Abstract
The liquid crystalline chromosomes of dinoflagellates are the alternative to the nucleosome-based organization of chromosomes in the eukaryotes. These nucleosome-less chromosomes have to devise novel ways to maintain active parts of the genome. The dinoflagellate histone-like protein HCc3 has significant sequence identity with the bacterial DNA-binding protein HU. HCc3 also has a secondary structure resembling HU in silico. We have examined HCc3 in its recombinant form. Experiments on DNA-cellulose revealed its DNA-binding activity is on the C-terminal domain. The N-terminal domain is responsible for intermolecular oligomerization as demonstrated by cross-linking studies. However, HCc3 could not complement Escherichia coli HU-deficient mutants, suggesting functional differences. In ligation assays, HCc3-induced DNA concatenation but not ring closure as the DNA-bending HU does. The basic HCc3 was an efficient DNA condensing agent, but it did not behave like an ordinary polycationic compound. HCc3 also induced specific structures with DNA in a concentration-dependent manner, as demonstrated by atomic force microscopy (AFM). At moderate concentration of HCc3, DNA bridging and bundling were observed; at high concentrations, the complexes were even more condensed. These results are consistent with a biophysical role for HCc3 in maintaining extended DNA loops at the periphery of liquid crystalline chromosomes.
INTRODUCTION
The binding of basic nuclear proteins to DNA is a central feature of chromosomal organization. The protein/chromatin DNA ratios range from 1:1 in the eukaryotes (1) to ∼1:1.75 in the prokaryotes (2), reflecting the generally higher level of chromatin organization in eukaryotic chromosomes. The dinoflagellate nucleus represents a major exception to this rule, with its permanently condensed chromosomes throughout the cell cycle but a protein/chromatin DNA ratio of ∼1:10 (3), which is even lower than that of the prokaryotes. Previous studies have also demonstrated that there are no nucleosomes in dinoflagellate chromosomes, with a corresponding lack of histones (4,5).
Under conventional transmission electron microscopy, dinoflagellate chromosomes are uniquely characterized by periodic structures, including bands, arches and crenulations (6–8). The geometry of the dinoflagellate chromosomes, constructed from TEM cross sections, corresponds to a twisted nematic or cholesteric form of liquid crystal DNA (9,10). Freeze-fracture-etch replicas and thin sections of in vitro concentrated DNA solutions are similar to the TEM images of dinoflagellate chromosomes (11), providing strong support for the cholesteric DNA packing model. The strong birefringence of the dinoflagellate chromosomes confirm the highly compact and ordered structure of a liquid crystal (12,13).
A group of acid soluble proteins were identified from the chromatin of the dinoflagellate Crypthecodinium cohnii by means of 2D electrophoresis analysis (5). Four isoforms of C. cohnii histone-like proteins (HCc) were later identified and named form HCc1 to HCc4 (14,15). Recent amino acid sequence analysis revealed that the HCcs share higher homologies with bacterial HU than to histone H1, and have no significant similarities to any of the other eukaryotic core histones (14). Interestingly, similar histone-like proteins were identified also in other human parasites of the intra-kingdom Alveolata (16).
HU protein is one of the well-characterized members of the prokaryotic IHF/HU family of small basic hetero- or homodimeric DNA-binding proteins. It binds to DNA non-specifically and introduces negative supercoils within the circular molecule (17,18). HU has been demonstrated to bend DNA in vitro (19). It has also been shown that HU has an important role in the initiation of replication (20), site-specific DNA rearrangements (21), DNA-strand transfer (22) and gene regulation (23). These diverse processes share a common feature in that the DNA needs to be locally bent, and these situations appear to be promoted by HU. Binding of HU to DNA has two architectural effects, depending on HU concentration: bending (or stabilizing already bent) DNA; or stiffening DNA (24). The HU protein functions in a dimeric form (25). The – 3D structure of Bacillus stearothermophilus HU has been determined by NMR (26) and X-ray crystallography (27). It appears that the functional unit is a wedge-shaped dimer composed of two monomers, which have a hydrophobic interaction between their N-terminal (N-ter) α-helices, and the two C-terminal (C-ter) anti-parallel β-sheet arms bind to the DNA double strand through an interaction with the minor groove.
The exact functions of HCcs are still unknown, but immunocytochemical studies suggested a possible role in gene transcription regulation (15) and maintenance of chromosome structure (28). In non-dividing cells, the HCc protein is located on extra-chromosomal loops and chromosomal nucleofilaments dispersed in the nucleoplasm. In mitotic cells, from prophase to early telophase, HCc protein was found to be homogeneously distributed throughout each entire dividing chromosome. HCc protein has also been detected in the nucleolar organizing region and the fibrillo-granular region, the two subcompartments of the permanently observable nucleolus (15,28). Furthermore, a histone-like protein identified from the dinoflagellate Lingulodinium polyedrum can be acetylated, and histone acetyltransferase-like activities have been detected in this species (29).
Despite the basic nature of HCc3 and its homology with bacterial HU, the structural basis for the functional properties of this protein is unknown. In the present study, we demonstrate that the HCc protein is a DNA condensation protein that may be involved in the modulation of chromosomal organization in dinoflagellates.
MATERIALS AND METHODS
Preparation of recombinant HLP proteins
The DNA fragments encoding HCc3 (GenBank accession: AY128510.1), its N- and C-ter domains (HCc3 aa 1–54 and HCc3 aa 55–113, respectively) were amplified by PCR. Restriction sites BamHI and HindIII were assigned to the 5′ and 3′ primers, respectively, for the in-frame insertion of the PCR fragments into the expression vector pQE30 or pQE70 (Qiagen Corporation, Valencia, CA, USA). The proteins were expressed as N-ter 6× His-tagged polypeptides (pQE30 constructs) or untagged protein (pQE70 constructs). The plasmid constructs were re-sequenced to confirm their identity. The recombinant proteins were over-expressed and purified by using the QIA expressionist protein expression and purification system (Qiagen Corporation) under the denaturing conditions indicated in the manufacturer's protocol. The purified protein was desalted by dialysis against 1× PBS. Polyclonal antibodies were produced against the recombinant full-length HCc3 and affinity-purified.
DNA-cellulose binding assay of recombinant HCc3
DNA-cellulose (native calf thymus DNA; Pharmacia) of 0.2 ml bed volume was pre-equilibrated with a basal buffer (20 mM Tris–Cl at pH 7.6; 1 mM EDTA; 5 mM β-mercaptoethanol; 150 mM NaCl). Protein samples were also diluted in the same basal buffer to a final volume of 1 ml before loading onto the DNA-cellulose column. The flow-through fraction was collected. The column was then washed with 0.5 ml of the same buffer. To test the affinity of the protein to the DNA-cellulose, the column was further washed with the same buffer (0.5 ml each), but using an increasing gradient of NaCl concentrations (200, 400, 600, 800 and 1000 mM). Proteins in all fractions were collected by TCA/acetone precipitation, and then analyzed on a 15% SDS-PAGE gel.
Electrophoretic mobility shift assay (EMSA)
In each reaction of 20 μl in volume, DNA (100 ng) was mixed with HCc3 protein at different dimer/bp ratios in a buffer containing 1× TAE. The mixtures were allowed to stand at room temperature for 20 min. Then electrophoresis loading buffer (6×) was added to each of the mixture to a final concentration of 1×. Samples were then loaded and resolved on a 0.8% TAE agarose gel. After electrophoresis, the gel was stained with EtBr.
