Abstract
TopoisomeraseII (Topo II) is a major component of chromosomal scaffolds and essential for mitotic chromosome condensation, but the mechanism of this action remains unknown. Here, we used an in vitro chromatin reconstitution system in combination with atomic force and fluorescence microscopic analyses to determine how Topo II affects chromosomal structure. Topo II bound to bare DNA and clamped the two DNA strands together, even in the absence of ATP. In addition, Topo II promoted chromatin compaction in a manner dependent on histone H1 but independent of ATP. Histone H1-induced 30-nm chromatin fibers were converted into a large complex by Topo II. Fluorescence microscopic analysis of the Brownian motion of chromatin stained with 4′,6-diamidino-2-phenylindole showed that the reconstituted chromatin became larger following the addition of Topo II in the presence but not the absence of histone H1. Based on these findings, we propose that chromatin packing is triggered by histone H1-dependent, Topo II-mediated clamping of DNA strands.
INTRODUCTION
High-order packing of genomic DNA into eukaryotic interphase nuclei is necessary for the maintenance of genomic information and the complex regulation of gene activities. Chromosomal packing is also critical during cell division because an aberration in the packing of the genome results in defects in the formation and correct separation of sister chromatids.
Many structural studies have been carried out to investigate the molecular mechanism of the higher order genome packing. Nanoscale observations using electron microscopy and atomic force microscopy (AFM) have revealed that the nucleosome, which forms a ‘beads-on-a-string’ structure with linker DNA, is the fundamental structural unit of the chromosome (1–3). These nucleosomal arrays are packed into 30-nm fibers with the help of linker-histone H1 (4–7). Although the mechanisms of how structures larger than 30-nm fibers are generated remain controversial, it has been proposed that chromatin fibers form loop structures that are attached to an axis called a scaffold or matrix. Indeed, DNA loops extending from an axis have been observed in histone-depleted metaphase chromosomes (8).
Biochemical studies on interphase nuclei have suggested that genomic DNA contains regions associated with the scaffold/matrix approximately every 100 kb, a region called the scaffold/matrix attachment region (SAR/MAR) (9). Some of the components associated with this scaffold/matrix region have been identified (10). For example, the metaphase scaffold/matrix contains the condensin complex and topoisomerase II (Topo II) (11–14). Both of these components are essential for mitotic chromosome condensation (15–17) and are located on the axis of condensed chromosomes (18). The interphase scaffold/matrix, on the other hand, contains RNA and ribonucleoprotein as major components (19,20). Also, NuMA, actin, DNA and RNA polymerases, histone acetyl transferase and Topo II are now considered to be components of the interphase scaffold/matrix.
Interestingly, Topo II is a component of the scaffold at both mitosis and interphase. Topo II catalyzes the decatenation of two DNA strands using energy from the hydrolysis of ATP (21). Topo II is a homodimer with a ring-like structure (22,23). In a mutant strain of fission yeast lacking functional Topo II, chromosomes cannot condense (15). In addition, Topo II-immunodepleted mitotic frog extracts cannot induce chromatin condensation in nuclei from HeLa cells or chicken erythrocytes (24).
In previous studies, we established an in vitro chromatin reconstitution system, wherein beads-on-a-string nucleosome arrays are produced upon removal of salt by dialysis (3) and 30-nm chromatin fibers can be generated by the addition of histone H1 (7). Here, we conducted molecular imaging analyses on the interaction between Topo II and in vitro-reconstituted chromatin fibers. We found that, in the absence of ATP, Topo II promotes further folding of 30-nm fibers but not of beads-on-a-string arrays. This means that linker-histone H1 is crucial for the formation of 30-nm chromatin fibers and that the subsequent function of Topo II is essential for achieving higher order structures.
MATERIALS AND METHODS
Materials
The plasmids used for chromatin reconstitution were kind gifts from Dr W. De Laat (Erasmus University Medical Center, Department of Cell Biology, Netherlands). The entire β-globin gene (∼170 kb) was cloned into the pCYPAC2 vector, producing a 185-kb plasmid (25). The locus control region (LCR) of the human β-globin gene (21.5 kb) was isolated by SalI/ClaI digestion and subcloned into pBR322, yielding a 26-kb plasmid.
The plasmids to be tested for the Topo II binding were constructed as follows: The 6-kb LCR region of the human β-globin gene including nuclease hypersensitive sites 4 and 5 was amplified by PCR using human genomic DNA as a template and then cloned into pT7Blue-2 (Merck, Germany), yielding pBGa. The region between nuclease hypersensitive sites 4 and 5 contains the SAR/MAR sequence (26).
Core histones were purified from HeLa cells according to the method developed by O’Neill et al. (27), with slight modifications. The cells were harvested, washed with PBS and lysed with L-buffer (140 mM NaCl, 10 mM Tris-HCl, pH 7.5 and 0.5% Triton X-100). Nuclei were isolated by low-speed centrifugation and washed three times with W-buffer (350 mM NaCl and 10 mM Tris-HCl, pH 7.5). The nuclei were then treated with micrococcal nuclease (40 units/mg of DNA) at 37°C for 15 min in D-buffer (10 mM Tris-HCl pH 7.5, 1.5 mM MgCl2, 1 mM CaCl2, 0.25 M sucrose and 0.1 mM phenylmethylsulfonyl fluoride. The reaction was stopped by the addition of EGTA to a final concentration of 2 mM, and the nuclei were sedimented by centrifugation at 10 000 × g for 5 min. The pellet was resuspended in N-buffer (10 mM Tris-HCl, pH 6.8, 5 mM EDTA, and 0.1 mM phenylmethylsulfonyl fluoride), and dialyzed against N-buffer overnight at 4°C. The sample was centrifuged at 10 000 × g for 10 min, and the soluble chromatin supernatant was redialyzed against HA-buffer (0.1 M NaPO4, pH 6.7 and 0.63 M NaCl) and mixed with hydroxyapatite resin (Bio-Rad). After batch binding at 4°C for 1 h, the resin was packed into a column and washed with five volumes of HA-buffer. The core histones were eluted with E-buffer (0.1 M NaPO4, pH 6.7 and 2 M NaCl). The eluate was applied to a gel filtration column (HiPrep 16/60 S-200; Amersham Biosciences) to separate the octamer from the H3-H4 tetramer, H2A-H2B dimer and other contaminants.
Histone H1 was purified from HeLa cells according to the method developed by Mirzabekov et al. (28) with slight modifications. The cells were harvested, washed with PBS, lysed in 140 mM NaCl, 10 mM Tris-HCl (pH 7.5) and 0.5% Triton X-100, and washed three times with the same buffer without the detergent. The nuclei were collected, washed with a buffer of 0.35 M NaCl and 10 mM Tris-HCl (pH 7.5), resuspended in 5% CCl3COOH and rotated at 4°C for 80 min. After centrifugation at 4000 × g for 15 min, the soluble histone H1-containing supernatant was dialyzed against 10 mM HCl containing 2 mM β-mercaptoethanol. The dialyzed sample was lyophilized and stored at −80°C. For chromatin reconstitution, the lyophilized protein was resuspended in 10 mM Tris-HCl (pH 7.5), 1 mM EDTA, 500 mM NaCl, 0.05% NP-40 and 20% glycerol.
