Abstract
Phosphodiesterase 11A (PDE11A) is a recently identified family of cAMP and cGMP hydrolyzing enzymes. Thus far, a single splice variant designated as PDE11A1 has been reported. In this study, we identify and characterize two additional splice variants of PDE11A, PDE11A2 and PDE11A3. The full-length cDNAs are 2,141 bp for PDE11A2 and 2205 bp for PDE11A3. The ORF of PDE11A2 predicts a protein of 576 aa with a molecular mass of 65.8 kDa. The ORF of PDE11A3 predicts a protein of 684 aa with a molecular mass of 78.1 kDa. Comparison of the PDE11A2 sequence with that of PDE11A1 indicates an additional 86 aa at the N terminus of PDE11A2. Part of this sequence extends the potential cGMP binding region (GAF domain) present in PDE11A1. Compared with PDE11A2, PDE11A3 has an additional 108 N-terminal amino acids. Sequence analysis of PDE11A3 indicates the presence of another GAF domain in this region. This diversification of regulatory sequences in the N-terminal region of PDE11A splice variants suggests the interesting possibility of differential regulation of these enzymes. Recombinant PDE11A2 and -A3 proteins expressed in the Baculovirus expression system have the ability to hydrolyze both cAMP and cGMP. The Km values for cAMP hydrolysis are 3.3 μM and 5.7 μM for PDE11A2 and PDE11A3, respectively. The Km values for cGMP hydrolysis are 3.7 μM and 4.2 μM for PDE11A2 and PDE11A3, respectively. Both PDEs showed a Vmax ratio for cAMP/cGMP of approximately 1.0. PDE11A2 is sensitive to dipyridamole, with an IC50 of 1.8 μM, and to zaprinast, with an IC50 of 28 μM. PDE11A3 demonstrated similar pattern of inhibitor sensitivity with IC50 values of 0.82 and 5 μM for dipyridamole and zaprinast, respectively.
Cyclic nucleotide phosphodiesterases (PDEs) regulate the intracellular levels of cAMP and cGMP. Therefore, PDEs contribute to the regulation of multiple physiological processes, including vascular resistance, cardiac output, visceral motility, immune response (1), inflammation (2), neuroplasticity, vision (3), and reproduction (4). As the involvement of cAMP in the regulation of these systems has been known for many years, it is not surprising that cAMP-specific PDEs have been found to play a critical role in their regulation. Examples include PDE7A and PDE4, which have been shown to be important players in the immune response and inflammation, respectively (1, 5).
Recently, nitric oxide (NO) has attracted much attention as a critical mediator of several physiological responses, including the phenomena listed above. Interestingly, one of the widely recognized intracellular events triggered by NO is the activation of guanylyl cyclase leading to increased cGMP levels. Because cGMP is degraded by cGMP-hydrolyzing PDEs, these enzymes are thought to play an important role in the regulation of responsiveness to NO. For example, PDE5, a cGMP-specific phosphodiesterase, is important for control of NO-mediated penile erection (6, 7). Other cGMP-degrading PDEs include PDE2, -6, -9, and -10. Although the roles of PDE2, -9, and -10 remain obscure, PDE6 is a critical component of phototransduction in the retina (8).
PDEs are not only an off switch for cGMP-mediated signaling, but they also can be the effectors of this second messenger. For example, PDE2, -5, -6, and -10 contain regulatory cGMP binding sites (9), by which cGMP can modify their activity. This provides a potential autoregulatory feedback mechanism for the cGMP circuit and also affects cAMP signaling as PDE2 and -10 can hydrolyze cAMP.