Chemical induced cross-linking of recombinant HCc3
Proteins were diluted in an incubation buffer (1× PBS, 0.2% β-mercaptoethanol) to a final concentration of 0.2 μM. After incubation at 25°C for 30 min, the cross-linker ethylene glyco-bis (succinic acid N-hydroxysuccinimide ester) (EGS) was added to a final concentration of 1 mM, followed by incubation at 25°C in the dark for 1 h. The reactions were stopped by adding Tris–Cl (pH 7.4) to a final concentration of 20 mM and then further incubated for 30 min. The proteins were then resolved on a 15% SDS-PAGE gel containing 5 M urea. After SDS-PAGE, the proteins were blotted on to a PVDF membrane by semi-dry transfer and immuno-detected with HCc3 antiserum.
T4-ligase mediated DNA circularization assay
One of the major properties of IHF/HU proteins is their ability to bend DNA, which is reflected in the induction of DNA circularization in the presence of T4 ligase (30). DNA was incubated with or without histone-like proteins in 20 μl of reaction mixture containing 1× T4 DNA ligase buffer (New England Biolab, Beverly, MA, USA) and 5 mM ATP for 15 min at 28°C. Then ligase (10 U) was added to the mixture and further incubated at 28°C for 15 min. The reactions were terminated by adding SDS and sodium acetate (pH 5.0) to final concentrations of 1% and 0.3 M, respectively. The DNA was then phenol–chloroform extracted and collected by ethanol precipitation. If samples were to be treated with Exonuclease III (Exo III) digestion, an equal volume of 2× digestion mixture (2× NEBuffer 1 and 1 U/μl Exonuclease III; New England Biolab) was added to the reaction and the digestion occurred at 37°C for 1 h, prior to the addition of stop solution. DNA samples were finally resuspended in 1× DNA loading buffer and analyzed on 8% TAE-PAGE gel or 0.8% TAE agarose gel, and stained with 0.1% ethidium bromide.
Atomic force microscopy (AFM)
Atomic force microscopy analysis of the DNA–HCc3 complex was done by using 3-aminopropyl-triethoxysilane (APTES) vapor-treated mica (AP-mica) for sample immobilization (31), with minor modifications to the procedure. With the vapor method, 30 μl of APTES (Sigma, St. Louis, MO, USA) was placed at the bottom of a 1.5-ml Eppendorf tube. The tube cap was closed for 10 min at room temperature to create an APTES atmosphere. A piece of freshly cleaved mica (0.8 × 0.8 cm2; Muscovite mica, V-1; Electron Microscopy Science, Fort Washington, PA, USA), kept from contact with the APTES solution, was placed in the APTES containing tube with the cap closed. The mica was then incubated at room temperature for 2 h.
Samples for examination were prepared by mixing DNA and HCc3 at appropriate dimer/bp molar ratios in the AFM buffer (20 mM Tris–Cl at pH 7.5, 10 mM MgCl2, 100 mM NaCl and 10 mM EDTA) and incubated at room temperature for 15 min. The final concentration of DNA was ∼5 ng/μl. Ten microliters (10 μl) of sample was dropped onto the AP-mica and was incubated for 10 min at room temperature. After adsorption, the mica was washed thoroughly in deionized water (18.2 MΩ cm; Millipore, Bedford, MA, USA), blotted at the edge, and dried with compressed nitrogen.
All samples were stored temporarily in a desiccator before AFM imaging. Imaging was carried out on a NanoScope IV STM/AFM (Digital Instruments, Santa Barbara, CA, USA) in the Tapping-Mode using the commercially available Tapping-mode Etched Silicon Probe (TESP; Veeco Instruments Inc., Plainview, NY USA). Images were analyzed with the program WsXM 4.0 Develop 7.0 Scanning Probe Microscopy Software (Nanotec Electronica, Madrid, Spain; http://www.nanotec.es).
With the AFM conditions we used, free DNA molecules present the average contour length of 0.926 μm (SD 0.107 μm; n = 39), which matched the expected length of a linear 2.8 kb DNA (0.924 μm for 0.33 nm/bp). Their average height was ∼0.65 nm (SD 0.15 nm; n = 173). This is consistent with the reported values for AFM measurement of DNA (∼0.44–1.0 nm) (32,33).
RESULTS
HCc3 exhibits structural features of prokaryotic HU/IHF proteins in in silico and in vitro studies
Computational prediction of HCc3 secondary structure
The HCc3 is a 113 aa protein (14). The results of amino acid scale-based prediction methods (34–37) strongly suggest the presence of three α-helical structures within the regions of aa 1–20, 21–40 and 41–60 (α0, α1 and α2; Figure 1) and one C-ter α-helix (α3). Four β-strands are postulated for HCc3 (β1, β2, β3′ and β3), one more (β3′) than the three in Bacillus HU. More than 60% of the amino acid residues of HCc3 within the alignment frame are homologous or identical to those of the B. stearothermophilus DNA-binding protein (GenBank accession: 1311345), of which the 3D structure has been deduced (26,27). The arrangements of α-helices and β-strands in both HCc3 and the B. stearothermophilus DNA-binding protein are also very similar. Based on the similarities between the bacterial HU protein and HCc3 (both primary and secondary structures), it is likely that HCc3 may also possess a similar folding pattern and possibly bind the DNA molecule in a similar way, but a high-resolution structure analysis will need to be performed for final confirmation.
Figure 1.
In silico analyses of HCc3 primary sequence. Amino acid sequence alignment of HCc3 protein with B. stearothermophilus HU protein (1HUE). Sequence alignment was done using the program ‘T-Coffee’ (84). Sequence homology is indicated below the sequences: asterisks indicate identical residues; colons indicate ‘strong’ chemical homology; periods indicate ‘weaker’ chemical homology. Secondary structures of 1HUE (27) and HCc3 (as predicted data) are indicated by colors (red for α-helices; green for β-strands).
HCc3 is a DNA-binding protein, with the C-terminal domain for DNA binding and the N-terminal domain for dimerization
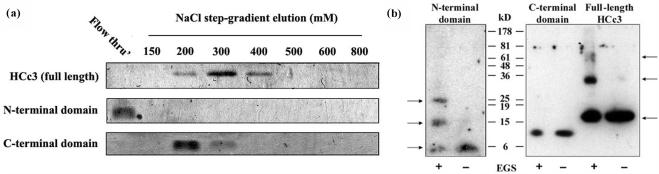
To test the model structure predicted by in silico analysis, a DNA-cellulose binding assay and a chemical induced cross-linking assay were performed (Figure 2). The results of the DNA-cellulose flow-through assay (Figure 2a) show that the HCc3 protein as a whole had a moderate affinity for native DNA, in that it was eluted from DNA-cellulose at ∼300 mM NaCl, which is consistent with the reported result of Vernet et al. (5). The C-ter domain showed a slightly lower affinity toward DNA-cellulose, and was eluted at 200 mM NaCl. The N-ter domain showed no DNA-binding activity in this experiment. The result demonstrates that the DNA-binding property of HCc3 can be attributed to the C-ter domain.
Figure 2.
Functional mapping of HCc3 domains. (a) Binding of HCc3 and truncated domains on DNA-cellulose. The result is shown as the NaCl elution profiles. The lane marked ‘Flow thru’ indicates the unbound fraction. The gels were Coomassie Blue stained and only the corresponding bands are shown here. (b) Association of HCc3 molecules in free solution was assayed in the cross-linking experiment. HCc3 and its truncated forms (N- and C-terminal domains) were treated with or without cross-linker EGS. Protein samples (estimated 1 pmol per lane) were resolved with 15% urea-SDS-PAGE and detected with immunoblotting. The arrows indicate the associated complexes as monomers, dimers and tetramers of the full-length protein and N-terminal domain.