Chromatin reconstitution
Equal amounts (0.5 μg) of the purified DNA and the histone octamer were mixed in Hi-buffer (10 mM Tris-HCl, pH 7.5, 2 M NaCl, 1 mM EDTA, 0.05% NP-40 and 5 mM β-mercaptoethanol), and placed in a dialysis tube (total volume, 50 μl). The dialysis was started with 150 ml of Hi-buffer by stirring at 4°C. Lo-buffer (10 mM Tris-HCl, pH 7.5, 1 mM EDTA, 0.05% NP-40 and 5 mM 2-mercaptoethanol) was added to the dialysis buffer at a rate of 0.46 ml/min, and simultaneously, the dialysis buffer was pumped out at the same speed with a peristaltic pump so that the final dialysis buffer contained 50 mM NaCl after 20 h. The sample was collected from the dialysis tube and stored at 4°C until use.
For experiments in which reconstitution with histone H1 was performed, histone H1 was added after the salt dialysis was completed (at a NaCl concentration of 50 mM). The molar ratio of histone H1 to histone octamer was 1:1. After standing on ice for 30 min, the sample was used for the Topo II treatment.
Topo II binding
Topo II used in this study was topoisomerase IIα (TopoGEN, Port Orange, FL). Various amounts of Topo II were mixed with 80 ng of supercoiled pBGa in 50 mM NaCl buffer (10 mM Tris-HCl, pH 7.5, 1 mM EDTA, 50 mM NaCl, 0.05% NP-40 and 5 mM β-mercaptoethanol) at 37°C for 30 min. In some experiments, a 1/10 volume of ATP-Mg buffer (0.3 mM ATP and 12.5 mM MgCl2) was added to the reaction mixture. The reaction samples were diluted 10-fold by adding HMgG buffer (10 mM HEPES, pH 7.5, 2 mM MgCl2 and 0.3% glutaraldehyde) and kept for 30 min at room temperature.
The reconstituted chromatin sample with or without H1 was treated with Topo II at 37°C for 30 min. After incubation, the sample was analyzed by AFM.
AFM observation
The reconstituted chromatin solution was fixed with 0.3% glutaraldehyde for 30 min at room temperature and then dropped onto a freshly cleaved mica surface that had been pretreated with 10 mM spermidine. After 10 min at room temperature, the mica was washed with water and dried under nitrogen gas. AFM observation was performed with a Nanoscope IIIa or IV (Digital Instruments) in air in the tapping mode. The cantilever (OMCL-AC160TS-W2; Olympus) was 129 μm in length with a spring constant of 33–62 N/m. The scanning frequency was 2–3 Hz, and images were captured with the height mode in a 512 × 512-pixel format. The obtained images were plane-fitted and flattened using the software supplied in the imaging module. Except where noted, AFM images in this study are presented as surface plots, which are efficient for displaying the entire structure. In the Supplementary Data, all AFM images are shown using the top view with a height scale.
Pull-down assay
The cDNA encoding histone H1 was a kind gift from Michael J. Hendzel (University of Alberta, Canada). The plasmid for overexpressing of histone H1 with an N-terminal GST tag was constructed as follows: the ORF of the histone H1 gene was isolated by EcoRI digestion and inserted into pGEX5X-2, yielding pGSTH1.
Plasmids pGSTH1 and pGEX5X-2 were introduced into Escherichia coli and grown in 30 ml of LB. During the log phase of growth, the medium was adjusted by the addition of isopropyl-β-d-thiogalactopyranoside (IPTG) into the culture medium (30 ml of LB) to the final concentration of 1 mM. After 4 h, the cells were harvested and resuspended in 1.5 ml of 50 mM NaCl buffer (as described in the ‘Topo II binding’ section). The suspended cells were treated with 0.2 mg/ml lysozyme for 30 min at 4°C. The cells were then sonicated, and the cell lysate was centrifuged at 11 000 × g for 30 min. The supernatant was collected and used for purification of the fusion proteins.
The supernatant (300 μl) was mixed with 30 μl of glutathione-Sepharose 4B beads (GE Healthcare Bioscience) equilibrated with 50 mM NaCl buffer at 4°C for 1.5 h. The beads were collected by gentle centrifugation and washed five times with 300 μl of 50 mM NaCl buffer.
After incubating the GST- or GST-H1-bound beads at 4°C for 1.5 h with 200 ng of Topo II in 80 μl 50 mM NaCl buffer, the supernatant was collected as the unbound fraction. The beads were then washed five times with 100 μl of 50 mM NaCl buffer, and proteins remaining bound were used for Western blot analyses.
Fluorescence observation in solution and statistical analysis
The Brownian motion of chromatin was examined by fluorescence microscopy as described previously (29). The reconstituted chromatin samples were treated with HMgG buffer (see the ‘Topo II binding’ section) and stained with 0.1 μM 4′,6-diamidino-2-phenylindole (DAPI). DNA staining was observed under UV irradiation at 365 nm using a Carl Zeiss Axiovert 135 TV microscope equipped with a ×100 oil immersion objective lens. Images were recorded on video tape at 30 frames per second through a high-sensitivity EB-CCD camera with an image-processing system (Hamamatsu Photonics). When we observed the chromatin complex existing in the bulk buffer solution (not attached to a coverglass), the complex moved by Brownian motion. To evaluate the hydrodynamic radius of the chromatin in bulk solution, we measured the Brownian motion of individual chromatin complexes as described previously (30). Due to a blurring of the fluorescent images of individual chromatin complexes, the observed DAPI signals were slightly (∼0.3 μm) larger than the actual sizes (31). We plotted the location of the center of mass of the DAPI signal every 0.03 s and determined the time dependence of the mean-square displacement for each complex. From these values, we evaluated the diffusion constant, D, as the projection onto the 2D plane:
where r is the 2D spatial position of the center of the mass of a chromatin complex, and the symbol 〈 〉 denotes a time average of squared displacements. We also calculated the hydrodynamic radius, RH, which is a measure of the chromatin conformation, according to the following equation:
where kB is the Boltzmann constant and η is the viscosity of the solvent.
RESULTS
Sequence-independent action of Topo II on bare DNA
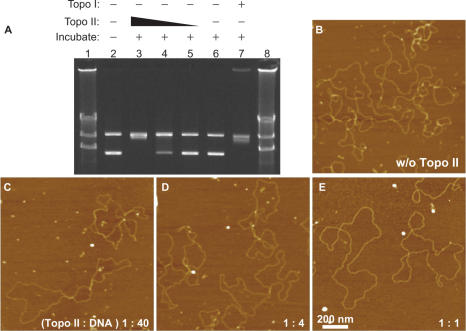
To understand how Topo II alters the molecular architecture of chromosomes, we first used agarose gel electrophoresis to examine the structure of bare DNA in the presence of Topo II. Mixing Topo II with a 9-kb supercoiled plasmid harboring the SAR/MAR region of the β-globin gene in the presence of ATP and MgCl2 altered the topology of the supercoiled plasmid (lanes 3–5 in Figure 1A). Similar results were observed when topoisomerase I was added instead of Topo II in the absence of ATP (lane 7). For 80 ng of plasmid DNA, 80 ng (lane 3), but not 2 (lane 5) or 20 ng (lane 4), of Topo II was sufficient to completely remove the supercoiling.
Figure 1.