The superfamily of cGMP PDEs has been recently extended by the identification of a family designated PDE11 (10). Thus far, one gene of this family has been cloned, named PDE11A. PDE11A is a cAMP- and cGMP-hydrolyzing enzyme. It has a potential regulatory cGMP binding site (GAF domain after the proteins where it was found: cGMP-binding and stimulated PDEs, Anabaena Adenylyl cyclases and Escherichia coli FhlA protein) (11) in its N-terminal region. Because Northern blotting of PDE11A revealed the existence of multiple bands, we have hypothesized that PDE11A is regulated by alternative mRNA splicing (10). In concert with this idea, immunoblot analysis of PDE11A showed three distinct protein bands of 56, 65, and 78 kDa in human skeletal muscle (10). We have designated them as PDE11A1, -A2, and -A3, respectively. In this study, we report the cloning of the full-length cDNAs for PDE11A2 and -A3 as well as the characterization of the recombinant proteins encoded by these cDNAs. In comparison to PDE11A1, the A2 and A3 variants have N-terminal regions that are extended by 86 and 194 aa for PDE11A2 and PDE11A3, respectively. As the catalytic, C-terminal regions of all three variants are identical, Km values for cAMP and cGMP hydrolysis are also similar. Therefore, it can be speculated that the diverse N-terminal region of PDE11A isoforms may confer differential regulation of the very similar catalytic activity. In concert with this hypothesis, PDE11A3 has another potential GAF domain not present in either the A1 or A2 variants of PDE11A.
Materials and Methods
Expressed Sequence Tag (EST) Database Search.
The EST database (dBEST) was searched with the published amino acid sequences of mammalian PDEs. Members of all 10 identified PDE families were used for this search, which was performed by using the program blast (Basic Local Alignment Search Tool). All clones identified in the EST database then were used as a query to search GenBank and determine whether they represented known or unknown PDEs.
Primers.
All primers were designed by using the program amplify (freeware by William Engels, University of Wisconsin, Madison). The primer sequences were as follows: 11 Strt1, GAAAAGTCCACTGTGTGTTGGGAATAG; 11 End3, GTAAGTAGGCCGACTGTCCACAGG; PDE11 cDNA synthesis primer, AGATATCTGGTCTGCCTCTGC; 11 End3.2, AGGTCAGTCTCTTCTTCAAAGAGG; and PDE11screen, ACTCCTAGAGGACATCGAATCACCAGTGG.
Cloning of the Full-Length cDNAs for PDE11A2 and -A3.
To obtain the full-length cDNAs of PDE11A2 and -A3, the rapid amplification of cDNA ends (RACE) technique was used as described (12). A Marathon Ready cDNA library from human testis (CLONTECH) was used as a template for PDE11A2 amplifications. PDE11A3 RACE reactions were carried out by using cDNA extended from poly(A)+ RNA of human testis. Advantage Polymerase PCR mix was purchased from CLONTECH. Reactions were carried out as described (12). All PCR products were subcloned into the TA Topo pCRII vector (Invitrogen) according to the manual (Topo TA Cloning, version E). The oligonucleotide probe for minilibrary screening was end labeled with [α32P]ATP (Amersham Pharmacia) by using the 5′ DNA Terminus Labeling System (Life Technologies, Grand Island, NY) according to the manufacturer's protocol. Plasmid DNA was prepared by using the SNAP kit (Invitrogen).
DNA Sequencing.
For sequencing, plasmid DNA was prepared taking advantage of the SNAP kit (Invitrogen). Sequencing was done with an ABI PRISM dye terminator cycle sequencing kit (Perkin–Elmer). Sequencing products were purified by using Centri-sep columns (Princeton Separations, Adelphia, NJ). Sequences were assembled with the program sequencher 3.0 (Gene Codes, Ann Arbor, MI).
The ORFs of the PDE11A2 and -A3 cDNAs were subcloned into the pFastBac1 baculovirus vector and expressed in SF9 cells as described (10, 13).
Kinetics and Inhibitor Studies.
