Abstract
KH (hnRNP K homology) domains, consisting of ∼70 amino acid residues, are present in a variety of nucleic-acid-binding proteins. Among these are poly(C)-binding proteins (PCBPs), which are important regulators of mRNA stability and posttranscriptional regulation in general. All PCBPs contain three different KH domains and recognize poly(C)-sequences with high affinity and specificity. To reveal the molecular basis of poly(C)-sequence recognition, we have determined the crystal structure, at 1.6 Å resolution, of PCBP2 KH3 domain in complex with a 7-nt DNA sequence (5′-AACCCTA-3′) corresponding to one repeat of the C-rich strand of human telomeric DNA. The domain assumes a type-I KH fold in a βααββα configuration. The protein–DNA interface could be studied in unprecedented detail and is made up of a series of direct and water-mediated hydrogen bonds between the protein and the DNA, revealing an especially dense network involving several structural water molecules for the last 2 nt in the core recognition sequence. Unlike published KH domain structures, the protein crystallizes without protein–protein contacts, yielding new insights into the dimerization properties of different KH domains. A nucleotide platform, an interesting feature found in some RNA molecules, was identified, evidently for the first time in DNA.
INTRODUCTION
The family of poly(C)-binding proteins (PCBPs), which is comprised of five members in mammalian cells, namely hnRNP K and PCBP1–4 (also known as αCP1–4; PCBP1 and PCBP2 are also known as hnRNP E1 and E2, respectively), has a wealth of functions attributed to them (1,2). Among these are transcriptional and translational silencing and enhancement. Furthermore, they play a role in mRNA stabilization and splicing and seem to be linked to apoptotic and developmental pathways and the regulation of viral function (3).
Examples of different PCBP functions have been intensely studied, with the effects on mRNA stability being among the earliest to be investigated. It has been noted that the high stability of α-globin mRNA, needed for optimal translation, is linked to the binding of PCBPs to pyrimidine-rich sequences within the 3′-UTR that leads to the formation of a so-called α-complex (3,4). Similar stabilizing effects through PCBP binding can be observed for the mRNAs of collagen-α1 (5,6), tyrosine hydroxylase (7), erythropoietin (8) and others. Diminution of the amount of expressed protein, which interestingly is also mediated by PCBPs, is achieved by translational silencing, as seen in 15-lipoxygenase mRNA (9–11).
PCBPs are also connected to developmental processes and viral replication. It was shown that the 5′-UTR of the Poliovirus genomic RNA harbors two binding sites for PCBP1 or PCBP2. Binding of PCBP2 to one of the sites, a C-rich internal bulge sequence known as loop B RNA within the internal ribosomal entry site (IRES) element, is required for cap-independent translation of the viral RNA (12–16). Binding of PCBP1 or 2 to the other site, a C-rich loop in the stem-loop B domain within a so-called cloverleaf-like RNA structure located at the very 5′-end of the 5′-UTR, stabilizes the genomic RNA, inhibits translation and switches the viral genomic RNA to a template for RNA replication (14,17). Unlike PCBP-binding sites within 3′UTRs of the cellular mRNAs, the two viral C-rich recognition sequences are presented in the context of a structured RNA, as demonstrated by our NMR structures of two sequence variants of the loop B RNA (18).
A splice variant of PCBP4 (termed MCG10) has been shown to promote apoptosis and cell cycle arrest in G2-M phase upon forced expression in human cells, possibly via interaction between PCBP4 and the C-rich RNA template sequence of human telomerase. Since its expression is normally induced in a p53-dependent manner, it represents a potential mediator of p53 tumor suppression (19). The functional roles of PCBPs in RNA regulation will certainly expand further. It was shown, for example, that PCBP2 associated with 160 mRNA species in vivo in a human hematopoietic cell line (20), suggesting a general significance of PCBPs in posttranslational regulation.
Additionally, PCBPs are able to bind to DNA sequences as well and seem to play a role in the more proximal events of gene expression, namely transcriptional regulation. For example, transcriptional activation can be observed upon binding of hnRNP K to the promoter of the human c-myc gene and the SV40 early promoter (21,22), whereas transcriptional silencing is seen during the repression of thymidine kinase, supposedly achieved through hnRNP K-mediated inhibition of the binding of other transcription factors to the promoter of the gene (23).
Despite this wealth of function, a unifying theme of PCBPs is their ability to interact tightly with poly(C) sequences of both RNA and DNA. This ability is conferred by the presence of three copies of a conserved RNA-binding motif termed KH domain (hnRNP K homology domain), whereby different nucleic-acid-binding specificities can be observed for different KH domains. The common arrangement of these domains within the primary sequence of the PCBPs is the close spacing of two domains at the N-terminus and a C-terminal KH domain that is separated from the previous two by a linker of variable length (see Figure 1A) (1). Considering the plethora of different functions established for the PCBPs, it remains a striking question as to how these diverse functions can be traced back to the interplay between the different KH domains, regulating nucleic-acid-binding specificity as well as the potential interaction with different downstream protein partners. This question gains even more gravity considering the strong sequence conservation between the different KH domains present in PCBPs (see Figure 1B), suggesting similar behavior.
Figure 1.
(A) Schematic diagram of the domain structure of human poly(C) binding protein-2 (PCBP2). Similar domain structures are observed in other members of the PCBP family. (B) Sequence alignment of the three KH domains from different PCBPs. Alignments were carried out using the program ClustalX. The sequence shown for PCBP2 KH3 corresponds to residues 285–359 in the full-length protein. The actual construct for crystallization has an artificial lysine before Ala285. Secondary structures of the KH3 domain were based on the crystal structure.