The self-association of these proteins was characterized by means of a chemical fixation of recombinant proteins using the homobifunctional amino-reactive reagent: (succinic acid N-hydroxysuccinimide ester) EGS. Chemical fixation of HCc3 in solution with EGS resulted in products with apparent molecular weights of (approximately) 15, 30 and 60 kDa, corresponding to the monomer, dimer and tetramer, respectively (Figure 2b), suggesting the ability of self-association in free solution. Estimating by the signal intensity, we found that the monomers were the dominant molecular species, with the levels of dimers and tetramers being successively lower. HCc3 obviously does not form trimers, because no corresponding band was detected. This suggests that, in free solution and in the absence of DNA, HCc3 can already exist in multimeric forms. Although no higher order oligomers could be observed, we cannot completely rule out this possibility due to detection limitations. Similarly, the HCc3 N-ter formed EGS cross-linking products with a pattern similar to that of the full-length protein (monomer/∼6 kD, dimer/∼15 kD and tetramer/∼25 kD, but no trimer) under the same experimental conditions. The HCc3 C-ter showed no intermolecular association in free solution. This also excludes the possibility that the fixation of self-associated complexes is a result of non-specific association. The result demonstrates that HCc3 is able to form multimers of even numbers and the N-ter domain is responsible for this property.
The N-terminal domain modulates DNA binding of HCc3
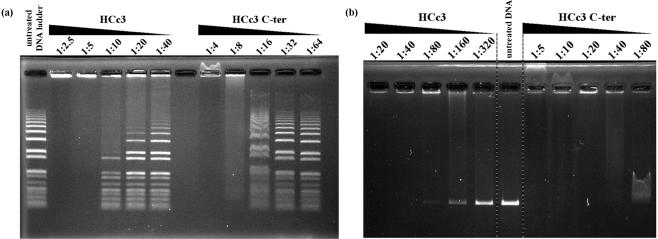
We further examined the DNA-binding properties of HCc3 and its C-ter DNA-binding domain with an electrophoretic mobility shift assay (EMSA) (Figure 3). With increasing concentration, the proteins were examined with either a linear DNA ladder marker (Figure 3a) or linear DNA of single length (2.8 kb EcoR I cut plasmid; Figure 3b). In general, both HCc3 and the C-ter can cause dosage-dependent aggregation of DNA, with HCc3 being about two to four times more sensitive by comparing the threshold of aggregation for each protein species. However, the two proteins differ in several aspects that suggest they bind and/or aggregate DNA with different mechanisms.
Figure 3.
Results of EMSA demonstrating different patterns of DNA band-shift in the presence of HCc3 and its C-terminal domain (C-ter) at different dimer/bp ratios. (a) EMSA with linear DNA ladder (1 kb plus; Invitrogen, Carlsbad, CA, USA). (b) EMSA with purified 2.8 kb linear DNA. The gel was EtBr stained.
In the presence of HCc3, free form DNAs that showed no band shifting was observed at low-HCc3 concentrations. DNAs of higher molecular weight were more susceptible to aggregation (Figure 3a; left side), as longer DNA molecules have a greater chance to recuit more proteins. In contrast, C-ter caused band shifting of un-aggregated DNAs, which was independent of DNA size and all the DNAs shared the same level of shifting within the same sample (Figure 3a; right side). The presence of free DNAs and absence of their shifting with the full-length HCc3 at low to moderate concentration suggests a dynamic equilibrium between the free and DNA-bound HCc3 protein (in dimeric or tetrameric form). That the binding of HCc3 dimer (or tetramer) per se to DNA may have a bias in favor of dissociation, especially under the diluted conditions of electrophoresis.
When the C-ter is at very high concentrations (dimer/bp ratio >1:20; Figure 3b, lanes 7–8) the DNA moved backward. This indicates a switching of electrostatic charge (from −ve to +ve) of the DNA–protein complex, as C-ter molecules continuously bind to the DNA. Moreover, the reentry of the sample into the gel matrix suggests the resolubilization of the DNA-protein aggregate, which is a very common feature of flexible, cationic DNA aggregating agents (more details in Discussion section). In contrast, HCc3 formed substantial aggregates with DNA such that sample reentry and reversed mobility were not observed, as confirmed by reverse electrophoresis under the same conditions (data not shown). Also, there was apparently no charge switching in the case of HCc3-DNA at super-high protein concentration, since the EtBr-stained bands stuck to the anode side of the loading well (Figure 3a, lanes 1–2; Figure 3b, lanes 1–3).
The C-ter domain we used in this work is considered as a polycationic polypeptide chain (16 basic residues on each polypeptide) with no defined conformation (i.e., flexible) (26). Consistent with the reentrant condensation/solubilization behavior, the C-ter-induced DNA condensation can be regarded as a result of electrostatic interactions between the counterions (DNA/protein). However, as suggested by the EMSA result, the full-length HCc3 protein may bind and aggregate DNA in a very different mode, which cannot be solely explained through the mechanism of electrostatic interactions. These result strongly implicate the N-ter domain as having an influence on the ability of the C-ter domain to effectively bind DNA, though the precise mechanism through which the N-ter domain modulates C-ter domain DNA binding is still unknown.
Functional tests of HCc3
HCc3 is not functionally equivalent to prokaryotic HU
Null mutations of both the hupA and hupB genes in Escherichia coli result in cells that have a cold-sensitive phenotype (38). Because structural homology suggests the functional similarity between HCc3 and HU, we wanted to see if expression of HCc3 could complement the cold-shock sensitivity of the E. coli HU deficient mutant JR1672. Expression of HCc3 could not rescue the cells from the stress of cold shock (see Supplementary Figure 1). Repeated experiments yielded similar results.
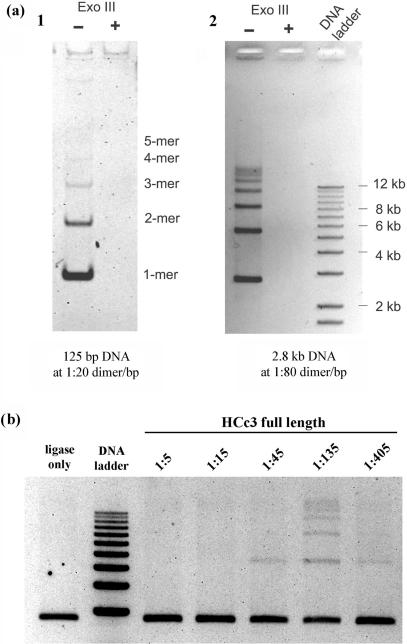
The T4-ligase-mediated DNA ligation assay was performed to test if a short-length DNA fragment could self-ligate (in the presence of T4 DNA ligase) and circularize when its termini came in close proximity as a result of protein-mediated DNA bending. Closed circular DNA should show slower agarose gel mobility than its linear counterpart and should be resistant to Exo III digestion. Usually, small DNA fragments (≤125 bp) are used because their termini cannot come in contact unaided, due to the natural rigidity of DNA molecules (19,30). However, unlike HU that promoted DNA circularization (19), HCc3 promoted concatenation of DNA fragments (Figure 4a). The Exo III sensitivity of the reaction products also suggests that these DNA products are in linear form (Figure 4a). This DNA concatenating effect of HCc3 can also be demonstrated with DNA samples of different lengths, from 125-bp (Figure 4a, left panel), 2.7-kb (Figure 4a, right panel) to 8.9-kb (data not shown). A bacteria (E. coli JM109) transformation test was also performed to see whether HCc3 can inhibit DNA circularization. Ligation of Hind III cut pUC18 plasmid DNA was performed in the presence or absence of HCc3 as described above. Then, the resulting ligation products were used to transform E. coli strain JM109 cells. Assuming the transformation efficiency for linear DNA was negligible, the results show that HCc3 suppresses transformation efficiency as a result of DNA circularization inhibition, in a dosage-dependent manner (see Supplementary Figure 3).
Figure 4.