PDE assays were carried out according to the method of Beavo and Hansen (14). The reactions were performed in a buffer containing 40 mM Mops (pH 7.5), 0.8 mM EGTA, 15 mM Mg acetate, 0.2 mg/ml BSA, and 50,000 cpm of [3H]cAMP/cGMP. Final reaction volume was 250 μl. The amount of enzyme was adjusted to a level that hydrolyzed no more than 30% of the substrate even at the lowest substrate concentration tested. For kinetic studies, cAMP/cGMP concentrations varied between 0.1 and 62.5 μM. For inhibitor studies involving PDE11A2 and PDE11A3, 12.5 nM cAMP was used. At this concentration of substrate, the IC50 value approximates the Ki. All kinetic and inhibitor studies were done in triplicate and repeated three times with homogenates from two independent batches of PDE11A2 or -A3. PDE activity was defined as the difference between activity in Sf-9 cells infected with PDE11A2 or -A3 recombinant viruses (harvested 3 days postinfection) and uninfected Sf-9 cells. 3-Isobutyl-1-methylxanthine (IBMX), erythro-9-[3-(2-hydroxynonyl)]adenine, and dipyridamole were obtained from Sigma. Zaprinast was a gift from May & Baker (Dagenham, U.K.). Rolipram was obtained from Biomol (Plymouth Meeting, PA). SCH51866 was a gift from Schering–Plough Research Institute, and sildenafil was a gift from Pfizer Global Research and Development, (Sandwich, U.K.). Enoximone was a gift from Merrell Dow Research Institute, Cincinnati.
Results
Cloning and Sequencing of PDE11A2 and -A3.
Recently, a PDE family, PDE11, has been discovered. The first identified member of this family, PDE11A, seemed to occur in several splice variant forms as shown by both Northern and Western blotting. However, only one cDNA, encoding the splice variant PDE11A1, has been isolated. Alternative splicing might lead to the generation of proteins with distinct properties. Therefore, it is crucial for understanding the role(s) of these gene products to identify and characterize all of its splice variants. For this reason, we set out to clone and study other splice variants of PDE11A.
Cloning and Sequencing of PDE11A2.
The EST database, dBEST, was searched with all published amino acid sequences representing the mammalian PDE superfamily. Using the amino acid sequence of bovine PDE5 as a query, an EST (clone AI025081) that was derived from a human testis cDNA library was identified. A GenBank search revealed that clone AI025081 was not identical to any of the known PDEs, including the recently identified PDE11A sequence derived from skeletal muscle. To obtain the full-length cDNA of PDE11A from testis, we designed primers based on the nucleotide sequence of AI025081 for nested 5′ and 3′ RACE reactions. The template for these reactions was a cDNA library prepared from a human testis. Both 5′ and 3′ RACE reactions resulted in multiple clones, which were sequenced. Alignment of the nucleotide sequences with the EST AI025081 produced a contiguous sequence. This sequence appeared to include the full 5′ end of the translated coding region, as indicated by the presence of four in-frame stop codons 5′ to the start methionine. It also encompassed the 3′ end of the coding region as revealed by the presence of the poly(A) tail.
To verify the cloning results obtained by RACE, we isolated a single clone containing the full-length cDNA of PDE11A2. For this purpose, we designed primers 11Strt1 (mapping at the 5′ end of the putative cDNA) and 11End3 [mapping just 5′ from the poly(A) tail]. Three independent PCRs were performed by using the Marathon Ready cDNA library from a human testis as a template. The resulting clones contained a cDNA with a sequence identical to the contiguous sequence obtained by alignment of the RACE clones, indicating that a full-length cDNA for this gene had been isolated.