Topo II decatenates DNA in the presence of ATP. (A) DNA samples (80 ng) were treated with or without topoisomerases, separated by agarose gel electrophoresis and stained with ethidium bromide. Lanes 1 and 8, λDNA digested by HindIII as a size marker. Lane 2, untreated 9-kb plasmid. The lower band indicates the supercoiled form, and the upper band indicates the nicked relaxed form. Lanes 3–5, plasmids treated with Topo II in the presence of ATP and 80 ng (lane 3), 20 ng (lane 4) or 2 ng (lane 5) of Topo II at 37°C for 30 min. Lane 6, plasmid incubated with Topo II (80 ng) at 37°C for 30 min in the absence of ATP. Lane 7, plasmid treated with topoisomerase I. (B–E) AFM analysis of plasmid (80 ng) treated in the presence of ATP and MgCl2 and without (B) or with 80 ng (E), 20 ng (D), or 2 ng (C) of Topo II. The mixed weight ratio of Topo II versus DNA is indicated in the lower right corner of the images. In each condition, 18 individual 2 × 2-μm areas were scanned, and at least 59 molecules of plasmid were analyzed.
Next, we examined samples of the reactions containing different amounts of Topo II by AFM (Figure 1B–E). As seen by agarose gel electrophoresis (Figure 1A), supercoiling of the plasmids was observed in the absence of Topo II (Figure 1B). Supercoiling was progressively lost as the amount of Topo II was increased, and the plasmids became completely relaxed in the presence 80 ng of Topo II (Figure 1C–E).
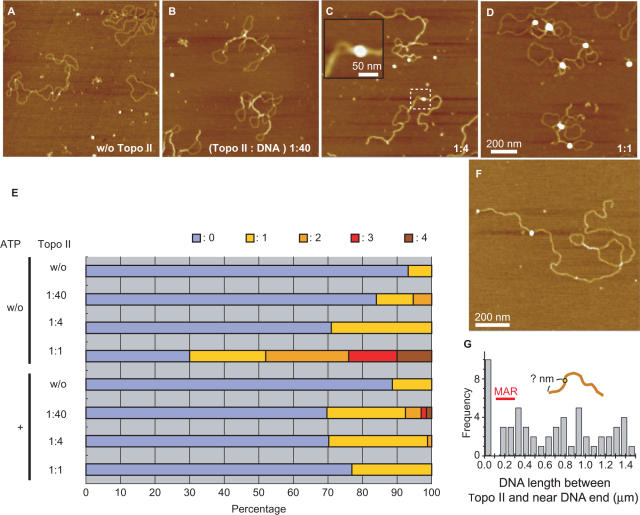
Topo II, however, did not mediate relaxation of the supercoiled plasmids in the absence of ATP (Figure 2A–D), although formed regions where two DNA strands ran in parallel or crossed over. As seen in the enlarged image (Figure 2C), two DNA strands were attached to each other and clamped by a particle with a size (36.6 ± 5.1 nm) similar to a Topo II dimer (23). These results demonstrated that Topo II can clamp two DNA strands in the absence of ATP. The number of Topo II particles (Topo II dimers) on the DNA molecules is shown in Figure 1E. As the amount of Topo II was increased, the number of Topo II–DNA complexes increased to ∼70%, with ∼30% of the DNA molecules remaining free of Topo II. Although we mixed 18 Topo II dimers per plasmid molecule in the experiments shown in Figures 1E and 2D, less than four Topo II dimers were associated with each plasmid (Figure 2E). These results suggest that Topo II binds weakly to bare DNA.
Figure 2.
Topo II clamps two strands together in the absence of ATP. (A–D) AFM analysis of plasmid (80 ng) treated in the absence of ATP and MgCl2 and without (A) or with 80 ng (D), 20 ng (C) or 2 ng (B) of Topo II. The mixed weight ratio of Topo II versus DNA is indicated in the lower right corner of the images. In each condition, 18 individual 2 × 2-μm areas were scanned, and at least 65 molecules of plasmid were analyzed. The inset in (C) shows an enlarged image in which Topo II binding to the DNA can be observed. The apparent diameter of Topo II was 36.6 ± 5.1 nm (mean ± SD; n = 63), which is ∼5 nm larger than the apparent size of the nucleosome. This apparent enlargement is due to a tip effect of the AFM imaging. According to electron microscopic observation (46), the diameter of Topo II dimer is ∼15 nm (9-nm large and 6-nm small domains), which is 4 nm larger than the real diameter (11 nm) of the nucleosome (47). (E) Concentration dependence of Topo II binding. In the AFM images, the number of bound Topo II molecules was counted per plasmid and plotted on a histogram. The plasmid used in this experiment was purified using a QIAGEN-tip 100 column, but it was impossible to remove all the contamination. The results show that a few plasmids possessed small particles even in the absence of Topo II. (F) AFM image of Topo II binding to linearized DNA containing the MAR sequence. (G) Frequency of Topo II binding to DNA containing a MAR sequence. The MAR sequence is 0.1–0.3 μm from the end of the DNA as shown in the illustration.
We next examined the sequence dependency of Topo II–DNA binding. We mixed Topo II with the 9-kb linear plasmid DNA containing the SAR/MAR sequence and analyzed the interaction between Topo II and DNA by AFM (Figure 2F). A histogram of the frequency of Topo II binding to the DNA (Figure 2G) suggests that it is independent of the SAR/MAR sequence. We previously reported that AFM can detect the sequence-specific binding of transcription factors, including AP2 (32) and Bach1 (33). Therefore, the present results suggest that Topo II has little if any sequence specificity.
Topo II promotes chromatin compaction in a histone H1-dependent manner
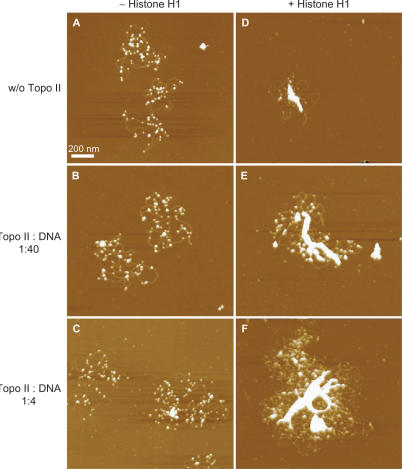
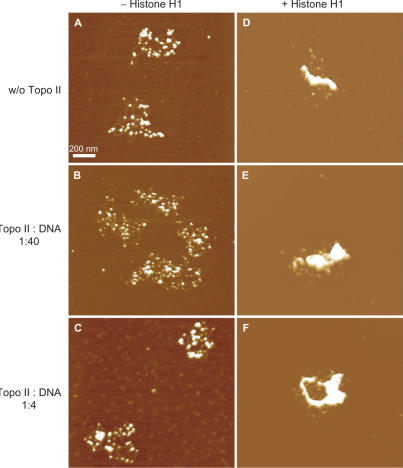
When we used a 26-kb supercoiled plasmid (21.5-kb LCR fragment of the β-globin locus in pBR322) for reconstitution of nucleosomal fibers, ∼50 nucleosomes were formed on each plasmid DNA (Figure 3A). The number of nucleosomes is below saturation because, in vivo, a 26-kb DNA should contain 104–130 nucleosomes. As previously described (3), however, this efficiency (50 nucleosome per 26-kb DNA) is the upper limit for reconstitution by salt dialysis, which does not require other proteins such as histone chaperons. We added various amounts of Topo II to these reconstituted ‘beads-on-a-string’ nucleosomes. We did not detect any structural changes (Figure 3B and C), although Topo II could bind weakly to the DNA (Figures 1 and 2). In contrast, a large complex was formed in the absence of ATP when Topo II was added to 30-nm chromatin fibers (7) that had been reconstituted from the 26-kb nucleosomes by the addition of histone H1 (Figures 2D–F).