To reveal the molecular details of sequence-specific nucleic acid recognition and protein–protein interactions entailing PCBP KH domains, we have so far employed both NMR and X-ray crystallography (24,25; Du et al., manuscript submitted). We report in this paper the high-resolution crystal structure of the PCBP2 KH3 domain in complex with a 7 nt C-rich DNA sequence corresponding to one repeat of the C-rich strand of human telomeric DNA. At a resolution of 1.6 Å, this represents the highest resolution structure of a KH domain–nucleic acid interaction so far. The structure reveals a complex combination of molecular interactions involved in specific recognition of the DNA sequence in unprecedented detail. While similarities to the other previously reported PCBP KH domain–nucleic acid crystal structures (26) are apparent, the current structure has a number of features that have not been described in any of the previous complexes; some of these features are likely to be important functionally.
MATERIALS AND METHODS
Cloning
The gene that encodes the KH3 domain of human PCBP2 was amplified by PCR using appropriate primers and a plasmid containing the gene for full-length PCBP2. The amplified gene was cloned between Nde I and Xho I sites of the plasmid vector pET 24a. The cloned plasmid was transformed into a BL21(DE3) strain of Escherichia coli (Stratagene). The protein was over-expressed with a sequence of MKH6K attached to the N-terminus of the native sequence (Ala285-Thr359).
Sample preparation and crystallization
N-terminal His-tagged PCBP2 KH3 was over-expressed in the BL21(DE3) strain of E. coli (Stratagene). For Se-Met-labeled protein, the bacteria were grown in M9 minimal medium until they reached an OD600 of 0.6–0.8, whereupon leucine, isoleucine, lysine, phenylalanine, threonine and valine were added to the culture to inhibit methionine biosynthesis. After 30 min, l-seleno-methionine (50 mg/l) was added, followed by IPTG (isopropyl-β-d-thiogalactopyranoside) to a final concentration of 0.06 mM to induce expression of the Se-Met-labeled protein at 12°C overnight. Cells were harvested and resuspended in 200-mM NaCl and 20-mM HEPES, pH 7.5 and lysed by sonication. After purification by Ni-NTA resin (QIAGEN), the His-tag was removed by the TAGzyme system from QIAGEN, and the sample was concentrated to a final concentration of about 4 mg/ml protein with the help of an Amicon Ultra-5K centrifugal device. Crystals of the PCBP2 KH3–DNA (5′-AACCCTA-3′) complex formed from a solution with a 1:1.2 protein:DNA ratio and were obtained by hanging-drop vapor diffusion against 2 M ammonium sulfate, 80 mM lithium sulfate, 100 mM CAPS, pH 8.18 and 5% glycerol at 22°C. Orthorhombic crystals grew to useful size within about 5 days with diffraction to 1.55 Å. The crystals are in space group R32 (a = 81.07 Å, b = 81.07 Å, c = 87.82 Å), with one protein–DNA complex per asymmetric unit.
Data collection, structure determination and refinement
A single SAD data set was collected at the peak wavelength of the selenium K absorption edge from a single frozen selenomethionine-containing crystal using Beamline 8.3.1 of the Advanced Light Source (ALS) at Berkeley National Laboratory. A native data set was also collected on the same crystal with longer exposure time (5 s). The SAD data set and native data set diffract to 1.75 and 1.55 Å resolution, respectively. Diffraction intensities were integrated and reduced by using the program DENZO and were scaled using SCALEPACK (27). The selenium atom was located using CNS (28). An interpretable electron density map at 1.6 Å was obtained after solvent flattening. The model was built by MOLOC (29) and Coot (30). Refinement was performed using Refmac5 within the CCP4 program suite (31) to an R factor of 20.5% (Rfree = 24.4%). The final model includes the protein residues 286–359 (PCBP2 numbering is used) and one DNA molecule with 7 nt. Analysis of the structure shows that all parameters are well within expected values at this resolution (see Table 1). The coordinates for the PCBP2 KH3–DNA complex have been deposited in the Protein Data Bank (pdb code 2P2R). Structure figures were generated using PyMol (W. L. DeLano, The PyMOL Molecular Graphics System (2002) on the World Wide Web http://www.pymol.org).
Table 1.
Crystallographic refinement statistics
| Crystal data | KH3–DNA |
|---|---|
| Space group | R32 |
| Unit cell dimensions (Å) | a = 81.07 |
| b = 81.07 | |
| c = 87.82 | |
| za | 1 |
| X-ray data collection statistics | |
| Wavelength (Å) (SeMet SAD Peak) | 0.979462 |
| Resolution (Å) | 60.0 – 1.55 |
| Observed reflections | 272 583 |
| Unique reflections | 15 516 |
| Completeness (last shell) (%) | 98.5 (84.66) |
| Rmerge (%)b (last shell) | 3.3 (38.2) |
| I/σ (last shell) | 22.8 (7.3) |
| Phasing and refinement statistics | |
| Resolution (Å) | 40.0 – 1.6 |
| Reflections in working set | 13 842 |
| Reflections in test set (5%) | 723 |
| Rcryst (%)c | 20.5 |
| Rfree (%)c | 24.4 |
| RMSD bonds (Å) | 0.009 |
| RMSD angles (°) | 1.334 |
| Mean B-factors (Å2) | 28.9 |
az is the number of equivalent structures per asymmetric unit.
bRmerge = ∑|Ihkl − < Ihkl > |/∑Ihkl, where Ihkl is the measured intensity of hkl reflection and <Ihkl> is the mean of all measured intensity of hkl reflection.
cRcryst = ∑hkl||Fobs| − |Fcalc||∑hkl|Fobs|, where Fobs is the observed structure factor amplitude and Fcalc is the structure factor calculated from model. Rfree is computed in the same manner as is Rcryst, with the test set of reflections (10%).