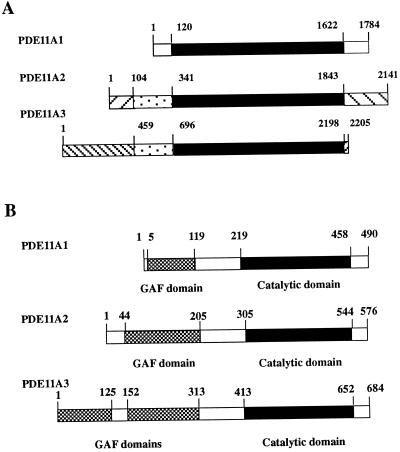
DNA sequence analysis revealed that this clone contains an ORF that is identical with PDE11A1, with the exception of an N-terminal stretch of 86 aa not present in PDE11A1. In addition, there were differences detected in the 3′ untranslated regions (UTR) of both cDNAs. Altogether, it seems that this clone represents a splice variant of PDE11A. The full-length cDNA is 2,141 bp in length, and the ORF predicts a protein of 576 aa with a molecular mass of 65.8 kDa. Because Western blot analysis with an antibody specific for PDE11A1 revealed the existence of additional bands, including one of about 65 kDa and designated PDE11A2, our clone is likely to represent this protein. Therefore, we have tentatively named this clone PDE11A2. PDE11A2 has a consensus PDE catalytic domain that is located between amino acids 305 and 544. At the N terminus, PDE11A2 contains a single GAF domain situated between amino acids 44 and 205. Compared with PDE11A1, the GAF domain of PDE11A2 has an additional 47 aa at the N terminus (Fig. 1B).
Figure 1.
(A) Similarities and differences between cDNAs for PDE11A splice variants. Region of identity shared by all three variants is in black. Sequences present in both PDE11A2 and PDE11A3 are dotted. Other patterns indicate sequences unique for each of the splice variants. Numbers depict positions in the nucleotide sequences. (B) Structure of PDE11A splice variants. Numbers indicate amino acid residues. Filled bars indicate catalytic domains. Hatched bars indicate GAF domains.
Cloning and Sequencing of PDE11A3.
To identify other possible PDE11A splice variants, the cDNA sequences were extended from testis poly(A)+ RNA by using the 5′ RACE system. A PDE11A-specific primer was used for first-strand cDNA synthesis. The primers used for PCR were the adapter primer supplied in the 5′ RACE kit and the 11End3.2 primer. The resulting 5′ RACE products were size selected above 1 kb and used to generate a minilibrary of approximately 2,000 clones in the TA cloning vector (pCR2.1-TA; Invitrogen). This minilibrary was screened by using an end-labeled oligonucleotide PDE11 screening as a probe. Eight positive colonies were picked, and all clones that extended the PDE11A2 sequence were verified by sequencing multiple independent PCR products.
DNA sequence analysis revealed that this clone contains an ORF including a large portion identical with PDE11A1 and -A2 sequences. In addition, it shared 76 aa with PDE11A2 in its N-terminal region. The unique sequences extended the N-terminal part of the PDE by 194 aa not present in PDE11A1 and 108 not found in PDE11A2 (Fig. 1B). In addition, there were unique sequences detected in the 3′ UTR region. Altogether, it seems that this clone represents another splice variant of PDE11A distinct from PDE11A1 and -A2. The full-length cDNA is 2,205 bp in length, and the ORF predicts a protein of 684 aa with a molecular mass of 78.1 kDa. Because Western blot analysis with an antibody specific to PDE11A C-terminal regions revealed the existence of additional bands including one of about 78 kDa and designated PDE11A3, this clone is likely to represent this protein. Therefore, we named this clone PDE11A3. PDE11A3 has a consensus PDE catalytic domain that is located between amino acids 413 and 652. At the N terminus, PDE11A3 contains two GAF domains situated between amino acids 1 and 125 and 152 and 313 (Fig. 1B).
Catalytic Properties of PDE11A2 and -A3.