Figure 3.
In the absence of ATP, Topo II induces small aggregates, but large aggregates are induced in the presence of histone H1. (A, D) Nucleosomal fibers were reconstituted from the 26-kb plasmid and core histones by the salt dialysis method and then incubated without Topo II and in the absence (A) or presence (D) of histone H1. (B, C, E and F) The reconstituted nucleosomal fibers containing (E and F) or lacking (B and C) histone H1 were treated with Topo II in the absence of ATP and MgCl2. Nucleosomal fiber containing 80 ng DNA was mixed with 2 ng (B and E) or 20 ng (C and F) of Topo II. The mixed weight ratio of Topo II versus DNA is indicated on the left side of the images. In each condition, at least three 12.5 × 12.5-μm areas were scanned, and 10–30 of the reconstituted chromatin complexes were scanned a second time at a higher magnification.
Nucleosome–nucleosome interactions are strongly affected by the salt environment. This accounts for the fact that the addition of ATP and MgCl2 to the reconstituted system caused the formation of small aggregates of nucleosomes (Figure 4A) that were not significantly affected by the addition of Topo II (Figure 4B and C). Again, in this system, large complexes were formed only from 30-nm chromatin fibers (Figure 4D–F).
Figure 4.
Topo II induces some aggregation in the presence of ATP, but greater compaction occurs only in the presence of histone H1. (A and D) Nucleosomal fibers were reconstituted from 26-kb plasmid and core histones by the salt dialysis method and incubated without Topo II and in the absence (A) or presence (D) of histone H1. (B, C, E and F) The reconstituted nucleosomal fibers containing (E and F) or lacking (B and C) histone H1 were treated with Topo II in the presence of ATP and MgCl2. Nucleosomal fiber containing 80 ng DNA was mixed with 2 ng (B and E) or 20 ng (C and F) of Topo II. The mixed weight ratio of Topo II versus DNA is indicated on the left of the images. In each condition, three 12.5 × 12.5-μm areas were scanned, and 10–30 of the reconstituted chromatin complexes were scanned a second time at higher magnification.
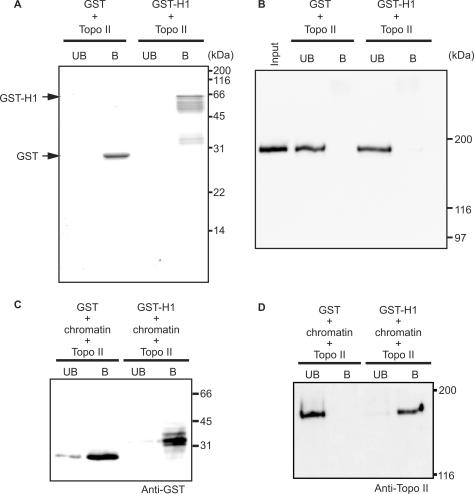
The finding that histone H1 is essential for Topo II-induced chromatin compaction (Figures 3 and 4) raised the possibility that the compaction is caused by a direct interaction between Topo II and histone H1. To test this possibility, we performed a pull-down assay using GST-H1. GST-H1 was expressed in E. coli and trapped on glutathione-Sepharose beads. After the beads were mixed with Topo II, the unbound and the bound fractions were separated by centrifugation. As shown in Figure 5A and B, Topo II was present in the unbound fraction; however, when Topo II was mixed with GST-H1 and reconstituted chromatin, Topo II was present in the bound fraction (Figure 5C and D). These results suggest that the chromatin compaction by Topo II is not due to a direct interaction between Topo II and histone H1 but rather an interaction between Topo II and H1-containing chromatin.
Figure 5.
Topo II does not bind directly to histone H1. (A and B) GST- or GST-H1-bound glutathione-Sepharose beads were mixed with Topo II and separated into unbound (lane UB) and bound fractions (lane B). (A) Polyacrylamide gel stained with Coomassie brilliant blue. The 26- and 59-kDa bands correspond to GST and GST-H1, respectively. (B) Immunoblotting using an anti-Topo II antibody. The input lane contained only Topo II protein at the same concentration as in the pull-down experiment. The 170-kDa bands contain Topo II. (C and D) GST and GST-histone H1 were purified and then mixed with reconstituted chromatin. Next, Topo II was added, and the samples were treated with glutathione-Sepharose beads and fractionated into unbound (lane UB) and bound (lane B) fractions. (C) Immunoblotting using anti-GST antibody. Because the experiment shown here needed more steps than those shown in panels (A and B), GST-H1 was degraded and appeared as a ∼33 kDa band. (D) Immunoblotting using an anti-Topo II antibody.
In the nucleus, chromatin fibers are held by scaffold/matrix structures that occur every several tens or hundreds of kilobases (9). We therefore monitored the behavior of chromatin fibers longer than 100 kb. When we mixed Topo II with 30-nm fibers reconstituted on a 186-kb plasmid DNA containing the entire β-globin gene, the fibers became tangled and formed a partial aggregate (Figure 6B). Addition of excess Topo II caused the formation of large aggregates (Figure 6C). These aggregates consisted of a bead unit with a diameter of 30.5 ± 7.3 nm (mean ± SD; n = 50; Figure 6C inset and D).
Figure 6.
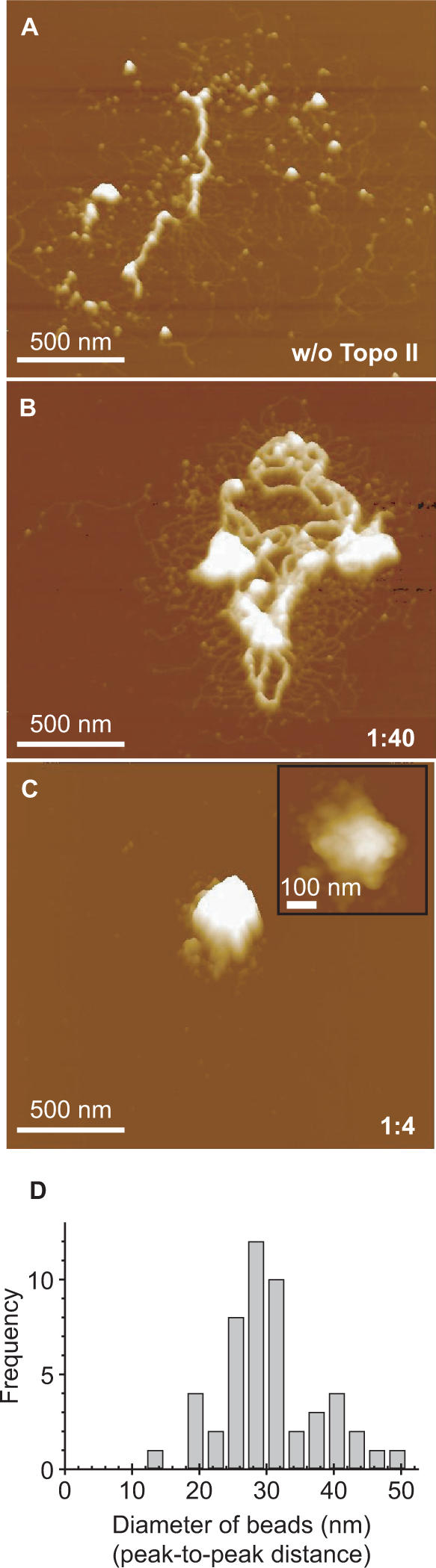
Compaction of 186-kb chromatin by Topo II. (A) AFM of nucleosomal fibers (beads-on-a-string) reconstituted from the 186-kb plasmid and treated with histone H1 to induce the assembly of 30-nm fibers. (B and C) AFM of chromatin fibers containing 80 ng DNA treated with 2 ng (B) or 20 ng (C) of Topo II. The mixed weight ratio of Topo II versus DNA is indicated in the right lower corner of the images. The inset shows an enlarged view in which the globular surface of the complex can be observed. Except for the inset of panel (C), which is a top view, all other images are surface plots. (D) The diameters of the globules on the surface of the complex were measured and are plotted as a histogram.
