drs. Alan E. Tomkinson and Aziz Sancar (University of North Carolina at Chapel Hill, NC, USA) for providing the baculovirus vectors. We appreciate Dr Howard Lieberman (Columbia University, NY, USA) for kindly supplying the 9-1-1 proteins for the initial work and thank Dr Susan Wallace (University of Vermont, VT, USA) for the DNA substrate. This work was supported by National Institute of Health grants CA78391 to AL, CA81063 to SM, and CA102271 and P01ES06676 to TKH.
Funding to pay the Open Access publication charges for this article was provided by CA78391.
Conflict of interest statement. None declared.
REFERENCES
- 1.Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 2.Krokan HE, Nilsen H, Skorpen F, Otterlei M, Slupphaug G. Base excision repair of DNA in mammalian cells. FEBS Lett. 2000;476:73–77. doi: 10.1016/s0014-5793(00)01674-4. [DOI] [PubMed] [Google Scholar]
- 3.Mol CD, Parikh SS, Putnam CD, Lo TP, Tainer JA. DNA repair mechanisms for the recognition and removal of damaged DNA bases. Annu. Rev. Biophys. Biomol. Struct. 1999;28:101–128. doi: 10.1146/annurev.biophys.28.1.101. [DOI] [PubMed] [Google Scholar]
- 4.Zharkov DO, Shoham G, Grollman AP. Structural characterization of the Fpg family of DNA glycosylases. DNA Repair (Amst) 2003;2:839–862. doi: 10.1016/s1568-7864(03)00084-3. [DOI] [PubMed] [Google Scholar]
- 5.Morland I, Rolseth V, Luna L, Rognes T, Bjoras M, Seeberg E. Human DNA glycosylases of the bacterial Fpg/MutM superfamily: an alternative pathway for the repair of 8-oxoguanine and other oxidation products in DNA. Nucleic Acids Res. 2002;30:4926–4936. doi: 10.1093/nar/gkf618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takao M, Kanno S, Kobayashi K, Zhang QM, Yonei S, van der Horst GT, Yasui A. A back-up glycosylase in Nth1 knock-out mice is a functional Nei (endonuclease VIII) homologue. J. Biol. Chem. 2002;277:42205–42213. doi: 10.1074/jbc.M206884200. [DOI] [PubMed] [Google Scholar]
- 7.Hazra TK, Kow YW, Hatahet Z, Imhoff B, Boldogh I, Mokkapati SK, Mitra S, Izumi T. Identification and characterization of a novel human DNA glycosylase for repair of cytosine-derived lesions. J. Biol. Chem. 2002;277:30417–30420. doi: 10.1074/jbc.C200355200. [DOI] [PubMed] [Google Scholar]
- 8.Hazra TK, Izumi T, Boldogh I, Imhoff B, Kow YW, Jaruga P, Dizdaroglu M, Mitra S. Identification and characterization of a human DNA glycosylase for repair of modified bases in oxidatively damaged DNA. Proc. Natl. Acad. Sci. USA. 2002;99:3523–3528. doi: 10.1073/pnas.062053799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bandaru V, Sunkara S, Wallace SS, Bond JP. A novel human DNA glycosylase that removes oxidative DNA damage and is homologous to Escherichia coli endonuclease VIII. DNA Repair (Amst) 2002;1:517–529. doi: 10.1016/s1568-7864(02)00036-8. [DOI] [PubMed] [Google Scholar]
- 10.Dou H, Mitra S, Hazra TK. Repair of oxidized bases in DNA bubble structures by human DNA glycosylases NEIL1 and NEIL2. J. Biol. Chem. 2003;278:49679–49684. doi: 10.1074/jbc.M308658200. [DOI] [PubMed] [Google Scholar]
- 11.Rosenquist TA, Zaika E, Fernandes AS, Zharkov DO, Miller H, Grollman AP. The novel DNA glycosylase, NEIL1, protects mammalian cells from radiation-mediated cell death. DNA Repair (Amst) 2003;2:581–591. doi: 10.1016/s1568-7864(03)00025-9. [DOI] [PubMed] [Google Scholar]