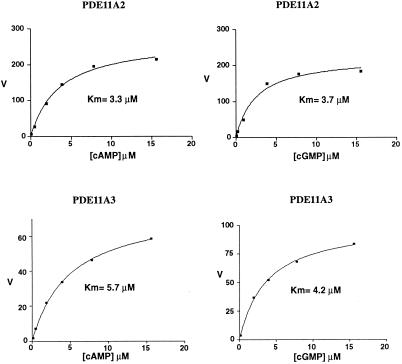
The full-length PDE11A2 and -A3 proteins were expressed in Sf-9 cells by using a Baculovirus expression system. At 0.14 μM cAMP/cGMP, extracts from the PDE11A2-expressing cells showed a 13-fold increase in cAMP/cGMP hydrolyzing PDE activity as compared with extracts from nonexpressing cells. These findings suggested that PDE11A2 is a dual substrate enzyme similar to PDE11A1. To further characterize the enzymatic properties of PDE11A2, more detailed kinetic studies were performed. The Km of PDE11A2 for cAMP was 3.3 μM and for cGMP was 3.7 μM, as represented by the average of five separate experiments with two independent enzyme preparations (Fig. 2). The Vmax ratio for cAMP/cGMP was 0.90. Analysis of PDE11A3 recombinant protein revealed very similar catalytic properties with Km values of 5.7 and 4.2 μM for cAMP and cGMP, respectively. The Vmax ratio for cAMP/cGMP was 0.99.
Figure 2.
PDE11A2 and PDE11A3 kinetics. PDEs were expressed in Sf-9 cells and activities measured in extracts as described in Materials and Methods. The concentration range of cAMP/cGMP is indicated on the x axis (μM). On the y axis, the velocity (V) of cAMP/cGMP hydrolysis was plotted (pmol/min/ml). Km values were determined by nonlinear regression fits of a single site binding site model using the graphpad prism program.
Inhibitors of PDE11A2 and -A3.
One approach to identification of the physiological role(s) for PDE11A is to apply specific drugs blocking its activity. Therefore, we set out to determine which, if any, of the several commonly available inhibitors of other PDEs could interfere with the activity of PDE11A2 and PDE11A3 expressed in Sf-9 cells. The spectrum of inhibitors tested in this study included nonselective compounds as well as selective inhibitors targeting members of other known PDE families. The results of the inhibitor studies are summarized in Table 1. The ability of PDE11A2 to hydrolyze cAMP was inhibited by dipyridamole, a PDE5, PDE6, PDE7, PDE8, and PDE10 inhibitor. The IC50 of dipyridamole for PDE11A2 was 1.8 μM. PDE11A2 was also moderately sensitive to zaprinast, a PDE5 and PDE6 inhibitor with an IC50 of 28 μM and IBMX with IC50 of 80 μM. Several other inhibitors used in this study, including sildenafil (Table 1), did not affect PDE11A2 hydrolysis of cAMP, even when applied at concentrations exceeding the IC50 values for other PDEs by 100-fold. PDE11A3 demonstrated a very similar profile of effective inhibitors (Table 1) with dipyridamole, zaprinast, and IBMX inhibiting half of the activity at concentrations of 0.82, 5, and 25 μM, respectively.
Table 1.
Inhibitor studies of PDE11A2 and PDE11A3 expressed in Sf-9 cells
| Inhibitor | PDE Selectivity (IC50) | IC50 (n = 3) for PDE 11A2 | IC50 (n = 3) for PDE 11A3 |
|---|---|---|---|
| IBMX | Nonselective (2–50 μM) | 80 ± 43 μM | 25 ± 10 μM |
| Papaverine | Nonselective (5–25 μM) | >100 μM | ND |
| EHNA | PDE 2 (1.0 μM) | >100 μM | ND |
| Rolipram | PDE 4 (2.0 μM) | >200 μM | ND |
| Dipyridamole | PDE 5 (0.9 μM) | 1.8 ± 1.1 μM | 0.82 ± 0.1 μM |
| PDE 6 (0.38 μM) | |||
| PDE 7 (9.0 μM) | |||
| PDE 8 (4.5 μM) | |||
| PDE 10 (1.1 μM) | |||
| SCH 51866 | PDE 1 & 5 (0.1 μM) | >100 μM | ND |
| PDE 7 (35 μM) | |||
| PDE 9 (1.5 μM) | |||
| PDE 10 (1.0 μM) | |||
| Enoximone | PDE 3 (1.0 μM) | >100 μM | ND |
| Sildenafil | PDE 5 (3.9 nM) | >500 nM | ND |
| Ro 201724 | PDE 4 (2.0 μM) | >200 μM | ND |
| Zaprinast | PDE 5 (0.76 μM) | 28 ± 7.6 μM | 5.0 ± 1.4 μM |
| PDE 6 (0.15 μM) | |||
| Pentoxifylline | Nonselective (45–150 μM) | >100 μM | ND |
ND, not determined. Conditions for inhibitor studies are described in Materials and Methods.