Fluorescence microscopy reveals Topo II-dependent compaction of 30-nm chromatin fibers
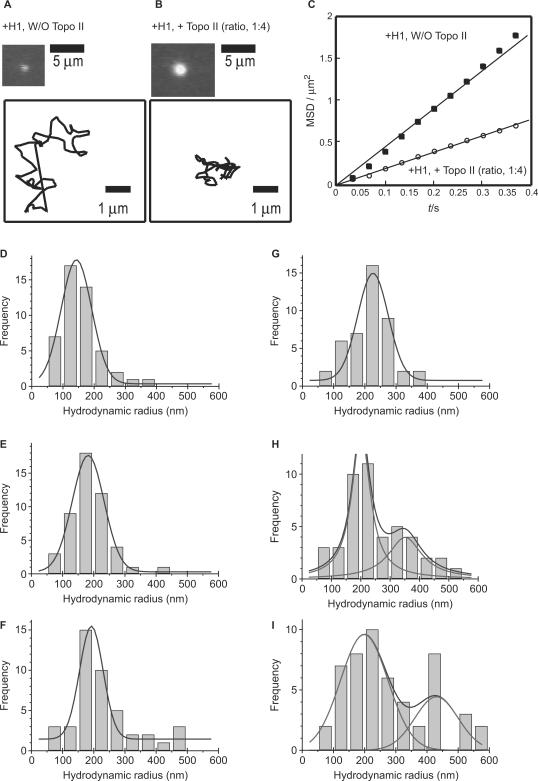
To determine the level of compaction, we stained the chromatin complexes with DAPI and determined their hydrodynamic radii by fluorescence microscopy (Figure 7). The individual 26-kb chromatin was observed as a small dot moving in the buffer (Figure 7A). We calculated the hydrodynamic radius, which is a measure of the size of the complex, from measurements of the Brownian motion. The hydrodynamic radius of the reconstituted nucleosomal array in the absence of both histone H1 and Topo II was 143.2 ± 48.8 nm (Figure 7D). When histone H1 was added into the nucleosomal array, the hydrodynamic radius significantly increased to 226.1 ± 97.9 nm (Figure 7G). This is due to the formation of the 30-nm chromatin fibers (Figure 3D). In the presence of 2 ng Topo II, the radius was 182.0 ± 51.1 nm (Figure 7E), and with 20 ng Topo II, it was 192.2 ± 37.8 nm (Figure 7F), indicating that addition of Topo II alone did not have much of an effect on the hydrodynamic radius of the nucleosomal array. Addition of Topo II to the H1-promoted chromatin fiber (containing 30-nm fibers) resulted in much greater increase in the hydrodynamic radius to 300–400 nm (Figure 7H and I). These findings indicate that Topo II mediates assembly of H1-containing 30-nm fibers, which increases the size of the cluster. This agrees well with the AFM images, wherein the size of the cluster increased several fold upon the addition of Topo II (Figure 3D–F).
Figure 7.
Analysis of the Brownian motion of DAPI-stained reconstituted chromatin in solution. (A and B) Differences between H1-containing reconstituted chromatin in the presence (B) and absence of Topo II (A). Top, fluorescent microscope images; bottom, trails of Brownian motion during 3 s. (C) Mean square displacement (MSD) of the H1-containing chromatin without and with Topo II. (D–I) Histograms of hydrodynamic radii of the chromatin complexes without (D–F) and with H1 (G–I). Reconstituted chromatin containing 80 ng of DNA was treated with 2 ng (E and H) or 20 ng of Topo II (F and I).
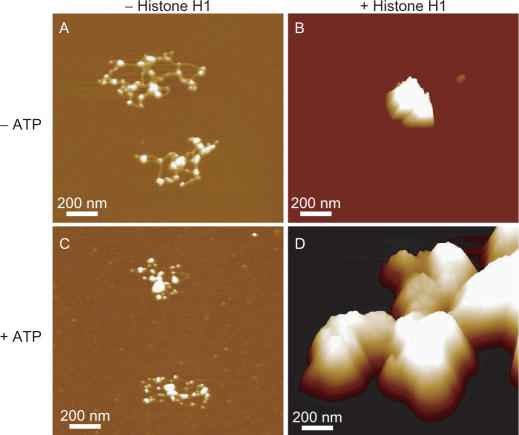
Even when we used one Topo II molecule per 250-bp DNA, which is more than the physiological ratio, beads-on-a-string nucleosomal fibers lacking H1 were not compacted in the absence or presence of ATP (Figure 8A and C). In contrast, 30-nm chromatin fibers containing H1 were highly condensed so that individual nucleosomes and 30-nm fibers could not be identified (Figure 8B and D). The volume calculated from the AFM image in Figure 8B was 8.66 × 106 nm3, which was 60 times larger than that of the chromatin complex in Figure 3D and corresponds to an assembly of ∼60 26-kb chromatin complexes. The volume calculated from the AFM image of Figure 8D was 2.93 × 108 nm3, which was 1000 times larger than that of the chromatin complex in panel Figure 4E and corresponds to an assembly of ∼1000 26-kb chromatin complexes. This suggests that Topo II clamps two independent DNA molecules and forms a large complex. Thus, both fluorescence microscopy and AFM demonstrated that Topo II induces chromatin compaction only in the presence of histone H1-promoted 30-nm fibers.
Figure 8.
Larger amounts of Topo II induce higher order assembly of H1-induced 30-nm fibers independent of ATP. AFM of nucleosomal fibers reconstituted by the salt dialysis method from 26-kb plasmid and core histones incubated without (A and C) or with (B and D) histone H1 and Topo II. The reconstituted nucleosomal fibers were treated with Topo II in the absence (A and B) or presence (C and D) of ATP and MgCl2. The nucleosomal fibers containing 80 ng DNA were mixed with 80 ng of Topo II.