RESULTS
Overall protein structure
The protein–DNA complex crystallized in space group R32 with only one protein–DNA complex per asymmetric unit. Electron density is present for residues 286–359 in the 76-amino-acid construct as well as all 7 residues of the DNA molecule and 86 water molecules. The overall structure of the complex is depicted in Figure 2A.
Figure 2.
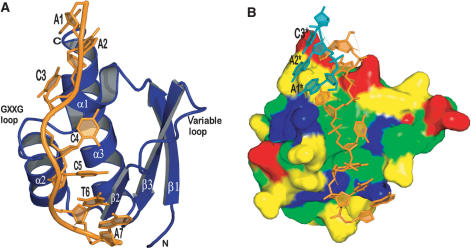
Overall structure of the PCBP2 KH3–DNA complex. (A) Structure of the complex in one asymmetric unit. The DNA and protein are colored orange and deep blue, respectively. (B) Surface representation of the protein showing the nucleic-acid-binding groove. Positively charged, negatively charged, uncharged hydrophilic and hydrophobic residues are colored blue, red, yellow and green, respectively. The floor of the nucleic-acid-binding groove is mainly defined by hydrophobic residues. The DNA in the complex is colored orange. The first three residues from a symmetry-related DNA (labeled A1*, A2* and C3*) are shown in dark teal and are stacking on top of the complex DNA. See text for details.
The PCBP2 KH3 adopts a typical type-I KH domain fold (32–36) of a β1-α1-α2-β2-β3-α3 configuration. The three β-strands (residues 289–295, 320–323 and 331–338) form an antiparallel β-sheet with a left-handed twist and a spatial order of β1-β3-β2, which is packed against the three α-helices (residues 297–303, 308–317 and 341–356). The invariable GXXG loop, here Gly304-Arg305-Gln306-Gly307, is located between α1 and α2; the variable loop Ala324-Ser330 lies between β2 and β3. The core of the protein consists almost exclusively of hydrophobic residues, ensuring a tight fold as well as forming the hydrophobic floor of a narrow groove where DNA binding can take place (see Figure 2B). The groove is defined by the juxtaposition of helices α1 and α2, the connecting GRQG loop and strands β2 and β3 together with the connecting variable loop; residues contacting the DNA emanate from all of these structural elements.
Crystal contacts and overall DNA structure
Interestingly, no protein–protein contacts were observed in the crystal. Instead, crystal contacts were solely formed by base stacking interactions of DNA molecules from adjacent asymmetric units. Ade1 of the heptanucleotide stacks on Cyt3 of a symmetry-related DNA and vice versa, leading to the formation of quite an interesting structure entailing six nucleotide residues (see Figure 2B). A remarkable feature of this structure is the arrangement of Ade2 and Cyt3. Those two consecutive bases were found to be coplanar with the N3 position of Ade2 forming an intramolecular hydrogen bond with the N4 amino group of Cyt3. This feature is reminiscent of the adenosine platform motif that was first observed in the crystal structure of the P4–P6 domain of the group I self-splicing intron (37). Other types of platforms have been reported subsequently. A conserved AU platform important for protein recognition was reported in the ribosomal protein S8-RNA complex (38). The same type of platform also occurs in the HIV-1 RNA packaging signal (39). A GU platform in a GUA triple in the sarcin/ricin loop of 23S large ribosomal subunit RNA has also been documented (40). To our knowledge, however, no AC platform has been reported for DNA so far, and platform-like structures have been limited to examples derived solely from RNA.
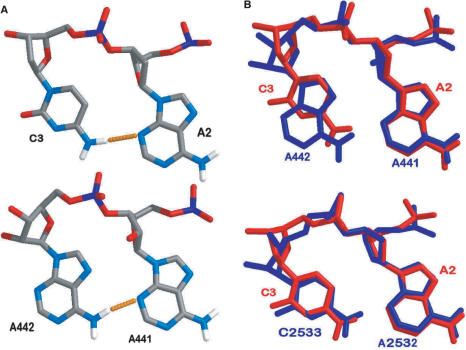
To illustrate the close relationship between our DNA structure and previously documented RNA platform structures, we have compared it with an AA and an AC platform found in the crystal structure of the 50S ribosomal subunit of Haloarcula Marismortui (41) (see Figure 3). The overall geometry is very similar; the overlay (see Figure 3B) in particular exhibits the similarity between the different platform structures.
Figure 3.
(A) Comparison between the structure of Ade2 and Cyt3 of our crystallization DNA (top) and Ade 441 and Ade 442 from the crystal structure of the 50S ribosomal subunit of Haloarcula marismortui (bottom). Hydrogen bonds are depicted as yellow dashed bars, atoms are color-coded light blue, red, grey, white and dark blue for nitrogen, oxygen, carbon, hydrogen and phosphorous, respectively. (B) Top: Overlay of the structure of Ade2 and Cyt3 of our crystallization DNA (red) and Ade 441 and Ade 442 from the crystal structure of the 50S ribosomal subunit of H. marismortui (blue). Bottom: Overlay of our DNA (red) and Ade2532 and Cyt 2533 from the crystal structure of the 50S ribosomal subunit of H. marismortui (blue).