- 12.Klungland A, Rosewell I, Hollenbach S, Larsen E, Daly G, Epe B, Seeberg E, Lindahl T, Barnes DE. Accumulation of premutagenic DNA lesions in mice defective in removal of oxidative base damage. Proc. Natl. Acad. Sci. USA. 1999;96:13300–13305. doi: 10.1073/pnas.96.23.13300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takao M, Kanno S, Shiromoto T, Hasegawa R, Ide H, Ikeda S, Sarker AH, Seki S, Xing JZ, Le XC, et al. Novel nuclear and mitochondrial glycosylases revealed by disruption of the mouse Nth1 gene encoding an endonuclease III homolog for repair of thymine glycols. EMBO J. 2002;21:3486–3493. doi: 10.1093/emboj/cdf350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shinmura K, Tao H, Goto M, Igarashi H, Taniguchi T, Maekawa M, Takezaki T, Sugimura H. Inactivating mutations of the human base excision repair gene NEIL1 in gastric cancer. Carcinogenesis. 2004;25:2311–2317. doi: 10.1093/carcin/bgh267. [DOI] [PubMed] [Google Scholar]
- 15.Vartanian V, Lowell B, Minko IG, Wood TG, Ceci JD, George S, Ballinger SW, Corless CL, McCullough AK, Lloyd RS. The metabolic syndrome resulting from a knockout of the NEIL1 DNA glycosylase. Proc. Natl. Acad. Sci. USA. 2006;103:1864–1869. doi: 10.1073/pnas.0507444103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bartek J, Lukas C, Lukas J. Checking on DNA damage in S phase. Nat. Rev. Mol. Cell Biol. 2004;5:792–804. doi: 10.1038/nrm1493. [DOI] [PubMed] [Google Scholar]
- 17.Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 18.Zhou BB, Elledge SJ. The DNA damage response: putting checkpoints in perspective. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- 19.Burtelow MA, Roos-Mattjus PM, Rauen M, Babendure JR, Karnitz LM. Reconstitution and molecular analysis of the hRad9-hHus1-hRad1 (9-1-1) DNA damage responsive checkpoint complex. J. Biol. Chem. 2001;276:25903–25909. doi: 10.1074/jbc.M102946200. [DOI] [PubMed] [Google Scholar]
- 20.Shiomi Y, Shinozaki A, Nakada D, Sugimoto K, Usukura J, Obuse C, Tsurimoto T. Clamp and clamp loader structures of the human checkpoint protein complexes, Rad9-Rad1-Hus1 and Rad17-RFC. Genes Cells. 2002;7:861–868. doi: 10.1046/j.1365-2443.2002.00566.x. [DOI] [PubMed] [Google Scholar]
- 21.Venclovas C, Thelen MP. Structure-based predictions of Rad1, Rad9, Hus1 and Rad17 participation in sliding clamp and clamp-loading complexes. Nucleic Acids Res. 2000;28:2481–2493. doi: 10.1093/nar/28.13.2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bermudez VP, Lindsey-Boltz LA, Cesare AJ, Maniwa Y, Griffith JD, Hurwitz J, Sancar A. Loading of the human 9-1-1 checkpoint complex onto DNA by the checkpoint clamp loader hRad17-replication factor C complex in vitro. Proc. Natl. Acad. Sci. USA. 2003;100:1633–1638. doi: 10.1073/pnas.0437927100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ellison V, Stillman B. Biochemical characterization of DNA damage checkpoint complexes: clamp loader and clamp complexes with specificity for 5′ recessed DNA. PLoS. Biol. 2003;1:E33. doi: 10.1371/journal.pbio.0000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Majka J, Burgers PM. Yeast Rad17/Mec3/Ddc1: a sliding clamp for the DNA damage checkpoint. Proc. Natl. Acad. Sci. USA. 2003;100:2249–2254. doi: 10.1073/pnas.0437148100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dahm K, Hubscher U. Colocalization of human Rad17 and PCNA in late S phase of the cell cycle upon replication block. Oncogene. 2002;21:7710–7719. doi: 10.1038/sj.onc.1205872. [DOI] [PubMed] [Google Scholar]