HCc3 promotes ligase-mediated DNA concatenation. All images are presented as negative images. (a) Concatenation of 125-bp (left panel) and 2.8-kb (right panel) DNA fragments to a higher level in the presence of HCc3 at respective dimer/bp ratios. The linearity of the resulting products was confirmed by Exo III digestion (marked with ‘+/−’ signs). The 125-bp product was resolved on a 8% non-denaturing polyacrylamide gel, and the 2.8-kb product on a 1% TAE agarose gel. (b) Variations in intensity of DNA concatenation in the presence of HCc3 at different dimer/bp ratios. The marker bands indicate DNA sizes from 3 to 12 kb at 1-kb increments.
We also examined the extent of DNA concatenation with increasing HCc3 concentrations (Figure 4b). DNA concatenation peaked at around 1:135 dimer/bp, but was completely inhibited at higher HCc3 concentrations (1:5 and 1:15).
AFM analysis of HCc3–DNA complexes
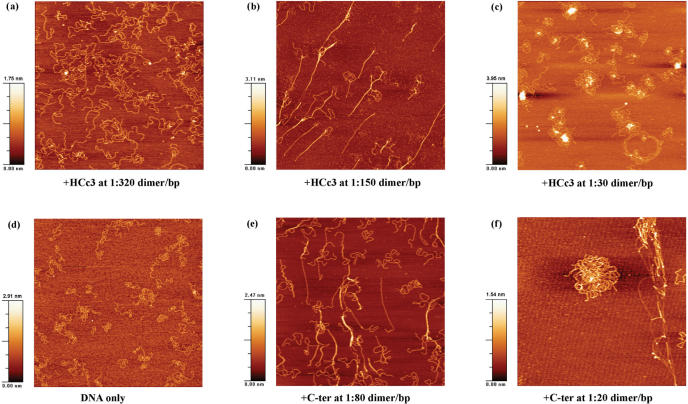
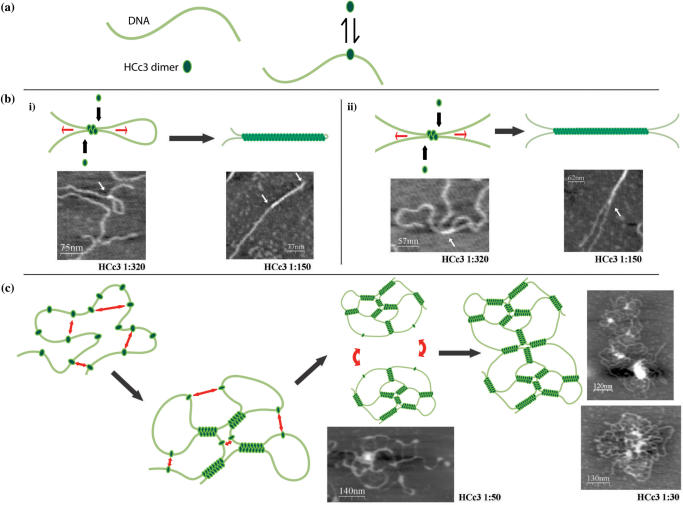
Atomic force microscopy is a useful tool for biological research to image DNA and DNA–proteins complexes (24,39,40). At moderate protein concentrations, where the conditions corresponded to the respective aggregation thresholds based on the EMSA result (Figure 2), both HCc3 and C-ter caused intermolecular bundling of DNA strands (Figure 5b, c). However, the DNA strands are more extended in the sample with HCc3. Also, HCc3 complexes are characterized by the presence of some structures with a direct fold-back of DNA without any hairpin looping [Figure 6b(i), right panel], which is an unusual conformation for DNA as a semi-rigid polymer.
Figure 5.
AFM images of DNA–protein complexes at different dimer/bp ratios. All images are 2.5 × 2.5 μm2 in dimension.
Figure 6.
A diagram showing the mechanism of HCc3-induced DNA condensation. (a) DNA strand is represented by a gray line. The HCc3 dimer is represented by a green oval (left panel). HCc3 dimer is in equilibrium of free solution form and DNA-bound form (right panel). (b) A diagram showing the DNA looping and bundling mediated by HCc3 oligomerization at moderate protein concentrations. DNA-bound HCc3 dimers bridge two DNA strands through the interaction of N-terminal domain. Or, alternatively, a single HCc3 tetratmer bridge up two DNA strands. As a result a ‘seeding’ point is formed. Starting from this seeding point, newly jointed HCc3 dimers sit along side from this point and results in extension of oligomer along the two DNA strands, which is indicated by the arrows. Synergistic interactions provided by both the oligomerizing N-terminal domain and the DNA strands may be responsible for this cooperative process. Eventually, ‘tightened’ loop (i) or extend DNA bundling (ii) formed. (c) At high HCc3 concentration, a single DNA strand may acquire multiple molecules of HCc3 dimers. Intramolecular attraction (indicated by two-headed arrows) results in multiple ‘seeding’ points, and thus multiple looping and protein oligomer extension. Continuous formation of intramolecular ‘seeding’ points and oligomer extension result in a condensed, network-like foci. This process may occur at intermolecular level, and results in larger scale HCc3–DNA complex. Selected AFM images of corresponding HCc3 concentrations are placed beside the diagrammatic models for comparison.
At high protein concentrations corresponding to the formation of totally retarded aggregates in the EMSA, both HCc3 and C-ter formed ‘flower’ shaped complexes with DNA (Figure 5c, f). Numerous unresolvable DNA molecules were involved in each individual complex, but the C-ter complex was apparently higher in DNA content and larger in size (∼50% in diameter). A disintegrated C-ter complex could also be observed (Figure 5f; upper right corner). The lateral, extended linear structures in both cases are almost certainly naked DNA, since they have the height (∼0.6 nm) similar to the control sample of pure DNA (Figure 5d). However, it cannot be excluded that single protein molecules or dimers are bound to the DNA, but which are undistinguishable due to their small size. The lateral, linear structures of HCc3 complexes are arranged more like a DNA ‘network’ and are less dense than that of the C-ter complexes. HCc3 complexes are also characterized by the presence of highly condensed foci at the core of the complexes, ranging from 2 to 8 nm in height (Figure 5c). Some less integrated cores show a ‘branch’-like structure (Figure 6c). These foci are believed to be aggregates of HCc3 protein. Unlike the more discrete C-ter complexes, the HCc3 complexes tend to associate with each other, forming clustered complexes involving multiple foci.
Although these images are only snapshots of the HCc3–DNA complexes at fixed concentrations, they probably represent a continuum of dynamic HCc3–DNA structures that occur along with changes in HCc3 concentration.
DISCUSSION
Despite being eukaryotes according to molecular phylogenetic analysis (41), dinoflagellates are known to have no histones or the corresponding nucleosomes (42). Little is known about their organization of permanently condensed chromatin (43) or the associated histone-like proteins (5,44). Histones are the molecular solution to chromosomal DNA compaction in eukaryotes, and probably enabled the divergence of the eukaryotes from the prokaryotes. It is generally believed that the core histones evolved from archaeal histones (45,46) and that linker histones were of eubacterial origin (47). Amino acid sequence analysis revealed low level of homology between dinoflagellate HLPs and histone H1 (14,47). In this work, both in silico and in vitro analyses suggest that HCc3 and HU share structural homology. HCc3 is essentially a dimeric DNA-binding protein, with its DNA-binding domain located in the C-ter region, and the dimerization domain in the N-ter region. However, HCc3 is functionally distinct from bacterial HU, despite their structural similarities: first, HCc3 failed to complement the E. coli HU-deficient mutant; secondly, HCc3 did not promote DNA bending. The in silico derived wedge-like structure of HCc3 provides the only structural basis for a DNA-binding protein and suggests a phylogenetic relationship with HU, but may not be a critical factor in determining its specific biophysical or biochemical properties.