Overview of the DNA binding
Recognition of the DNA is achieved by the combination of a set of different interactions including hydrogen bonds, electrostatic interactions, van der Waals contacts and shape complementarities. The DNA orients in such a way that its 5′- and 3′-ends contact the C and the N-terminal regions of PCBP2 KH3 domain, respectively. This relative spatial relationship between a KH domain and its nucleic acid target is conserved among all reported KH domain-DNA/RNA complex structures. The phosphate backbone of the DNA is located mainly on the left ridge of the binding groove [as seen by looking at the DNA with the 5′ end on top (see Figure 2A)]. Only a few close contacts of positively charged residues namely Arg305 and Arg314 with the backbone can be found. This finding is somewhat different from our previous structure of PCBP2 KH1 in complex with the same DNA (24), where the left ridge of the nucleic-acid-binding groove is predominantly occupied by four positively charged amino acids (three lysines and an arginine). In the current case, however, the backbone is mainly contacted via a set of hydrogen bonds, both directly and water-mediated. Three phosphate backbones of the DNA are involved. These are: Cyt4 O2P contacting the Gln306 amide, a Cyt3 O1P water 12 Gly300 amide bridge and Ade7 O1P, which is bridged by water 40 and water 22 to the Ile321 carbonyl and amide, respectively (see Figure 4).
Figure 4.
Stereo view of the PCBP2 KH3–DNA complex structure. The dense network of hydrogen bonds (yellow dashed bars) involved in DNA–protein interaction is shown. Red spheres represent structured water molecules that participate in the hydrogen-bonding network. The DNA and protein are colored orange and deep blue, respectively. Protein residues that participate in hydrogen bond interaction with the DNA are shown in stick representation.
As shown in Figure 2B, a cluster of hydrophobic isoleucine residues 299, 303, 310 and 323 defines the floor of the binding groove, which contacts the riboses and bases of Cyt4 and Cyt5, the central two residues of the core-recognition motif of the DNA. Residues at these positions are highly conserved among the KH domains (see Figure 1). An Ile to Asn mutation in the second KH domain of FMRP, at a position corresponding to Ile310 in PCBP2 KH3, leads to a particularly severe case of Fragile-X mental retardation.
The variable loop remains remarkably open upon DNA binding. Only the first two residues of the loop, Ala324 and Asn325, participate in the dense hydrogen-bonding network of DNA–protein interaction (see next section for details).
Specific recognition of the crystallization DNA
Ade1 does not interact with the protein directly. The base of Ade1 is sandwiched between the base of Ade2 from the same DNA on one side and the base of Cyt3 from a symmetry-related DNA on the other side (see Figure 2B).
Ade2 lies on top of helix α1; its base is stacked with that of Ade1. The N7 position of Ade2 forms a water-mediated hydrogen bond with the side-chain of Asp297. The N3 position of Ade2 forms an intramolecular hydrogen bond with the N4 amino group of Cyt3. The bases of Ade2 and Cyt3 are coplanar, but their Watson-Crick functional groups are not facing each other (see Figures 2A and 5A).
Figure 5.
Stereo views of detailed hydrogen-bonding interactions between the DNA and PCBP2 KH3. Coloring schemes are identical to those in Figure 3. 2Fo-Fc electron density map contoured at 1σ is shown in green for the DNA and light blue for the protein side chains that participate in intermolecular hydrogen bonds. (A), (B), (C) and (D) are for hydrogen bonds to Ade2/Cyt3, Cyt4, Cyt5 and Thy6/Ade7, respectively.
The base of Cyt3 also lies on top of helix α1, inside a cleft formed between the invariable GRQG loop and helix α3. It is stacked with the Gly300-Cys301 peptide plane on one side and the base of Ade1 from a symmetry-related molecule on the other (see Figure 2B). All three of the Watson-Crick positions of Cyt3 are involved in intra- or intermolecular hydrogen bonds. Besides the intramolecular bond mentioned previously, the O2 and N3 groups of Cyt3 are contacted by the side chain of Lys309 from helix α2 (see Figure 5A). Formation of these hydrogen bonds makes the placement of a cytosine residue at this position more favorable than other residue types.
Cyt4 is the only nucleotide in the complex that has no stacking interaction with other bases. It occupies a center position of the nucleic-binding groove, with its ribose and hydrophobic edge of the base involved in hydrophobic contacts with the hydrophobic floor of the groove (see Figure 2B) and its Watson-Crick functional groups pointing to the right side of the groove (see Figure 5B). Here, it gets contacted by Arg333 that is conserved among all PCBP KH domains (see Figure 1B). Similar to other PCBP KH domain–DNA/RNA complex structures (24,26), the side chain of Arg333 reaches out from β3 to form two hydrogen bonds with the O2 and N3 acceptors of the base. The extended conformation of the Arg gets stabilized by two hydrogen bonds to the backbone carbonyl of Asn324. The base of Cyt4 is further contacted at its N4 amino group by an intramolecular hydrogen bond to the O1P phosphate group of Cyt3 and a water molecule that bridges to the side chain of Arg333 and the backbone carbonyl of Asn296 (see Figure 5B). This set of hydrogen bonds is only compatible with a Cytosine at this position.
Base-stacking interaction is observed among the last three residues (Cyt5, Thy6 and Ade7) of the crystallization DNA (see Figure 2A). In the vicinity of Cyt5 and Thy6, as many as seven structural water molecules could be found participating in a dense network of hydrogen bonds joining the DNA and the protein (see Figure 4).