Discussion
In this study, we describe the cloning and initial characterization of two splice variants of the recently identified PDE11A family. We have designated them PDE11A2 and PDE11A3 according to the current nomenclature of PDEs (15). The full-length cDNAs of PDE11A2 and PDE11A3 variants contain 2,141 bp and 2,205 bp, respectively. The ORF sequences predict proteins of 576 aa for PDE11A2 and 684 aa for PDE11A3. The predicted molecular masses are 65.8 and 78.1 kDa for PDE11A2 and PDE11A3, respectively. This is consistent with the observation of 65- and 78-kDa isoforms of PDE11A detected by Western blotting (10). The predicted amino acid sequences of PDE11A splice variants reveal a large region of identity that includes a PDE catalytic domain in the C-terminal part of the protein (amino acid positions 219–458 in the PDE11A1). The differences between the splice variants emerge in both N- and C-terminal regions (Fig. 1A). At the 5′ ends of the cDNA nucleotide sequences, the variant regions encompass nucleotides 1–120 in PDE11A1, 1–341 in PDE11A2, and 1–696 in PDE11A3. These variances predict an additional 86 and 194 aa in the predicted protein sequence of PDE11A2 and -A3, respectively. The differences extend into the 5′ UTR region, suggesting that these variants of PDE11A result from the existence of at least two alternative first exons. Whether the mechanism to generate this variability of 5′ regions of PDE11A cDNAs is alternative splicing of pre-mRNA, usage of alternative promoters, or both is not clear at present.
Another variant region of PDE11A splice variants is located in the 3′ UTR region (nucleotides 1622–1784 in PDE11A1, 1843–2141 in PDE11A2, and 2198–2205 in PDE11A3) (Fig. 1A). As this area of mRNA frequently regulates the stability of the messenger molecule, it is tempting to speculate that expression of PDE11A isoforms may be subject to differential regulation by distinct rates of degradation of their mRNAs.
Comparison of the predicted amino acid sequences of PDE11A1, -A2, and -A3 shows that PDE11A3 contains an additional GAF domain (Fig. 1B). Also, the single GAF domain present in PDE11A2 is larger by 47 aa than that of PDE11A1 (Fig. 1B). Therefore, an interesting possibility exists that the regulatory functions of the different GAF domains of the PDE11A variants are not identical. While these studies were in progress, Omori and colleagues reported another PDE11A splice variant designated PDE11A4 as well as the same PDE11A3 described in this report (K. Omori, ref. 25). Similar to PDE11A3, the A4 variant also contains tandem arranged GAF domains. Interestingly, tandem GAF domains are observed in the N-terminal regulatory regions of PDE2, -5, -6, and -10 (16–18). In the first three examples, the paired GAF domains bind cGMP. The regulatory consequences of the binding are different for each enzyme. In PDE2, cGMP bound to GAF domains increases PDE activity for both cAMP and cGMP (19). In PDE5 and -6, the GAF domains are thought to regulate the enzymes by promoting phosphorylation (20, 21) and by affecting interactions with transducin, respectively (22, 23). Therefore, the presence of repeated GAF domains in the PDE11A3 might provide the necessary structure to ensure effective cGMP binding and regulation. However, it should be mentioned that there is as yet little evidence for high-affinity cGMP binding by the tandem GAF domains of PDE10 (16). A possible explanation for this might be that the GAFa domain of PDE10 contains an atypical GAF domain consensus sequence (Fig. 3). All cGMP-binding GAF domains present in PDEs show the conserved consensus motif N(K/R)XnFX3DE (18). The same consensus also is observed in the GAF domains of PDE11. Although all GAF domains present in PDEs have so far been shown to bind cGMP, the possibility exists that there are other small molecules binding to these structures. For example, the GAF domain of E. coli FhlA, a member of the NtrC family of transcription factors, is able to bind formate (24).