However, HCc3 is functionally more similar to another group of small DNA-binding proteins, the H-NS-like proteins (40,48,49), in spite of lacking significant sequence homology. At the structural level, all H-NS like proteins are similar: the proteins can be divided into a C-ter DNA-binding domain and an N-ter oligomerization domain, and H-NS exists as a dimer. The dimer has the ability to self-associate and to form large oligomers (48). A characteristic feature of dimeric H-NS is the presence of two DNA-binding domains that can potentially interact with two DNA duplexes simultaneously and results in ‘bridges’ between adjacent DNA duplexes, as demonstrated by AFM studies (40). The results of our study suggest that HCc3 fits these criteria as a DNA-bridging agent. H-NS was also reported to promote DNA circularization, which suggests DNA-bending activity (50), but no such DNA-bending event was observed with HCc3.
It has been proposed that cation-induced DNA condensation in aqueous solution results from cation cross-linking: electrostatic bridging of adjacent helices by trivalent or higher valence cations (51). The phenomenon of reentrant DNA aggregation/resolubilization with increasing concentrations of polycationic substances (i.e., polycationic salts, polyamines or basic proteins) has also been described (52–55). The addition of polycations, including polyamines and basic proteins, to a DNA solution first leads to the precipitation of the DNA. DNA precipitation reaches a maximum at a critical polycation concentration, and then further addition of polycations resolubilizes the DNA precipitate. It is the correlative distribution of polycations along the long polyanionic DNA that promotes the precipitation of DNA (56,57). However, ‘charge fractionization’ can become possible as a result of over-screening (or excess binding) of the DNA molecule to an excess of strongly correlated polycations (53,56,58). Charge inversion occurs subsequently and thus promotes resolubilization. The molecular flexibility of the polycation is an essential criterion for charge fractionalization since it allows a correlative redistribution and partial detachment of charges. As revealed by our EMSA experiment, the HCc3 C-ter domain, behaved like a typical polycation in terms of reentrant condensation and charge inversion of DNA. HCc3 does not follow this rule, which suggests that the presence of N-ter domain inhibited charge fractionalization by maintaining the rigidity of the protein molecule.
Many substances can promote ligase-mediated DNA concatenation while inhibiting circularization. These include the inorganic multivalent cobalt (III) hexammine (59), polyamines (60) and the DNA condensing protein, RecA (61). In a typical ligation reaction involving ligase, the collision of intermolecular DNA termini is one of the limiting factors, especially in a diluted DNA solution. RecA protein, a DNA bundling and condensing protein (62–64), brings DNA termini into proximity as a result of DNA bundling and this may account for its DNA concatenating effect, as well as its inhibition of circularization. This bundling-aided concatenation is further confirmed in our work using HCc3 and C-ter. The maximum DNA concatenation that was achieved is consistent with the observation of DNA-bundling complexes images in AFM examination at similar moderate protein concentrations. Apart from DNA bundling, DNA concatenation may also be induced by protein-mediated stiffening of the DNA. Binding of HCc3 or C-ter generates isolated filaments apparently stiffer than bare DNA or protein-induced condensed DNA (Figure 5). This could be a result of a repulsive effect due to the correlative distribution of proteins. The measurement of DNA persistence length would be an index of stiffness in the presence/absence of protein, may provide an alternative explanation for the inhibition of DNA circularization.
The formation of flower shape complexes at higher concentrations of HCc3 or the C-ter is another interesting aspect. In fact, the formation of such a complex is a general event that can be observed with condensed DNA induced by many substances including divalent cation (64), polycations (65), cationic silanes (66), cationic lipids (67,68) or DNA-binding proteins (38,69). Obviously, the foci structure largely contains the DNA-aggregating constituents, regardless of the mechanism of aggregation. The lateral structures represent naked DNA, and possibly due to the partial removal of the aggregation agents during the washing steps (70). Obviously, the HCc3 complexes are more stable and resistant to washing, as the structures remained relatively compact and connected (Figure 5c), whereas the C-ter complex had no discrete foci and collapsed structure was observed (Figure 5f, upper-right corner). In fact, HCc3 showed a slightly higher affinity than the C-ter toward DNA in the DNA-cellulose binding assay (Figure 2a).
We can conclude from our results that HCc3 self-aggregation, DNA bridging and condensation are all processes correlated with the nature of the N-ter domain. First, it maintains the rigidity of the HCc3 molecule to avoid charge inversion, which causes resolubilization at high protein concentrations; second, it provides a bridging mechanism to hold together DNA strands, which means the protein becomes more efficient at DNA aggregation; third, it allows the HCc3 protein to work in a cooperative way by switching the dimeric molecules with moderate DNA affinity per se into stronger binding oligomeric clusters that resist disintegration. The oligmeric interaction may involve disulfide linkages formed by a Cys residue in the N-ter domain (helix α1) (5). Consistent with this, extraction of HCc proteins from the organism is more efficient with the addition of sulfhydryl reducing agents, such as β-mercaptoethanol (unpublished data).
A model is proposed for DNA condensation induced by HCc3 (Figure 6). HCc3 dimers (or perhaps tetramers) are confined in solution by anchoring on a DNA strand. The first DNA bridge is formed through the interaction between two bound HCc3 dimers. This first bridge leads to an increased chance of further DNA bridging in the neighboring and intervening sections, by reducing the ‘search space’ for additionally bound HCc3 proteins (i.e., cooperatively), and therefore results in DNA bundling. Longer DNA strands recruit more HCc3 dimers and thus are condensed more efficiently than the shorter ones. At high-HCc3 concentration, the local protein concentration is increased and mutual interactions take place in multiple dimensions between HCc3 molecules bound on the same DNA strand or nearby ones. This results in the secondary level of condensation (‘flower’ shape complex) with DNA loops and condensed foci. This simple mechanism for DNA compaction through DNA-bridging has been proposed for a number of other proteins, including H-NS (39), viral integration factor BAF (71), bacterial protein LrpC (72) and Dps (38). Interestingly, recent findings demonstrated that an HU mutant acquired DNA condensation activity as a result of gaining the oligomerization activity (73). This proposed mechanism is clearly distinct from other DNA-associating proteins, such as HU (24), SMC (74), IHF (75) and FIS (76), which are proposed to function through a wrapping mechanism (48).
We postulate that, through the regulation of HCc protein concentration and/or localization in the nucleus, the topological context required for gene transcription of ‘active chromatin’ during the dinoflagellate cell cycle is regulated. HCc may organize extra-chromosomal DNA loops into different topologies depending on its distribution and local concentration. At moderate concentration, HCc3 can potentially induce DNA looping with its DNA-bundling action. In a similar way, HCc3 may play an essential role in regulating gene transcription by assisting the looping out of DNA from the condensed dinoflagellate chromosome, to allow access to the transcription factory, i.e., nucleolus. During interphase, and within the NOR region, the HCc protein assists the looping out of DNA from the condensed chromosome for the assessibility of the transcriptional machinery (15). DNA looping is an essential aspect of gene transcriptional control, in both the prokaryotes and eukaryotes (77,78). DNA looping facilitates remote control of gene transcription by trans-acting enhancers. Also, there is increasing evidence that these dynamic interactions regulate the repositioning of genes to foci of active transcription within the nucleus (78). Alternatively, released DNA can be efficiently condensed with highly concentrated HCc proteins and thus result in fast retreat of the extended DNA loops. This is consistent with the previous observations of a homogeneous distribution of HCc proteins throughout each dividing chromosome in mitotic cells (28). This is possibly for the preparation of DNA chromosome segregation.