Specific recognition of Cyt5 is achieved by a set of hydrogen bonds involving the Watson-Crick positions similar to Cyt4. The side chain of Arg314 is protruding from the C-terminal end of helix α2 to donate two hydrogen bonds to the O2 position. The N4 amino group is involved in two hydrogen bonds: one to the backbone oxygen atom of Ile323 and one water mediated to the side chain of Asn325. Finally, the Cyt5 N3 gets bridged by a water molecule to the Ile323 backbone amide (see Figure 5C). This network of hydrogen-bonding interactions should allow only cytosine to be recognized at this position.
For Thy6, the O2 group and the two Watson-Crick positions N3 and O4 are all involved in direct or water-mediated hydrogen bonds (see Figure 5D). The O2 group forms a hydrogen bond to the side-chain of Lys322. N3 forms a hydrogen bond with water molecule #14 that bridges to the side-chain of Asn325 and the backbone carbonyl of Ile323. O4 is contacted by two water molecules 31 and 16, which bridge to O1P of Cyt5 and OD1 of Asn325, respectively.
Ade7 is the last nucleotide in the crystallization DNA. Two direct hydrogen bonds are observed from the side chains of Lys322 and Asn325 to N3 and N1 groups of Ade7, respectively (see Figure 5D).
DISCUSSION
Several other KH domain–nucleic acid co-crystal structures are available for comparison with our current structure. These include: the 2.4 Å structure of NOVA2 KH3 in complex with a SELEX RNA stem-loop (42); core recognition sequence 5′-UCAC-3′, the 1.8 Å structure of hnRNP K KH3 in complex with a 6-nt DNA (26); core recognition sequence: 5′-TCCC-3′, and the 1.7/2.1/2.6 Å structures of PCBP2 KH1 in complex with a 7-nt DNA and a 12-nt DNA/RNA, respectively (1, Du et al., in preparation); and core recognition sequences 5′-ACCC-3′ and 5′-CCCT/U-3′.
Although the sequences of the KH domains and the recognized DNA/RNA are different from case to case, a comparison of all these crystal structures clearly reveals some common structural features of KH domain–nucleic acid interaction. First of all, the structures of the KH domains are very similar to each other, with the possible exception of the variable loop. For PCBP2 KH1, several residues from the variable loop interact with the nucleic acid target; but for other KH domains, the variable loop has little contact with the DNA/RNA.
Secondly, a core DNA/RNA recognition motif consisting of four nucleotides is observed in all structures. Each of the core motifs assumes a similar conformation. The bases and riboses of corresponding nucleic acid residues are similarly oriented and occupy virtually the same location within the nucleic-acid-binding groove. Structural similarities of both the KH domain and the nucleic acid core recognition motif make it clear that each residue of the recognition motif can only contact a defined set of amino acid residues whose presence in the vicinity of the nucleotide is dictated by the conserved KH domain structure. The properties of those amino acid residues decide specificity of the recognition.
Except for the NOVA2 KH3–RNA complex, all of the crystal structures are of complexes between KH domains of the PCBP family proteins and nucleic acids. Although each of the core recognition motifs recognized by the PCBP KH domains contains a triple-C sequence, the placement of the triple-C sequence in the binding groove is not the same in every structure. The triple-C sequence can be found either at positions 1–3 or 2–4 of the tetranucleotide core recognition motif. Whatever the case is, the second and third positions are always occupied by cytosines. Specific hydrogen bonds for the recognition of these two cytosines seem conserved among all the structures. Two arginine side chains (Arg314 and Arg333 in PCBP2 KH3) are critical for the hydrogen-bonding networks. These two arginines are absolutely conserved among all the KH domains of the PCBP proteins. Every one of the PCBP KH domains should therefore be able to recognize at least two cytosines, at the second and third positions of the nucleic-acid-binding groove. Residues at the first and fourth positions of the tetranucleotide core recognition motif for PCBP KH domains are less restrictive in terms of the type of nucleotide that could be accommodated. At the first position, every nucleotide type except guanosine has been found; at the fourth position, all pyrimidine residues (T/U/C) are compatible. No conserved hydrogen-bonding interaction between the protein and the nucleic acid is observed at the first and fourth positions.
As the highest resolution KH domain–nucleic acid crystal structure, the current structure reveals how the DNA is recognized by the KH domain in unprecedented detail. Most outstandingly, six out of the seven nucleotides of the crystallization DNA form hydrogen bonds, either directly or water-mediated, with the protein. Furthermore, for all four nucleotides in the core recognition motif (Cyt3 to Thy6), all of the potential hydrogen-bond-forming functional groups of the bases (not just the Watson-Crick groups) are actually involved in hydrogen bonds (see Figure 5). Such a high degree of intermolecular hydrogen-bonding interaction is not observed in any of the previously reported KH domain–nucleic acid structures. The number of structured water molecules (at least 10) involved in the hydrogen bond network responsible for DNA–protein recognition has also not been seen before. The structure provides a good example to highlight the importance of water molecules in both specific and nonspecific protein–nucleic acid interactions. Defining these kinds of water molecules would be a very difficult, if not impossible, task for structure determination of protein–nucleic acid complexes by NMR.