- 26.Meister P, Poidevin M, Francesconi S, Tratner I, Zarzov P, Baldacci G. Nuclear factories for signaling and repairing DNA double strand breaks in living fission yeast. Nucleic Acids Res. 2003;31:5064–5073. doi: 10.1093/nar/gkg719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zou L, Liu D, Elledge SJ. Replication protein A-mediated recruitment and activation of Rad17 complexes. Proc. Natl. Acad. Sci. USA. 2003;100:13827–13832. doi: 10.1073/pnas.2336100100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Giannattasio M, Lazzaro F, Longhese MP, Plevani P, Muzi-Falconi M. Physical and functional interactions between nucleotide excision repair and DNA damage checkpoint. EMBO J. 2004;23:429–438. doi: 10.1038/sj.emboj.7600051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kai M, Wang TS. Checkpoint activation regulates mutagenic translesion synthesis. Genes Dev. 2003;17:64–76. doi: 10.1101/gad.1043203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 31.Wu X, Shell SM, Zou Y. Interaction and colocalization of Rad9/Rad1/Hus1 checkpoint complex with replication protein A in human cells. Oncogene. 2005;24:4728–4735. doi: 10.1038/sj.onc.1208674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lavin MF. The Mre11 complex and ATM: a two-way functional interaction in recognizing and signaling DNA double strand breaks. DNA Repair (Amst) 2004;3:1515–1520. doi: 10.1016/j.dnarep.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 33.Wang Y, Qin J. MSH2 and ATR form a signaling module and regulate two branches of the damage response to DNA methylation. Proc. Natl. Acad. Sci. USA. 2003;100:15387–15392. doi: 10.1073/pnas.2536810100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoshioka K, Yoshioka Y, Hsieh P. ATR kinase activation mediated by MutSalpha and MutLalpha in response to cytotoxic O6-methylguanine adducts. Mol. Cell. 2006;22:501–510. doi: 10.1016/j.molcel.2006.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chang DY, Lu AL. Interaction of checkpoint proteins Hus1/Rad1/Rad9 with DNA base excision repair enzyme MutY homolog in fission yeast, Schizosaccharomyces pombe. J. Biol. Chem. 2005;280:408–417. doi: 10.1074/jbc.M406800200. [DOI] [PubMed] [Google Scholar]
- 36.Shi G, Chang D-Y, Cheng CC, Guan X, Venclovas C, Lu A-L. Physical and functional interactions between MutY homolog (MYH) and checkpoint proteins Rad9-Rad1-Hus1. Biochem. J. 2006;400:53–62. doi: 10.1042/BJ20060774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Toueille M, El Andaloussi N, Frouin I, Freire R, Funk D, Shevelev I, Friedrich-Heineken E, Villani G, Hottiger MO, Hubscher U. The human Rad9/Rad1/Hus1 damage sensor clamp interacts with DNA polymerase beta and increases its DNA substrate utilisation efficiency: implications for DNA repair. Nucleic Acids Res. 2004;32:3316–3324. doi: 10.1093/nar/gkh652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Friedrich-Heineken E, Toueille M, Tannler B, Burki C, Ferrari E, Hottiger MO, Hubscher U. The two DNA clamps Rad9/Rad1/Hus1 complex and proliferating cell nuclear antigen differentially regulate flap endonuclease 1 activity. J. Mol. Biol. 2005;353:980–989. doi: 10.1016/j.jmb.