To execute the dual function of HCc proteins, a regulation system is essential, at least at the post-translational level, possibly by acetylation. As in the case of histones, acetylation removes positive charges, thereby reducing the affinity between the histones and the DNA (79). As in the case of bacterial HU, HCc proteins may also interact functionally with topoisomerases (80–82), which are also involved in DNA replication and condensation, and is persistently expressed throughout the cell cycle of C.cohnii (83). In the presence of topoisomerase II, local topological changes of DNA filaments can possibly be modulated through the assistance of HCc proteins, in a synergistic and cooperative manner.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
The present study was partly supported by a CERG grant (HKUST6096/02M)) from the Research Grant Council of Hong Kong and Grant HIA05/06.SC01 from University Grant Council to JTYW. We would like to thank Dr Xhie, Jie (Materials Characterization and Preparation Facility, The Hong Kong University of Science and Technology) and Mr Yeung, Man-nin (The Hong Kong Polytechnic University Materials Research Centre) for their help with AFM analyses. Thanks are extended to Mike Bennett for reading several versions of the manuscript. Funding to pay the Open Access publication charges for this article was provided by Grant HIA05/06.SC01.
Conflict of interest statement. None declared.
REFERENCES
- 1.Kellenberger E, Arnold-Schulz-Gahmen B. Chromatins of low-protein content: special features of their compaction and condensation. FEMS Microbiol. Lett. 1992;79:361–370. doi: 10.1111/j.1574-6968.1992.tb14064.x. [DOI] [PubMed] [Google Scholar]
- 2.Holck A, Lossius I, Aasland R, Haarr L, Kleppe K. DNA- and RNA-binding proteins of chromatin from Escherichia coli. Biochim. Biophys. Acta. 1987;908:188–199. doi: 10.1016/0167-4781(87)90058-3. [DOI] [PubMed] [Google Scholar]
- 3.Rizzo PJ, Jones M, Ray SM. Isolation and properties of isolated nuclei from the florida red tide dinoflagellate Gymnodinium breve (davis) J. Protozool. 1982;29:217–222. doi: 10.1111/j.1550-7408.1982.tb04014.x. [DOI] [PubMed] [Google Scholar]
- 4.Bodansky S, Mintz LB, Holmes DS. The mesokaryote Gyrodinium cohnii lacks nucleosomes. Biochem. Biophys. Res. Commun. 1979;88:1329–1336. doi: 10.1016/0006-291x(79)91126-4. [DOI] [PubMed] [Google Scholar]
- 5.Vernet G, Sala-Rovira M, Maeder M, Jacques F, Herzog M. Basic nuclear proteins of the histone-less eukaryote Crypthecodinium cohnii (pyrrhophyta): two-dimensional electrophoresis and DNA-binding properties. Biochim. Biophys. Acta. 1990;1048:281–289. doi: 10.1016/0167-4781(90)90068-d. [DOI] [PubMed] [Google Scholar]
- 6.de Haller G, Kellenberger E. Etude au microscope electronique des plasmas contenant de l’acide deoxyribonucleique III. Variations ultra-structurales des chromosomes d’amphidinium. J. Microsc. 1964;3:627–642. [Google Scholar]
- 7.Dodge JD. The chromosomes of dinoflagellates. Int. Rev. Cytol. 1985;94:5–20. [Google Scholar]
- 8.Kellenberger E. Organization of the genetic material of phage, bacteria and dinoflagellates. In: Today G, editor. Proceedings of the Eleventh International Congress of Genetics; Oxford, The Hague: Pergamon Press; 1964. pp. 309–321. [Google Scholar]
- 9.Gautier A, Michel-Salamin L, Tosi-Couture E, McDowall AW, Dubochet J. Electron microscopy of the chromosomes of dinoflagellates in situ: confirmation of Bouligand's liquid crystal hypothesis. J. Ultrastruct. Mol. Struct. Res. 1986;97:10–30. [Google Scholar]
- 10.Bouligand Y, Soyer MO, Puiseux-Dao S. The fibrillary structure and orientation of chromosomes in dinoflagellata. Chromosoma. 1968;24:251–287. doi: 10.1007/BF00336195. [DOI] [PubMed] [Google Scholar]
- 11.Rill RL, Livolant F, Aldrich HC, Davidson MW. Electron microscopy of liquid crystalline DNA: direct evidence for cholesteric-like organization of DNA in dinoflagellate chromosomes. Chromosoma. 1989;98:280–286. doi: 10.1007/BF00327314. [DOI] [PubMed] [Google Scholar]
- 12.Wong JT, Kwok AC. Proliferation of dinoflagellates: blooming or bleaching. Bioessays. 2005;27:730–740. doi: 10.1002/bies.20250. [DOI] [PubMed] [Google Scholar]
- 13.Livolant F, Leforestier A. Condensed phases of DNA: structures and phase transitions. Prog. Polym. Sci. 1996;21:1115–1164. [Google Scholar]
- 14.Wong JT, New DC, Wong JC, Hung VK. Histone-like proteins of the dinoflagellate Crypthecodinium cohnii have homologies to bacterial DNA-binding proteins. Eukaryot. Cell. 2003;2:646–650. doi: 10.1128/EC.2.3.646-650.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sala-Rovira M, Geraud ML, Caput D, Jacques F, Soyer-Gobillard MO, Vernet G, Herzog M. Molecular cloning and immunolocalization of two variants of the major basic nuclear protein (HCC) from the histone-less eukaryote Crypthecodinium cohnii (pyrrhophyta) Chromosoma. 1991;100:510–518. doi: 10.1007/BF00352201. [DOI] [PubMed] [Google Scholar]
- 16.Chan YH, Kwok AC, Tsang JS, Wong JTY. Alveolata histone-like proteins have different evolutionary origins. J. Evolution. Biol. 2006;19:1717–1721. doi: 10.1111/j.1420-9101.2006.01089.x. [DOI] [PubMed] [Google Scholar]
- 17.Broyles SS, Pettijohn DE. Interaction of the Escherichia coli HU protein with DNA. Evidence for formation of nucleosome-like structures with altered DNA helical pitch. J. Mol. Biol. 1986;187:47–60. doi: 10.1016/0022-2836(86)90405-5. [DOI] [PubMed] [Google Scholar]
- 18.Rouviere-Yaniv J, Yaniv M, Germond JE. E. coli DNA binding protein HU forms nucleosomelike structure with circular double-stranded DNA. Cell. 1979;17:265–274. doi: 10.1016/0092-8674(79)90152-1. [DOI] [PubMed] [Google Scholar]
- 19.Hodges-Garcia Y, Hagerman P, Pettijohn D. DNA ring closure mediated by protein HU. J. Biol. Chem. 1989;264:14621–14623. [PubMed] [Google Scholar]
- 20.Dixon NE, Kornberg A. Protein HU in the enzymatic replication of the chromosomal origin of Escherichia coli. Proc. Natl. Acad. Sci. USA. 1984;81:424–428. doi: 10.1073/pnas.81.2.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson RC, Bruist MF, Simon MI. Host protein requirements for in vitro site-specific DNA inversion. Cell. 1986;46:531–539. doi: 10.1016/0092-8674(86)90878-0. [DOI] [PubMed] [Google Scholar]
- 22.Lavoie BD, Chaconas G. A second high affinity HU binding site in the phage Mu transpososome. J. Biol. Chem. 1994;269:15571–15576. [PubMed] [Google Scholar]
- 23.Flashner Y, Gralla JD. DNA dynamic flexibility and protein recognition: differential stimulation by bacterial histone-like protein HU. Cell. 1988;54:713–721. doi: 10.1016/s0092-8674(88)80016-3. [DOI] [PubMed] [Google Scholar]