DISCUSSION
In the current study, we investigated the positive role of histone H1 in the compaction of chromatin complexes by Topo II. Chromatin complexes containing H1 are assembled by Topo II into higher order structures, whereas chromatin complexes lacking H1 are not compacted by Topo II. H1 can act in this regard because it is highly positively charged, which can allow the compaction via Topo II. The present AFM and fluorescence studies on the interaction between Topo II and DNA/nucleosomes revealed the following: (i) the DNA–Topo II interaction is not dependent on the SAR/MAR sequence; (ii) Topo II removes supercoiling of plasmid DNA in the presence of ATP as expected but holds the two DNA strands in place in the absence of ATP; and (iii) Topo II induces chromatin compaction only in the presence of histone H1, regardless of the presence of ATP.
Cooperativity of Topo II and H1 in chromatin compaction
Vologodskii and colleagues (34) have reported the images of DNA–Topo II complexes in the presence of ATP. Their findings were similar to our results in Figure 2C, which shows Topo II holding two DNA strands together. This complex may be an intermediate in the DNA decatenation reaction. According to our current findings, even in the absence of ATP, Topo II can mediate assembly of this complex. Topo II has been shown to induce chromatin compaction even in the absence of ATP in experiments using crude extract (35). These results suggest that the ability of Topo II to clamp two DNA strands together is important for its ability to induce chromatin compaction.
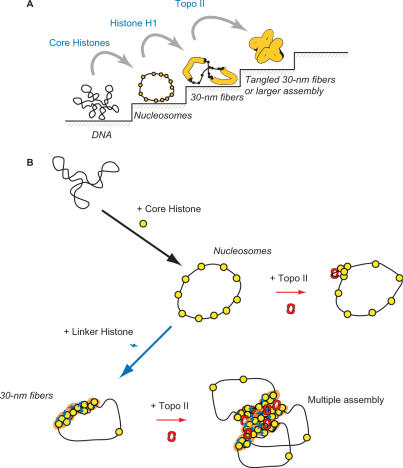
Our most interesting finding was that Topo II-induced compaction is dependent on histone H1 because it suggests that packing is stepwise or hierarchical, proceeding sequentially from DNA to nucleosomes, 30 nm fibers and loops attached to a scaffold (Figure 9A). Previous in vitro studies on Topo II-induced chromatin compaction have used intact chromatin, for example from HeLa nuclei (24) or microsurgically isolated mammalian chromatin (35). Our in vitro system using reconstituted nucleosome fibers and H1-containing chromatin fibers allowed us to demonstrate that Topo II-induced compaction requires H1-containing chromatin (Figures 3 and 4).
Figure 9.
Schematic illustrations of the molecular mechanisms of chromatin compaction by histone H1 and Topo II. (A) Stepwise packing of reconstituted chromatin. First, DNA forms a nucleosomal fiber with core histones, and histone H1 induces the formation of 30-nm fibers. Second, Topo II induces compaction when the 30-nm fiber is formed in the presence of histone H1. (B) Molecular mechanism of Topo II-induced chromatin compaction. Topo II clamping of two DNA strands induces a small amount of aggregation of beads-on-a-string nucleosomal fibers but promotes large-scale aggregation of 30-nm fibers.
We have reported that the 3D assembly of 30-nm fibers can be detected by the excess amount of H1 (7) and PC4 (positive coactivator 4) (36). A 1:1 molar ratio of H1 to core histone octamer promotes the formation of 30-nm fibers, and a 4:1 molar ratio induces the formation of large aggregates consisting of ∼90-nm bead units (7). PC4 induces irregularly sized (50–200 nm in diameter) nucleosome aggregates independent of the presence of H1 (36). The formation of these aggregates is clearly different from Topo II-induced compaction (Figure 9B).
Positive role of H1 on chromatin compaction by Topo II
In the presence of Topo II, the H1-containing 30-nm fibers are converted to larger 3D complexes, but the beads-on-a-string fibers fail to compact in the absence of H1 (Figure 9). H1 has two possible roles in compaction: (i) it can increase the contact area, strengthening the attraction between fibers, and (ii) it reduces the negative charge of the nucleosome complex, decreasing repulsion between fibers. At pH 7.0, DNA has negative charge of −2 e per base pair, and histone core and H1 proteins have positive charges of 147 and 54 e per respective molecule, respectively. An H1-containing nucleosome therefore has a smaller negative charge (−100 e) than an H1-free nucleosome (−150 e), suggesting that the strength of Coulombic repulsion is smaller when H1 is present. Chromatin formation and the interaction between nucleosomes have been discussed in terms of the charges [(37) and references therein]. Our studies in bulk solution by fluorescence microscopy (Figure 7) indicated that the 30-nm fibers, which are generated in the presence of H1, do not aggregate, suggesting the fiber has a charged colloidal nature.
Topo II appears to induce the parallel alignment of double-stranded DNA chains and to promote formation of thick filaments in the absence of ATP (Figure 2), indicating that Topo II mediates assembly of DNA strands independently of its enzymatic activity. It is reasonable that this interaction also occurs within DNA–histone complexes (i.e. nucleosomes). The marked difference in the effect of Topo II on reconstituted chromatin in the absence and presence of histone H1 can be attributed to the difference in the fundamental geometric structures containing the 30-nm fibers. In the absence of H1, each Topo II molecule mediates a weak attraction between the nucleosome cores. In the presence of H1, as the contact areas between the fibers are increased, the attraction between two clusters also increases due to the added attraction via Topo II. Therefore, the attraction between 30-nm fibers would be much stronger than the attraction between the pair of nucleosomes, and it could be enhanced by Topo II.
The free energy of the cluster of 30-nm fibers, which determines the stability, can be described with the following equation:
where N is the number of 30-nm fibers in the assembly, and a, b and c are positive constants and α and β are exponents. For simplicity, the fibers are assumed to have the same lengths. The first term in the equation corresponds to the stabilization due to the attractive interaction mediated by Topo II; the second is the surface energy and the exponent, where α is generally <1; and the third is derived from the destabilization due to the remaining negative charge in the assembly, where the exponent β is larger than 1. In an idealized system with a spherical symmetry, β is 5/3. For the assembly of nonspherical objects, β is somewhat smaller than 5/3 but is always >1. This equation predicts that the assembly of 30-nm fibers will stop at a finite size, which depends on the remaining negative charges and attractions, and it further implies the formation of a larger assembly when the remaining negative charges are further reduced.
In summary, in addition to favorable geometry of the attractive interaction between the 30-nm fibers, the reduction of the negative charge in the polynucleosome structure by H1 may decrease the instability due to electrostatic repulsion. Such a charge effect of H1 can also stabilize the assembly of 30-nm fibers.
Physiological significance of Topo II-induced chromatin compaction
The matrix attachment region (MAR) has been experimentally identified as a segment of genomic DNA that is resistant to digestion by nucleases (38) or that remains attached to the insoluble fraction of the nucleus after digestion with restriction endonucleases (39). A MAR has been located in the histone gene cluster (38) and the β-globin locus (39), which contain the consensus sequence for interaction with Topo II (38,40). In our experiments, Topo II did not show detectable affinity for the MAR region of the β-globin gene (Figure 2G). Based on these results, we concluded that the MAR region does not influence the effect of Topo II in the in vitro system. Mechanisms for loading Topo II on the MAR region must exist in vivo, and Topo II is expected to condense the chromatin with a help of other scaffold proteins such as the condensin complex. A comparison of in vitro and in vivo chromatin structures is needed to determine the significance of the MAR region in chromatin compaction by Topo II.