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 39.Wang W, Brandt P, Rossi ML, Lindsey-Boltz L, Podust V, Fanning E, Sancar A, Bambara RA. The human Rad9-Rad1-Hus1 checkpoint complex stimulates flap endonuclease 1. Proc. Natl. Acad. Sci. USA. 2004;101:16762–16767. doi: 10.1073/pnas.0407686101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smirnova E, Toueille M, Markkanen E, Hubscher U. The human checkpoint sensor and alternative DNA clamp Rad9-Rad1-Hus1 modulates the activity of DNA ligase I, a component of the long-patch base excision repair machinery. Biochem. J. 2005;389:13–17. doi: 10.1042/BJ20050211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang W, Lindsey-Boltz LA, Sancar A, Bambara RA. Mechanism of stimulation of human DNA ligase I by the Rad9-Rad1-Hus1 checkpoint complex. J. Biol. Chem. 2006;281:20865–20872. doi: 10.1074/jbc.M602289200. [DOI] [PubMed] [Google Scholar]
- 42.Gu Y, Parker A, Wilson TM, Bai H, Chang DY, Lu AL. Human MutY homolog (hMYH), a DNA glycosylase involved in base excision repair, physically and functionally interacts with mismatch repair proteins hMSH2/hMSH6. J. Biol. Chem. 2002;277:11135–11142. doi: 10.1074/jbc.M108618200. [DOI] [PubMed] [Google Scholar]
- 43.Parker A, Gu Y, Mahoney W, Lee S-H, Singh KK, Lu A-L. Human homolog of the MutY protein (hMYH) physically interacts with protein involved in long-patch DNA base excision repair. J. Biol. Chem. 2001;276:5547–5555. doi: 10.1074/jbc.M008463200. [DOI] [PubMed] [Google Scholar]
- 44.Bradford M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 45.Hazra TK, Mitra S. Purification and characterization of NEIL1 and NEIL2, members of a distinct family of mammalian DNA glycosylases for repair of oxidized bases. Methods Enzymol. 2006;408:33–48. doi: 10.1016/S0076-6879(06)08003-7. [DOI] [PubMed] [Google Scholar]
- 46.Towbin HT, Staehlin T, Gordon J. Eletrophoretic transfer of proteins from polyacrylamide gel to nitrocellulose sheets procedure. Proc. Natl. Acad. Sci. USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lu A-L, Tsai-Wu J-J, Cillo J. DNA determinants and substrate specificities of Escherichia coli MutY. J. Biol. Chem. 1995;270:23582–23588. doi: 10.1074/jbc.270.40.23582. [DOI] [PubMed] [Google Scholar]
- 48.Lu A-L, Li X, Gu Y, Wright PM, Chang D-Y. Repair of oxidative DNA damage. Cell Biochem. Biophy. 2001;35:141–170. doi: 10.1385/CBB:35:2:141. [DOI] [PubMed] [Google Scholar]
- 49.Mitra S, Izumi T, Boldogh I, Bhakat KK, Hill JW, Hazra TK. Choreography of oxidative damage repair in mammalian genomes. Free Radic. Biol. Med. 2002;33:15–28. doi: 10.1016/s0891-5849(02)00819-5. [DOI] [PubMed] [Google Scholar]
- 50.Mol CD, Izumi T, Mitra S, Tainer JA. DNA-bound structures and mutants reveal abasic DNA binding by APE1 and DNA repair coordination. Nature. 2000;403:451–456. doi: 10.1038/35000249. [DOI] [PubMed] [Google Scholar]
- 51.Wilson SH, Kunkel TA. Passing the baton in base excision repair. Nat. Struct. Biol. 2000;7:176–178. doi: 10.1038/73260. [DOI] [PubMed] [Google Scholar]
- 52.Frosina G, Cappelli E, Fortini P, Dogliotti E. In vitro base excision repair assay using mammalian cell extracts. Methods Mol. Biol. 1999;113:301–315. doi: 10.1385/1-59259-675-4:301. [DOI] [PubMed] [Google Scholar]