- 24.van Noort J, Verbrugge S, Goosen N, Dekker C, Dame RT. Dual architectural roles of HU: formation of flexible hinges and rigid filaments. Proc. Natl. Acad. Sci. USA. 2004;101:6969–6974. doi: 10.1073/pnas.0308230101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Drlica K, Rouviere-Yaniv J. Histone-like proteins of bacteria. Microbiol. Rev. 1987;51:301–319. doi: 10.1128/mr.51.3.301-319.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vis H, Boelens R, Mariani M, Stroop R, Vorgias CE, Wilson KS, Kaptein R. 1H, 13C, and 15N resonance assignments and secondary structure analysis of the HU protein from Bacillus stearothermophilus using two- and three-dimensional double- and triple-resonance heteronuclear magnetic resonance spectroscopy. Biochemistry. 1994;33:14858–14870. doi: 10.1021/bi00253a025. [DOI] [PubMed] [Google Scholar]
- 27.White SW, Wilson KS, Appelt K, Tanaka I. The high-resolution structure of DNA-binding protein HU from Bacillus stearothermophilus. Acta Cryst. 1999;D55:801–809. doi: 10.1107/s0907444999000578. [DOI] [PubMed] [Google Scholar]
- 28.Geraud M-L, Sala-Rovira M, Herzog M, Soyer-Gobillard M-O. Immunocytochemical localization of the DNA-binding protein HCC during the cell cycle of the histone-less dinoflagellate protoctista Crypthecodinium cohnii b. Biol. Cell. 1991;71:123–134. [Google Scholar]
- 29.Chudnovsky Y, Li JF, Rizzo PJ, Hastings JW, Fagan TF. Cloning, expression, and characterization of a histone-like protein from the marine dinoflagellate Lingulodinium polyedrum (Dinophyceae) J. Phycol. 2002;38:543–550. [Google Scholar]
- 30.Payet D. Use of DNA microcircles in protein-DNA binding studies. In: Travers A, Buckle M, editors. DNA-Protein Interactions: A Practical Approach. Hong Kong: Oxford University Press; 2000. p. 398. [Google Scholar]
- 31.Lyubchenko YL, Gall AA, Shlyakhtenko LS, Harrington RE, Jacobs BL, Oden PI, Lindsay SM. Atomic force microscopy imaging of double stranded DNA and RNA. J. Biomol. Struct. Dyn. 1992;10:589–606. doi: 10.1080/07391102.1992.10508670. [DOI] [PubMed] [Google Scholar]
- 32.Rippe K, Mucke N, Langowski J. Superhelix dimensions of a 1868 base pair plasmid determined by scanning force microscopy in air and in aqueous solution. Nucleic Acids Res. 1997;25:1736–1744. doi: 10.1093/nar/25.9.1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shanado Y, Kato J, Ikeda H. Escherichia coli HU protein suppresses DNA-gyrase-mediated illegitimate recombination and SOS induction. Genes Cells. 1998;3:511–520. doi: 10.1046/j.1365-2443.1998.00208.x. [DOI] [PubMed] [Google Scholar]
- 34.Karplus K, Hu B. Evaluation of protein multiple alignments by SAM-T99 using the balibase multiple alignment test set. Bioinformat. 2001;17:713–720. doi: 10.1093/bioinformatics/17.8.713. [DOI] [PubMed] [Google Scholar]
- 35.Chou PY, Fasman GD. Prediction of the secondary structure of proteins from their amino acid sequence. Adv. Enzymol. Relat. Areas Mol. Biol. 1978;47:45–148. doi: 10.1002/9780470122921.ch2. [DOI] [PubMed] [Google Scholar]
- 36.Levitt M. Conformational preferences of amino acids in globular proteins. Biochemistry. 1978;17:4277–4285. doi: 10.1021/bi00613a026. [DOI] [PubMed] [Google Scholar]
- 37.Deleage G, Roux B. An algorithm for protein secondary structure prediction based on class prediction. Protein Eng. 1987;1:289–294. doi: 10.1093/protein/1.4.289. [DOI] [PubMed] [Google Scholar]
- 38.Wada M, Kano Y, Ogawa T, Okazaki T, Imamoto F. Construction and characterization of the deletion mutant of hupA and hupB genes in Escherichia coli. J. Mol. Biol. 1988;204:581–591. doi: 10.1016/0022-2836(88)90357-9. [DOI] [PubMed] [Google Scholar]
- 39.Ceci P, Cellai S, Falvo E, Rivetti C, Rossi GL, Chiancone E. DNA condensation and self-aggregation of Escherichia coli Dps are coupled phenomena related to the properties of the N-terminus. Nucleic Acids Res. 2004;32:5935–5944. doi: 10.1093/nar/gkh915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dame RT, Wyman C, Goosen N. H-NS mediated compaction of DNA visualised by atomic force microscopy. Nucleic Acids Res. 2000;28:3504–3510. doi: 10.1093/nar/28.18.3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hinnebusch AG, Klotz LC, Blanken RL, Loeblich A.R., III. An evaluation of the phylogenetic position of the dinoflagellate Crypthecodinium cohnii based on 5S rRNA characterization. J. Mol. Evol. 1981;17:334–337. doi: 10.1007/BF01734355. [DOI] [PubMed] [Google Scholar]
- 42.Rizzo PJ. Comparative aspects of basic chromatin proteins in dinoflagellates. Biosystems. 1981;14:433–443. doi: 10.1016/0303-2647(81)90048-4. [DOI] [PubMed] [Google Scholar]
- 43.Soyer MO, Haapala OK. Structural changes of dinoflagellate chromosomes by pronase and ribonuclease. Chromosoma. 1974;47:179–192. doi: 10.1007/BF00331805. [DOI] [PubMed] [Google Scholar]
- 44.Rizzo PJ, Choi J, Morris RL. The major histone-like protein from the nonphotosynthetic dinoflagellate Crypthecodinium cohnii (pyrrophyta) is present in stationary phase cultures. J. Phycol. 1984;20:95–100. [Google Scholar]
- 45.Pereira SL, Reeve JN. Archaeal nucleosome positioning sequence from Methanothermus fervidus. J. Mol. Biol. 1999;289:675–681. doi: 10.1006/jmbi.1999.2812. [DOI] [PubMed] [Google Scholar]
- 46.Sandman K, Reeve JN. Structure and functional relationships of archaeal and eukaryal histones and nucleosomes. Arch. Microbiol. 2000;173:165–169. doi: 10.1007/s002039900122. [DOI] [PubMed] [Google Scholar]
- 47.Kasinsky HE, Lewis JD, Dacks JB, Ausio J. Origin of H1 linker histones. FASEB J. 2001;15:34–42. doi: 10.1096/fj.00-0237rev. [DOI] [PubMed] [Google Scholar]
- 48.Dame RT, Luijsterburg MS, Krin E, Bertin PN, Wagner R, Wuite GJ. DNA bridging: a property shared among H-NS-like proteins. J Bacteriol. 2005;187:1845–1848. doi: 10.1128/JB.187.5.1845-1848.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Luijsterburg MS, Noom MC, Wuite GJ, Dame RT. The architectural role of nucleoid-associated proteins in the organization of bacterial chromatin: a molecular perspective. J. Struct. Biol. 2006;156:262–272. doi: 10.1016/j.jsb.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 50.Spurio R, Falconi M, Brandi A, Pon CL, Gualerzi CO. The oligomeric structure of nucleoid protein H-NS is necessary for recognition of intrinsically curved DNA and for DNA bending. EMBO J. 1997;16:1795–1805. doi: 10.1093/emboj/16.7.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Widom J, Baldwin RL. Cation-induced toroidal condensation of DNA studies with Co3+(NH3)6. J. Mol. Biol. 1980;144:431–453. doi: 10.1016/0022-2836(80)90330-7. [DOI] [PubMed] [Google Scholar]
- 52.Pelta J, Livolant F, Sikorav JL. DNA aggregation induced by polyamines and cobalthexamine. J. Biol. Chem. 1996;271:5656–5662. doi: 10.1074/jbc.271.10.5656. [DOI] [PubMed] [Google Scholar]
- 53.Raspaud E, Pelta J, de Frutos M, Livolant F. Solubility and charge inversion of complexes of DNA and basic proteins. Phys. Rev. Lett. 2006;97:068103. doi: 10.1103/PhysRevLett.97.068103. [DOI] [PubMed] [Google Scholar]