The proteins of the PCBP family contain three KH domains (see Figure 1A). By determining the structures of PCBP2 KH1 and KH3 domains with the same human telomeric C-rich strand DNA (24), we show that different KH domains from the same protein can recognize the same DNA sequence. Based on the conservation of residues critical for specific recognition, it is most likely that the KH2 domain can also engage in specific nucleic acid interactions. Since most of the known PCBP targets contain tandem poly(C) sequences, it would be very interesting to see how these targets are recognized by the multiple KH domains from the same protein, and how the KH domains rearrange as a result of nucleic acid binding. Currently, it may be noted that although both KH1 and KH3 domains interact with the same DNA sequence, the KH3 domain forms more intermolecular hydrogen bonds with the recognized DNA. This may translate into a difference in the affinity of interaction. It is reasonable to speculate that the KH domain–nucleic acid interaction with higher affinity may serve to anchor the PCBP protein to the target DNA/RNA, allowing the other two KH domains to interact properly with their particular poly(C) motif within the tandem sequences.
Another potentially significant difference between the PCBP2 KH1 and KH3 domains revealed by our crystal structures is that the KH1 domain can form a presumably very stable homodimer via a protein interaction interface located on the molecular surface opposite to the nucleic-acid-binding groove (24); no protein–protein interaction is observed in the KH3 crystal structure, however. Such a difference in the ability of the KH domains to participate in protein–protein interactions is most likely dictated by different surface properties of the domains. As shown in Figure 6, the KH1 domain has a surface that shows strong hydrophobic properties, allowing extended hydrophobic interactions in the KH1 homodimer that buries 1188 Å of solvent-accessible surface area in each monomer and providing a strong driving force for formation of the dimer. A model of the PCBP2 KH2 domain also shows a similar continuous, extended hydrophobic surface, suggesting that the KH2 domain may also participate in protein–protein interactions similar to KH1. For the KH3 domain, the presence of a number of hydrophilic residues from α3, including Ser357, Ser356, Asn352, Gln348, Ser345, as well as Thr293 from β1 (see also Figure 1B), disrupts the hydrophobic surface as seen in the KH1 crystal structure and the KH2 model. Although the KH domains assume a similar overall structure and bind nucleic acids via a common binding groove, differences in specific protein–nucleic acid and protein–protein interactions may provide a molecular basis for differentiated functional roles of the KH domains.
Figure 6.
Surface representations of the three KH domains in the PCBP proteins. Left: crystal structure of the PCBP2 KH1 domain. Middle: a homologous model (built by the program Modeller based on the crystal structure of PCBP2 KH1) of the PCBP2 KH2 domain. Right: crystal structure of the PCBP2 KH3 domain from the present study. Positively charged, negatively charged, uncharged hydrophilic and hydrophobic residues are colored in blue, red, yellow and green, respectively. Note the large, continuous hydrophobic surface area (in green) of the KH1 and KH2 domains.
A very interesting feature seen in the current structure is the unusual DNA arrangement with Ade2 and Cyt3 in the form of an AC platform (see Figure 3). In structured RNA molecules, those secondary structure motifs serve to disrupt the regular helical geometry and are important to expose functional groups capable of forming hydrogen bonds, thus creating potential binding sites for ligands or other interaction partners. To our knowledge, this is the first time a platform structure has been reported for DNA. It remains to be elucidated whether this finding has functional implications and might occur in vivo in order to restructure DNA molecules upon PCBP binding, or whether it is merely an effect depending on the formation of crystal contacts via adjacent DNA molecules (see Figure 2B).
Unfortunately, crystals of a protein–RNA (5′-AACCCUA-3′) complex, grown under the same conditions as the protein–DNA complex, dissolved the instant the crystallization well was opened, so no diffraction data were collected. Consequently, it could not be determined if the AC platform is induced in RNA molecules upon PCBP2 KH3 binding as well.
Our crystal structures clearly show that both the KH1 and KH3 domains can bind the C-rich strand of human telomeric DNA repeat in vitro. A recent study performed in the lab of Elizabeth H. Blackburn shows that PCBP1 is one of the nucleic-acid-binding proteins present in the human telomere–telomerase complex (unpublished results, personal communication). Further studies are required to reveal whether the molecular interactions we depicted in our structures also occur in vivo and how PCBP proteins might be involved in telomere–telomerase regulation.
ACKNOWLEDGEMENTS
Partial support for this work was provided by National Institutes of Health grants AI46967 (T.L.J.) and GM51232 (R.M.S.). We thank Chris Waddling for managing the UCSF X-ray Crystallization Laboratory and Nick Ulyanov for helpful discussion. The coordinates for the PCBP2 KH3–DNA complex have been deposited in the Protein Data Bank (pdb code 2P2R). Funding to pay the Open Access publication charge was provided by NIH grant AI46967.
Conflict of interest statement. None declared.