- 53.Krokan HE, Standal R, Slupphaug G. DNA glycosylases in the base excision repair of DNA. Biochem. J. 1997;325:1–16. doi: 10.1042/bj3250001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Matsumoto Y, Kim K, Hurwitz J, Gary R, Levine DS, Tomkinson AE, Park MS. Reconstitution of proliferating cell nuclear antigen-dependent repair of apurinic/apyrimidinic sites with purified human proteins. J. Biol. Chem. 1999;274:33703–33708. doi: 10.1074/jbc.274.47.33703. [DOI] [PubMed] [Google Scholar]
- 55.Srivastava DK, Berg BJ, Prasad R, Molina JT, Beard WA, Tomkinson AE, Wilson SH. Mammalian abasic site base excision repair. Identification of the reaction sequence and rate-determining steps. J. Biol. Chem. 1998;273:21203–21209. doi: 10.1074/jbc.273.33.21203. [DOI] [PubMed] [Google Scholar]
- 56.Pascucci B, Strucki M, Jonsson ZO, Dogliotti E, Hubscher U. Long patch base excision repair with purified human proteins: DNA ligase I as patch size mediator for DNA polymerases d and e. J. Biol. Chem. 1999;274:33696–33702. doi: 10.1074/jbc.274.47.33696. [DOI] [PubMed] [Google Scholar]
- 57.Wiederhold L, Leppard JB, Kedar P, Karimi-Busheri F, Rasouli-Nia A, Weinfeld M, Tomkinson AE, Izumi T, Prasad R, Wilson SH, et al. AP endonuclease-independent DNA base excision repair in human cells. Mol. Cell. 2004;15:209–220. doi: 10.1016/j.molcel.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 58.Gu Y, Lu A-L. Differential DNA recognition and glycosylase activity of the native human MutY homolog (hMYH) and recombinant hMYH expressed in bacteria. Nucleic Acids Res. 2001;29:2666–2674. doi: 10.1093/nar/29.12.2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ohtsubo T, Nishioka K, Imaiso Y, Iwai S, Shimokawa H, Oda H, Fujiwara T, Nakabeppu Y. Identification of human MutY homolog (hMYH) as a repair enzyme for 2- hydroxyadenine in DNA and detection of multiple forms of hMYH located in nuclei and mitochondria. Nucleic Acids Res. 2000;28:1355–1364. doi: 10.1093/nar/28.6.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Slupska MM, Luther WM, Chiang JH, Yang H, Miller JH. Functional expression of hMYH, a human homolog of the Escherichia coli MutY protein. J. Bacteriol. 1999;181:6210–6213. doi: 10.1128/jb.181.19.6210-6213.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Das A, Hazra TK, Boldogh I, Mitra S, Bhakat KK. Induction of the human oxidized base-specific DNA glycosylase NEIL1 by reactive oxygen species. J. Biol. Chem. 2005;280:35272–35280. doi: 10.1074/jbc.M505526200. [DOI] [PubMed] [Google Scholar]
- 62.Warbrick E. PCNA binding through a conserved motif. BioEssays. 1998;20:195–199. doi: 10.1002/(SICI)1521-1878(199803)20:3<195::AID-BIES2>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 63.Zhang P, Mo JY, Perez A, Leon A, Liu L, Mazloum N, Xu H, Lee MY. Direct interaction of proliferating cell nuclear antigen with the p125 catalytic subunit of mammalian DNA polymerase d. J. Biol. Chem. 1999;274:266647–26653. doi: 10.1074/jbc.274.38.26647. [DOI] [PubMed] [Google Scholar]
- 64.Doublie S, Bandaru V, Bond JP, Wallace SS. The crystal structure of human endonuclease VIII-like 1 (NEIL1) reveals a zincless finger motif required for glycosylase activity. Proc. Natl. Acad. Sci. USA. 2004;101:10284–10289. doi: 10.1073/pnas.0402051101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Waga S, Hannon GJ, Beach D, Stillman B. The p21 inhibitor of cyclin-dependent kinases controls DNA replication by interaction with PCNA. Nature. 1994;369:574–578. doi: 10.1038/369574a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.