Proteomic analysis of the metaphase chromosome has shown that the molar ratio of Topo II to histone H4 is 0.72:100 (41). This corresponds to our experimental condition of a weight ratio of 1:40 Topo II to DNA in Figures 3E, 4E and 6B. Because this weight ratio corresponds to one Topo II molecule per 10 kb of DNA and given an in vivo efficiency of one nucleosome per 200 bp of DNA, it is expected that there is one Topo II molecule per 50 nucleosomes (100 molecules of histone H4). Therefore, our experimental system (1:40 weight ratio) may reflect the in vivo situation, although we cannot precisely determine the molar ratio in the reconstituted chromatin at a single-molecule level. In this regard, it is interesting that 30-nm fibers loop out from the condensed part (Figure 6B), which might reflect the highly condensed scaffold region and chromatin looping that has been previously observed by electron microscopy (42). One of the key results of our study is that Topo II does not convert 30-nm fibers into larger complexes. Even in the aggregated complex (Figure 6D), the structural units were 30 nm, indicating that the Topo II may mediate chromatin compaction simply by bringing the 30-nm fibers into contact. In other words, Topo II cannot induce larger chromosomal units such as 80-nm (43), 100-nm (44) and 300-nm (45) beads or fibers, which have been observed by electron microscopy.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by the Japanese Ministry of Education, Culture, Sports, Science and Technology (Grant-in-Aid for Scientific Research on Priority Areas to K.T. and K.Y.) and the Japan Society for the Promotion of Science (Grant-in-Aid for Young Scientists (B) to K.H.). Funding to pay the Open Access publication charge was provided by the Grant-in-Aid for Young Scientists (B) from the Japan Society for the Promotion of Science.
Conflict of interest statement. None declared.
REFERENCES
- 1.Germond JE, Hirt B, Oudet P, Gross-Bellark M, Chambon P. Folding of the DNA double helix in chromatin-like structures from simian virus 40. Proc. Natl. Acad. Sci. USA. 1975;72:1843–1847. doi: 10.1073/pnas.72.5.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leuba SH, Yang G, Robert C, Samori B, van Holde K, Zlatanova J, Bustamante C. Three-dimensional structure of extended chromatin fibers as revealed by tapping-mode scanning force microscopy. Proc. Natl. Acad. Sci. USA. 1994;91:11621–11625. doi: 10.1073/pnas.91.24.11621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hizume K, Yoshimura SH, Takeyasu K. Atomic force microscopy demonstrates a critical role of DNA superhelicity in nucleosome dynamics. Cell Biochem. Biophys. 2004;40:249–262. doi: 10.1385/CBB:40:3:249. [DOI] [PubMed] [Google Scholar]
- 4.Thoma F, Koller T, Klug A. Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin. J. Cell Biol. 1979;83:403–427. doi: 10.1083/jcb.83.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leuba SH, Bustamante C, van Holde K, Zlatanova J. Linker histone tails and N-tails of histone H3 are redundant: scanning force microscopy studies of reconstituted fibers. Biophys. J. 1998;74:2830–2839. doi: 10.1016/S0006-3495(98)77990-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leuba SH, Bustamante C, Zlatanova J, van Holde K. Contributions of linker histones and histone H3 to chromatin structure: scanning force microscopy studies on trypsinized fibers. Biophys. J. 1998;74:2823–2829. doi: 10.1016/S0006-3495(98)77989-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hizume K, Yoshimura SH, Takeyasu K. Linker histone H1 per se can induce three-dimensional folding of chromatin fiber. Biochemistry. 2005;44:12978–12989. doi: 10.1021/bi050623v. [DOI] [PubMed] [Google Scholar]
- 8.Paulson JR, Laemmli UK. The structure of histone-depleted metaphase chromosomes. Cell. 1977;12:817–828. doi: 10.1016/0092-8674(77)90280-x. [DOI] [PubMed] [Google Scholar]
- 9.Jackson DA, Dickinson P, Cook PR. The size of chromatin loops in HeLa cells. EMBO J. 1990;9:567–571. doi: 10.1002/j.1460-2075.1990.tb08144.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hancock R. A new look at the nuclear matrix. Chromosoma. 2000;109:219–225. doi: 10.1007/s004120000077. [DOI] [PubMed] [Google Scholar]
- 11.Berrios M, Osheroff N, Fisher PA. In situ localization of DNA topoisomerase II, a major polypeptide component of the Drosophila nuclear matrix fraction. Proc. Natl. Acad. Sci. USA. 1985;82:4142–4146. doi: 10.1073/pnas.82.12.4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gasser SM, Laroche T, Falquet J, Boy de la Tour E, Laemmli UK. Metaphase chromosome structure. Involvement of topoisomerase II. J. Mol. Biol. 1986;188:613–629. doi: 10.1016/s0022-2836(86)80010-9. [DOI] [PubMed] [Google Scholar]
- 13.Saitoh N, Goldberg IG, Wood ER, Earnshaw WC. ScII: an abundant chromosome scaffold protein is a member of a family of putative ATPases with an unusual predicted tertiary structure. J. Cell Biol. 1994;127:303–318. doi: 10.1083/jcb.127.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kimura K, Hirano T. ATP-dependent positive supercoiling of DNA by 13S condensin: a biochemical implication for chromosome condensation. Cell. 1997;90:625–634. doi: 10.1016/s0092-8674(00)80524-3. [DOI] [PubMed] [Google Scholar]
- 15.Uemura T, Ohkura H, Adachi Y, Morino K, Shiozaki K, Yanagida M. DNA topoisomerase II is required for condensation and separation of mitotic chromosomes in S. pombe. Cell. 1987;50:917–925. doi: 10.1016/0092-8674(87)90518-6. [DOI] [PubMed] [Google Scholar]
- 16.Saka Y, Sutani T, Yamashita Y, Saitoh S, Takeuchi M, Nakaseko Y, Yanagida M. Fission yeast cut3 and cut14, members of a ubiquitous protein family, are required for chromosome condensation and segregation in mitosis. EMBO J. 1994;13:4938–4952. doi: 10.1002/j.1460-2075.1994.tb06821.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hirano T, Mitchison TJ. A heterodimeric coiled-coil protein required for mitotic chromosome condensation in vitro. Cell. 1994;79:449–458. doi: 10.1016/0092-8674(94)90254-2. [DOI] [PubMed] [Google Scholar]
- 18.Maeshima K, Laemmli UK. A two-step scaffolding model for mitotic chromosome assembly. Dev. Cell. 2003;4:467–480. doi: 10.1016/s1534-5807(03)00092-3. [DOI] [PubMed] [Google Scholar]