REFERENCES
- 1.Makeyev AV, Liebhaber SA. The poly(C)-binding proteins: a multiplicity of functions and a search for mechanisms. RNA. 2002;8:265–278. doi: 10.1017/s1355838202024627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gamarnik AV, Andino R. Interactions of viral protein 3CD and poly(rC) binding protein with the 5' untranslated region of the poliovirus genome. J. Virol. 2000;74:2219–2226. doi: 10.1128/jvi.74.5.2219-2226.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weiss I, Liebhaber S. Erythroid cell-specific mRNA stability elements in the alpha 2-globin 3' nontranslated region. Mol. Cell. Biol. 1995;15:2457–2465. doi: 10.1128/mcb.15.5.2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chkheidze AN, Lyakhov DL, Makeyev AV, Morales J, Kong J, Liebhaber SA. Assembly of the alpha-globin mRNA stability complex reflects binary interaction between the pyrimidine-rich 3′ untranslated region determinant and poly(C) binding protein alpha CP. Mol. Cell. Biol. 1999;19:4572–4581. doi: 10.1128/mcb.19.7.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stefanovic B, Hellerbrand C, Holcik M, Briendl M, Aliebhaber S, Brenner D. Posttranscriptional regulation of collagen alpha1(I) mRNA in hepatic stellate cells. Mol. Cell. Biol. 1997;17:5201–5209. doi: 10.1128/mcb.17.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lindquist JN, Kauschke SG, Stefanovic B, Burchardt ER, Brenner DA. Characterization of the interaction between {alpha}CP2 and the 3′-untranslated region of collagen {alpha}1(I) mRNA. Nucleic Acids Res. 2000;28:4306–4316. doi: 10.1093/nar/28.21.4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paulding WR, Czyzyk-Krzeska MF. Regulation of tyrosine hydroxylase mRNA stability by protein-binding, pyrimidine-rich sequence in the 3′-untranslated region. J. Biol. Chem. 1999;274:2532–2538. doi: 10.1074/jbc.274.4.2532. [DOI] [PubMed] [Google Scholar]
- 8.Czyzyk-Krzeska MF, Bendixen AC. Identification of the poly(C) binding protein in the complex associated with the 3′ untranslated region of erythropoietin messenger RNA. Blood. 1999;93:2111–2120. [PubMed] [Google Scholar]
- 9.Ostareck DH, Ostareck-Lederer A, Wilm M, Thiele BJ, Mann M, Hentze MW. mRNA silencing in erythroid differentiation: hnRNP K and hnRNP E1 regulate 15-lipoxygenase translation from the 3′ end. Cell. 1997;89:597–606. doi: 10.1016/s0092-8674(00)80241-x. [DOI] [PubMed] [Google Scholar]
- 10.Ostareck DH, Ostareck-Lederer A, Shatsky IN, Hentze MW. Lipoxygenase mRNA silencing in erythroid differentiation: the 3′UTR regulatory complex controls 60S ribosomal subunit joining. Cell. 2001;104:281–290. doi: 10.1016/s0092-8674(01)00212-4. [DOI] [PubMed] [Google Scholar]
- 11.Ostareck-Lederer A, Ostareck DH, Standart N, Thiele BJ. Translation of 15-lipoxygenase mRNA is inhibited by a protein that binds to a repeated sequence in the 3′ untranslated region. EMBO J. 1994;13:1476–1481. doi: 10.1002/j.1460-2075.1994.tb06402.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blyn LB, Towner JS, Semler BL, Ehrenfeld E. Requirement of Poly(rC) binding protein 2 for translation of poliovirus RNA. J. Virol. 1997;71:6243–6246. doi: 10.1128/jvi.71.8.6243-6246.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blyn LB, Swiderek KM, Richards O, Stahl DC, Semler BL, Ehrenfeld E. Poly(rC) binding protein 2 binds to stem-loop IV of the poliovirus RNA 5′ noncoding region – identification by automated liquid chromatography tandem mass spectrometry. Proc. Natl Acad. Sci. USA. 1996;93:11115–11120. doi: 10.1073/pnas.93.20.11115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gamarnik AV, Andino R. Switch from translation to RNA replication in a positive-stranded RNA virus. Gene Dev. 1998;12:2293–2304. doi: 10.1101/gad.12.15.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gamarnik AV, Andino R. Two functional complexes formed by KH domain containing proteins with the 5′ noncoding region of poliovirus RNA. RNA. 1997;3:882–892. [PMC free article] [PubMed] [Google Scholar]
- 16.Parsley TB, Towner JS, Blyn LB, Ehrenfeld E, Semler BL. Poly (rC) binding protein 2 forms a ternary complex with the 5′-terminal sequences of poliovirus RNA and the viral 3CD proteinase. RNA. 1997;3:1124–1134. [PMC free article] [PubMed] [Google Scholar]
- 17.Andino R, Rieckhof GE, Baltimore D. A functional ribonucleoprotein complex forms around the 5′ end of poliovirus RNA. Cell. 1990;63:369–380. doi: 10.1016/0092-8674(90)90170-j. [DOI] [PubMed] [Google Scholar]
- 18.Du Z, Ulyanov NB, Yu J, Andino R, James TL. NMR structures of loop B RNAs from the stem-loop IV domain of the enterovirus internal ribosome entry site: a single C-to-U substitution drastically changes shape and flexibility of RNA. Biochemistry. 2004;43:5757–5771. doi: 10.1021/bi0363228. [DOI] [PubMed] [Google Scholar]