Figure 3.
Multiple sequence alignment of the PDE11A1, PDE11A2, and PDE11A3 GAF domains to GAF domains of other PDEs. The GAF domain boundaries were determined by the Simple Modular Architecture Tool (smart). For clarity of viewing, 35–95 nonhomologous amino acid residues from the N-terminal and C-terminal ends of the sequences delineated by smart have been omitted. Numbers in parentheses at the end of each sequence indicate the number of amino acids omitted. Stars mark the conserved sequence motif present in most GAF domains. PDE2, HSPDE2A (GenBank accession no. U67733); PDE5, HSPDE5A (AJ004865); PDE6, HSPDE6A, alpha, (NM-000440); PDE10, HSPDE10A (NM-006661); PDE11A1, HSPDE11A1 (AJ251509); PDE11A2, HSPDE11A2 (AF281865); PDE11A3, HSPDE11A3 (AJ278682).
Characterization of the catalytic properties of recombinant PDE11A2 and -A3, expressed in Sf-9 cells, revealed that, like PDE11A1, PDE11A2 and PDE11A3 could hydrolyze both cAMP and cGMP. Also, the Km values were similar for all three splice variants. This is consistent with the identity of the catalytic domains of the PDE11A isoforms. In addition to possessing similar catalytic properties, all three PDE11A splice variants share similar sensitivities to various inhibitors. In particular, dipyridamole and zaprinast are effective inhibitors of PDE11A2 and PDE11A3, as was observed for PDE11A1. Therefore, the function of the PDE11A heterogeneity is most likely for the generation of enzymes with the same catalytic, but distinct regulatory properties, subcellular localization, or stability.
In summary, we have cloned two splice variants of PDE11A, PDE11A2 and PDE11A3. Further investigation of the expression pattern of both variants as well as studies on their regulation will shed more light on the functional significance of PDE11A diversity.
Acknowledgments
This work was supported in part by National Institutes of Health Grants DK21723 and HL44948 and a grant from Pfizer Global Research.
Abbreviations
- PDE
phosphodiesterase
- RACE
rapid amplification of cDNA ends
- EST
expressed sequence tag
- IBMX
3-isobutyl-1-methylxanthine
- UTR
untranslated region
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AF281865 and AJ278682).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.200355397.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.200355397
References
- 1.Li L, Yee C, Beavo J A. Science. 1999;283:848–851. doi: 10.1126/science.283.5403.848. [DOI] [PubMed] [Google Scholar]
- 2.Dousa T P. Kidney Int. 1999;55:29–62. doi: 10.1046/j.1523-1755.1999.00233.x. [DOI] [PubMed] [Google Scholar]
- 3.Pfister C, Bennett N, Bruckert F, Catty P, Clerc A, Pages F, Deterre P. Cell Signaling. 1993;5:235–241. doi: 10.1016/0898-6568(93)90015-e. [DOI] [PubMed] [Google Scholar]
- 4.Jin S L, Richard F J, Kuo W P, D'Ercole A J, Conti M. Proc Natl Acad Sci USA. 1999;96:11998–12003. doi: 10.1073/pnas.96.21.11998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Torphy T J, Zhou H L, Foley J J, Sarau H M, Manning C D, Barnette M S. J Biol Chem. 1995;270:23598–23604. doi: 10.1074/jbc.270.40.23598. [DOI] [PubMed] [Google Scholar]