- 19.Fey EG, Krochmalnic G, Penman S. The nonchromatin substructures of the nucleus: the ribonucleoprotein (RNP)-containing and RNP-depleted matrices analyzed by sequential fractionation and resinless section electron microscopy. J. Cell Biol. 1986;102:1654–1665. doi: 10.1083/jcb.102.5.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nickerson J. Experimental observations of a nuclear matrix. J. Cell Sci. 2001;114:463–474. doi: 10.1242/jcs.114.3.463. [DOI] [PubMed] [Google Scholar]
- 21.Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell Biol. 2002;3:430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 22.Berger JM, Gamblin SJ, Harrison SC, Wang JC. Structure and mechanism of DNA topoisomerase II. Nature. 1996;379:225–232. doi: 10.1038/379225a0. [DOI] [PubMed] [Google Scholar]
- 23.Nettikadan SR, Furbee CS, Muller MT, Takeyasu K. Molecular structure of human topoisomerase II alpha revealed by atomic force microscopy. J. Electron Microsc. (Tokyo) 1998;47:671–674. doi: 10.1093/oxfordjournals.jmicro.a023641. [DOI] [PubMed] [Google Scholar]
- 24.Adachi Y, Luke M, Laemmli UK. Chromosome assembly in vitro: topoisomerase II is required for condensation. Cell. 1991;64:137–148. doi: 10.1016/0092-8674(91)90215-k. [DOI] [PubMed] [Google Scholar]
- 25.Patrinos GP, de Krom M, de Boer E, Langeveld A, Imam AM, Strouboulis J, de Laat W, Grosveld FG. Multiple interactions between regulatory regions are required to stabilize an active chromatin hub. Genes Dev. 2004;18:1495–1509. doi: 10.1101/gad.289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Drunen CM, Sewalt RG, Oosterling RW, Weisbeek PJ, Smeekens SC, van Driel R. A bipartite sequence element associated with matrix/scaffold attachment regions. Nucleic Acids Res. 1999;27:2924–2930. doi: 10.1093/nar/27.14.2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O’Neill TE, Roberge M, Bradbury EM. Nucleosome arrays inhibit both initiation and elongation of transcripts by bacteriophage T7 RNA polymerase. J. Mol. Biol. 1992;223:67–78. doi: 10.1016/0022-2836(92)90716-w. [DOI] [PubMed] [Google Scholar]
- 28.Mirzabekov AD, Pruss DV, Ebralidse KK. Chromatin superstructure-dependent crosslinking with DNA of the histone H5 residues Thr1, His25 and His62. J. Mol. Biol. 1990;211:479–491. doi: 10.1016/0022-2836(90)90366-T. [DOI] [PubMed] [Google Scholar]
- 29.Nakai T, Hizume K, Yoshimura SH, Takeyasu K, Yoshikawa K. Phase transition in reconstituted chromatin. Europhys. Lett. 2005;69:1024–1030. [Google Scholar]
- 30.Araki S, Nakai T, Hizume K, Takeyasu K, Yoshikawa K. Hydrodynamic radius of circular DNA is larger than that of linear DNA. Chem. Phys. Lett. 2006;418:255–259. [Google Scholar]
- 31.Yoshikawa K, Takahashi M, Vasilevskaya VV, Khokhlov AR. Large discrete transition in a single DNA molecule appears continuous in the ensemble. Phys. Rev. Lett. 1996;76:3029–3031. doi: 10.1103/PhysRevLett.76.3029. [DOI] [PubMed] [Google Scholar]
- 32.Nettikadan S, Tokumasu F, Takeyasu K. Quantitative analysis of the transcription factor AP2 binding to DNA by atomic force microscopy. Biochem. Biophys. Res. Commun. 1996;226:645–649. doi: 10.1006/bbrc.1996.1409. [DOI] [PubMed] [Google Scholar]
- 33.Yoshimura SH, Yoshida C, Igarashi K, Takeyasu K. Atomic force microscopy proposes a ‘kiss and pull’ mechanism for enhancer function. J. Electron Microsc. (Tokyo) 2000;49:407–413. doi: 10.1093/oxfordjournals.jmicro.a023822. [DOI] [PubMed] [Google Scholar]
- 34.Vologodskii AV, Zhang W, Rybenkov VV, Podtelezhnikov AA, Subramanian D, Griffith JD, Cozzarelli NR. Mechanism of topology simplification by type II DNA topoisomerases. Proc. Natl. Acad. Sci. USA. 2001;98:3045–3049. doi: 10.1073/pnas.061029098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bojanowski K, Maniotis AJ, Plisov S, Larsen AK, Ingber DE. DNA topoisomerase II can drive changes in higher order chromosome architecture without enzymatically modifying DNA. J. Cell Biochem. 1998;69:127–142. doi: 10.1002/(sici)1097-4644(19980501)69:2<127::aid-jcb4>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 36.Das C, Hizume K, Batta K, Kumar BR, Gadad SS, Ganguly S, Lorain S, Verreault A, Sadhale PP, et al. Transcriptional coactivator PC4, a chromatin-associated protein, induces chromatin condensation. Mol. Cell. Biol. 2006;26:8303–8315. doi: 10.1128/MCB.00887-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muhlbacher F, Holm C, Schiessel H. Controlled DNA compaction within chromatin: the tail-bridging effect. Europhys. Lett. 2006;73:135–141. [Google Scholar]
- 38.Gasser SM, Laemmli UK. The organisation of chromatin loops: characterization of a scaffold attachment site. EMBO J. 1986;5:511–518. doi: 10.1002/j.1460-2075.1986.tb04240.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jarman AP, Higgs DR. Nuclear scaffold attachment sites in the human globin gene complexes. EMBO J. 1988;7:3337–3344. doi: 10.1002/j.1460-2075.1988.tb03205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adachi Y, Kas E, Laemmli UK. Preferential, cooperative binding of DNA topoisomerase II to scaffold-associated regions. EMBO J. 1989;8:3997–4006. doi: 10.1002/j.1460-2075.1989.tb08582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Uchiyama S, Kobayashi S, Takata H, Ishihara T, Hori N, Higashi T, Hayashihara K, Sone T, Higo D, et al. Proteome analysis of human metaphase chromosomes. J. Biol. Chem. 2005;280:16994–17004. doi: 10.1074/jbc.M412774200. [DOI] [PubMed] [Google Scholar]
- 42.Marsden MP, Laemmli UK. Metaphase chromosome structure: evidence for a radial loop model. Cell. 1979;17:849–858. doi: 10.1016/0092-8674(79)90325-8. [DOI] [PubMed] [Google Scholar]
- 43.Kobori T, Kodama M, Hizume K, Yoshimura SH, Ohtani T, Takeyasu K. Comparative structural biology of the genome: nano-scale imaging of single nucleus from different kingdoms reveals the common physicochemical property of chromatin with a 40 nm structural unit. J. Electron Microsc. (Tokyo) 2006;55:31–40. doi: 10.1093/jmicro/dfi076. [DOI] [PubMed] [Google Scholar]
- 44.Belmont AS, Bruce K. Visualization of G1 chromosomes: a folded, twisted, supercoiled chromonema model of interphase chromatid structure. J. Cell Biol. 1994;127:287–302. doi: 10.1083/jcb.127.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rattner JB, Lin CC. Radial loops and helical coils coexist in metaphase chromosomes. Cell. 1985;42:291–296. doi: 10.1016/s0092-8674(85)80124-0. [DOI] [PubMed] [Google Scholar]
- 46.Schultz P, Olland S, Oudet P, Hancock R. Structure and conformational changes of DNA topoisomerase II visualized by electron microscopy. Proc. Natl. Acad. Sci. USA. 1996;93:5936–5940. doi: 10.1073/pnas.93.12.5936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.