- 19.Zhu J, Chen X. MCG10, a Novel p53 Target gene that encodes a KH domain RNA-binding protein, is capable of inducing apoptosis and cell cycle arrest in G2-M. Mol. Cell. Biol. 2000;20:5602–5618. doi: 10.1128/mcb.20.15.5602-5618.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Waggoner SA, Liebhaber SA. Identification of mRNAs associated with alphaCP2-containing RNP complexes. Mol. Cell. Biol. 2003;23:7055–7067. doi: 10.1128/MCB.23.19.7055-7067.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tomonaga T, Levens D. Activating transcription from single stranded DNA. Proc. Natl Acad. Sci. USA. 1996;93:5830–5835. doi: 10.1073/pnas.93.12.5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gaillard D, Cabannes E, Strauss F. Identity of the RNA binding protein K of hnRNP particles with protein H16, a sequence specific single strand DNA binding protein. Nucleic Acids Res. 1994;22:4183–4186. doi: 10.1093/nar/22.20.4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lau JS, Baumeister P, Kim E, Roy B, Hsieh TY, Lai M, Lee AS. Heterogeneous nuclear ribonucleoproteins as regulators of gene expression through interactions with the human thymidine kinase promoter. J. Cell Biochem. 2000;79:395–406. doi: 10.1002/1097-4644(20001201)79:3<395::aid-jcb50>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 24.Du Z, Lee JK, Tjhen R, Li S, Pan H, Stroud RM, James TL. Crystal structure of the first KH domain of human poly(C)-binding protein-2 in complex with a C-rich strand of human telomeric DNA at 1.7 Å. J. Biol. Chem. 2005;280:38823–38830. doi: 10.1074/jbc.M508183200. [DOI] [PubMed] [Google Scholar]
- 25.Du Z, Yu J, Chen Y, Andino R, James TL. Specific recognition of the C-rich strand of human telomeric DNA and the RNA template of human telomerase by the first KH domain of human poly(C)-binding Protein-2. J. Biol. Chem. 2004;279:48126–48134. doi: 10.1074/jbc.M405371200. [DOI] [PubMed] [Google Scholar]
- 26.Backe PH, Messias AC, Ravelli RB, Sattler M, Cusack S. X-ray crystallographic and NMR studies of the third KH domain of hnRNP K in complex with single-stranded nucleic acids. Structure (Camb.) 2005;13:1055–1067. doi: 10.1016/j.str.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 27.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 28.Brünger AT. X-PLOR version 3.843. New Haven, Connecticut: Yale University; 1996. [Google Scholar]
- 29.Gerber PR, Muller K. MAB: a generally applicable molecular force field for structural modeling in medicinal chemistry. J. Comput. Aided Mol. Des. 1995;9:251–268. doi: 10.1007/BF00124456. [DOI] [PubMed] [Google Scholar]
- 30.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Cryst. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 31.CCP4. The CCP4 suite: programs for protein crystallography. Acta Cryst. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 32.Musco G, Kharrat A, Stier G, Fraternali F, Gibson TJ, Nilges M, Pastore A. The solution structure of the first KH domain of FMR1, the protein responsible for the fragile X syndrome. Nat. Struct. Biol. 1997;9:712–716. doi: 10.1038/nsb0997-712. [DOI] [PubMed] [Google Scholar]
- 33.Musco G, Stier G, Joseph C, Morelli MAC, Nilges M, Gibson TJ, Pastore A. Three-dimensional structure and stability of the KH domain-molecular insights into the fragile X syndrome. Cell. 1996;85:237–245. doi: 10.1016/s0092-8674(00)81100-9. [DOI] [PubMed] [Google Scholar]
- 34.Baber JL, Libutti D, Levens D, Tjandra N. High precision solution structure of the C-terminal KH domain of heterogeneous nuclear ribonucleoprotein K, a c-myc transcription factor. J. Mol. Biol. 1999;289:949–962. doi: 10.1006/jmbi.1999.2818. [DOI] [PubMed] [Google Scholar]
- 35.Lewis HA, Chen H, Edo C, Buckanovich RJ, Yang YY, Musunuru K, Zhong R, Darnell RB, Burley SK. Crystal structures of Nova-1 and Nova-2 K-homology RNA-binding domains. Structure Fold Des. 1999;7:191–203. doi: 10.1016/S0969-2126(99)80025-2. [DOI] [PubMed] [Google Scholar]
- 36.Grishin NV. KH domain: one motif, two folds. Nucleic Acids Res. 2001;29:638–643. doi: 10.1093/nar/29.3.638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cate JH, Gooding AR, Podell E, Zhou K, Golden BL, Szewczak AA, Kundrot CE, Cech TR, Doudna JA. RNA tertiary structure mediation by adenosine platforms. Science. 1996;273:1696–1699. doi: 10.1126/science.273.5282.1696. [DOI] [PubMed] [Google Scholar]
- 38.Tishchenko S, Nikulin A, Fomenkova N, Nevskaya N, Nikonov O, Dumas P, Moine H, Ehresmann B, Ehresmann C, et al. Detailed analysis of RNA-protein interactions within the ribosomal protein S8-rRNA complex from the archaeon Methanococcus jannaschii. J. Mol. Biol. 2001;311:311–324. doi: 10.1006/jmbi.2001.4877. [DOI] [PubMed] [Google Scholar]
- 39.Amarasinghe GK, De Guzman RN, Turner RB, Summers MF. NMR structure of stem-loop SL2 of the HIV-1 psi RNA packaging signal reveals a novel A-U-A base-triple platform. J. Mol. Biol. 2000;299:145–156. doi: 10.1006/jmbi.2000.3710. [DOI] [PubMed] [Google Scholar]
- 40.Correll CC, Wool IG, Munishkin A. The two faces of the Escherichia coli 23 S rRNA sarcin/ricin domain: the structure at 1.11 A resolution. J. Mol. Biol. 1999;292:275–287. doi: 10.1006/jmbi.1999.3072. [DOI] [PubMed] [Google Scholar]
- 41.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 angstrom resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 42.Lewis HA, Musunuru K, Jensen KB, Edo C, Chen H, Darnell RB, Burley SK. Sequence-specific RNA binding by a Nova KH domain: implications for paraneoplastic disease and the fragile X syndrome. Cell. 2000;100:323–332. doi: 10.1016/s0092-8674(00)80668-6. [DOI] [PubMed] [Google Scholar]