- 54.Raspaud E, Olvera de la Cruz M, Sikorav JL, Livolant F. Precipitation of DNA by polyamines: a polyelectrolyte behavior. Biophys. J. 1998;74:381–393. doi: 10.1016/S0006-3495(98)77795-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nguyen TT, Rouzina I, Shklovskii BI. Reentrant condensation of DNA induced by multivalent counterions. J. Chem. Phys. 2000;112:2562–2568. [Google Scholar]
- 56.Grosberg AY, Nguyen TT, Shklovskii BI. Colloquium: the physics of charge inversion in chemical and biological systems. Rev. Mod. Phys. 2002;74:329–345. [Google Scholar]
- 57.Shklovskii BI. Wigner crystal model of counterion induced bundle formation of rodlike polyelectrolytes. Phys. Rev. Lett. 1999;82:3268–3271. [Google Scholar]
- 58.Zhang R, Shklovskii BI. Phase diagram of solution of oppositely charged polyelectrolytes. Physica A. 2005;352:216–238. [Google Scholar]
- 59.Rusche JR, Howard-Flanders P. Hexamine cobalt chloride promotes intermolecular ligation of blunt end DNA fragments by T4 DNA ligase. Nucleic Acids Res. 1985;13:1997–2008. doi: 10.1093/nar/13.6.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Takahashi M, Uchida T. Thermophilic HB8 DNA ligase: effects of polyethylene glycols and polyamines on blunt-end ligation of DNA. J. Biochem. 1986;100:123–131. doi: 10.1093/oxfordjournals.jbchem.a121684. [DOI] [PubMed] [Google Scholar]
- 61.Rusche JR, Konigsberg W, Howard-Flanders P. Isolation of altered RecA polypeptides and interaction with ATP and DNA. J. Biol. Chem. 1985;260:949–955. [PubMed] [Google Scholar]
- 62.West SC, Cassuto E, Mursalim J, Howard-Flanders P. Recognition of duplex DNA containing single-stranded regions by RecA protein. Proc. Natl. Acad. Sci. USA. 1980;77:2569–2573. doi: 10.1073/pnas.77.5.2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shi W-X, Larson RG. Atomic force microscopic study of aggregation of RecA-DNA nucleoprotein filaments into left-handed supercoiled bundles. Nano Lett. 2005;5:2476–2481. doi: 10.1021/nl051783v. [DOI] [PubMed] [Google Scholar]
- 64.Levin-Zaidman S, Frenkiel-Krispin D, Shimoni E, Sabanay I, Wolf SG, Minsky A. Ordered intracellular RecA-DNA assemblies: a potential site of in vivo RecA -mediated activities. Proc. Natl. Acad. Sci. USA. 2000;97:6791–6796. doi: 10.1073/pnas.090532397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fang Y, Spisz TS, Hoh JH. Ethanol-induced structural transitions of DNA on mica. Nucleic Acids Res. 1999;27:1943–1949. doi: 10.1093/nar/27.8.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hansma HG, Golan R, Hsieh W, Lollo CP, Mullen-Ley P, Kwoh D. DNA condensation for gene therapy as monitored by atomic force microscopy. Nucleic Acids Res. 1998;26:2481–2487. doi: 10.1093/nar/26.10.2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fang Y, Hoh JH. Cationic silanes stabilize intermediates in DNA condensation. FEBS Lett. 1999;459:173–176. doi: 10.1016/s0014-5793(99)01237-5. [DOI] [PubMed] [Google Scholar]
- 68.Dunlap DD, Maggi A, Soria MR, Monaco L. Nanoscopic structure of DNA condensed for gene delivery. Nucleic Acids Res. 1997;25:3095–3101. doi: 10.1093/nar/25.15.3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gershon H, Ghirlando R, Guttman SB, Minsky A. Mode of formation and structural features of DNA-cationic liposome complexes used for transfection. Biochemistry. 1993;32:7143–7151. doi: 10.1021/bi00079a011. [DOI] [PubMed] [Google Scholar]
- 70.Montigny WJ, Houchens CR, Illenye S, Gilbert J, Coonrod E, Chang YC, Heintz NH. Condensation by DNA looping facilitates transfer of large DNA molecules into mammalian cells. Nucleic Acids Res. 2001;29:1982–1988. doi: 10.1093/nar/29.9.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hansma HG, Bezanilla M, Zenhausern F, Adrian M, Sinsheimer RL. Atomic force microscopy of DNA in aqueous solutions. Nucleic Acids Res. 1993;21:505–512. doi: 10.1093/nar/21.3.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lee MS, Craigie R. A previously unidentified host protein protects retroviral DNA from autointegration. Proc. Natl. Acad. Sci. USA. 1998;95:1528–1533. doi: 10.1073/pnas.95.4.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tapias A, Lopez G, Ayora S. Bacillus subtilis LrpC is a sequence-independent DNA-binding and DNA-bending protein which bridges DNA. Nucleic Acids Res. 2000;28:552–559. doi: 10.1093/nar/28.2.552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kar S, Choi EJ, Guo F, Dimitriadis EK, Kotova SL, Adhya S. Right-handed DNA supercoiling by an octameric form of histone-like protein HU: Modulation of cellular transcription. J. Biol. Chem. 2006;281:40144–40153. doi: 10.1074/jbc.M605576200. [DOI] [PubMed] [Google Scholar]
- 75.Mascarenhas J, Volkov AV, Rinn C, Schiener J, Guckenberger R, Graumann PL. Dynamic assembly, localization and proteolysis of the Bacillus subtilis SMC complex. BMC. Cell Biol. 2005;6:28. doi: 10.1186/1471-2121-6-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dame RT, van Mameren J, Luijsterburg MS, Mysiak ME, Janicijevic A, Pazdzior G, van der Vliet PC, Wyman C, Wuite GJ. Analysis of scanning force microscopy images of protein-induced DNA bending using simulations. Nucleic Acids Res. 2005;33:e68. doi: 10.1093/nar/gni073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schneider R, Lurz R, Luder G, Tolksdorf C, Travers A, Muskhelishvili G. An architectural role of the Escherichia coli chromatin protein Fis in organising DNA. Nucleic Acids Res. 2001;29:5107–5114. doi: 10.1093/nar/29.24.5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Matthews KS. DNA looping. Microbiol. Rev. 1992;56:123–136. doi: 10.1128/mr.56.1.123-136.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.West AG, Fraser P. Remote control of gene transcription. Hum. Mol. Genet. 2005;14:R101–R111. doi: 10.1093/hmg/ddi104. [DOI] [PubMed] [Google Scholar]
- 80.Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes Develop. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 81.Malik M, Bensaid A, Rouviere-Yaniv J, Drlica K. Histone-like protein HU and bacterial DNA topology: suppression of an HU deficiency by gyrase mutations. J. Mol. Biol. 1996;256:66–76. doi: 10.1006/jmbi.1996.0068. [DOI] [PubMed] [Google Scholar]
- 82.Bensaid A, Almeida A, Drlica K, Rouviere-Yaniv J. Cross-talk between topoisomerase I and HU in Escherichia coli. J. Mol. Biol. 1996;256:292–300. doi: 10.1006/jmbi.1996.0086. [DOI] [PubMed] [Google Scholar]
- 83.Mak CKM, Hung VKL, Wong JTY. Type II topoisomerase activities in both G1 and G2/M phases of the dinoflagellate cell cycle. Chromosoma. 2005;114:420–431. doi: 10.1007/s00412-005-0027-3. [DOI] [PubMed] [Google Scholar]
- 84.Notredame C, Higgins DG, Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.