- 6.Ballard S A, Gingell C J, Tang K, Turner L A, Price M E, Naylor A M. J Urol. 1998;159:2164–2171. doi: 10.1016/S0022-5347(01)63299-3. [DOI] [PubMed] [Google Scholar]
- 7.Soderling S H, Beavo J A. Curr Opin Cell Biol. 2000;12:174–179. doi: 10.1016/s0955-0674(99)00073-3. [DOI] [PubMed] [Google Scholar]
- 8.Beavo J A. Physiol Rev. 1995;75:725–748. doi: 10.1152/physrev.1995.75.4.725. [DOI] [PubMed] [Google Scholar]
- 9.Conti M, Jin S L. Prog Nucleic Acid Res Mol Biol. 1999;63:1–38. doi: 10.1016/s0079-6603(08)60718-7. [DOI] [PubMed] [Google Scholar]
- 10.Fawcett L, Baxendale R, Stacey P, McGrouther C, Harrow I, Soderling S, Hetman J, Beavo J A, Phillips S C. Proc Natl Acad Sci USA. 2000;97:3702–3707. doi: 10.1073/pnas.050585197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aravind L, Ponting C P. Trends Biochem Sci. 1997;22:458–459. doi: 10.1016/s0968-0004(97)01148-1. [DOI] [PubMed] [Google Scholar]
- 12.Soderling S H, Bayuga S J, Beavo J A. J Biol Chem. 1998;273:15553–15558. doi: 10.1074/jbc.273.25.15553. [DOI] [PubMed] [Google Scholar]
- 13.Hetman J M, Soderling S H, Glavas N A, Beavo J A. Proc Natl Acad Sci USA. 2000;97:472–476. doi: 10.1073/pnas.97.1.472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hansen R S, Beavo J A. Proc Natl Acad Sci USA. 1982;79:2788–2792. doi: 10.1073/pnas.79.9.2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beavo J A, Conti M, Heaslip R J. Mol Pharmacol. 1994;46:399–405. [PubMed] [Google Scholar]
- 16.Soderling S H, Bayuga S J, Beavo J A. Proc Natl Acad Sci USA. 1999;96:7071–7076. doi: 10.1073/pnas.96.12.7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Charbonneau H, Prusti R K, LeTrong H, Sonnenburg W K, Mullaney P J, Walsh K A, Beavo J A. Proc Natl Acad Sci USA. 1990;87:288–292. doi: 10.1073/pnas.87.1.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McAllister-Lucas L M, Sonnenburg W K, Kadlecek A, Seger D, Trong H L, Colbran J L, Thomas M K, Walsh K A, Francis S H, Corbin J D, et al. J Biol Chem. 1993;268:22863–22873. [PubMed] [Google Scholar]
- 19.Stroop S D, Charbonneau H, Beavo J A. J Biol Chem. 1989;264:13718–13725. [PubMed] [Google Scholar]
- 20.Thomas M K, Francis S H, Corbin J D. J Biol Chem. 1990;265:14971–14978. [PubMed] [Google Scholar]
- 21.Wyatt T A, Naftilan A J, Francis S H, Corbin J D. Am J Physiol. 1998;274:H448–H455. doi: 10.1152/ajpheart.1998.274.2.H448. [DOI] [PubMed] [Google Scholar]
- 22.Cote R H, Bownds M D, Arshavsky V Y. Proc Natl Acad Sci USA. 1994;91:4845–4849. doi: 10.1073/pnas.91.11.4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamazaki A, Bondarenko V A, Dua S, Yamazaki M, Usukura J, Hayashi F. J Biol Chem. 1996;271:32495–32498. doi: 10.1074/jbc.271.51.32495. [DOI] [PubMed] [Google Scholar]
- 24.Korsa I, Bock A. J Bacteriol. 1997;179:41–45. doi: 10.1128/jb.179.1.41-45.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yuasa, A., Kotera, J., Fujishige, K., Michibata, H., Sasaki, T. & Omori, K. (2000) J. Biol. Chem., in press. [DOI] [PubMed]