Abstract
The solution structure of the C-terminal Domain V of the τ subunit of E. coli DNA polymerase III was determined by nuclear magnetic resonance (NMR) spectroscopy. The fold is unique to τ subunits. Amino acid sequence conservation is pronounced for hydrophobic residues that form the structural core of the protein, indicating that the fold is representative for τ subunits from a wide range of different bacteria. The interaction between the polymerase subunits τ and α was studied by NMR experiments where α was incubated with full-length C-terminal domain (τC16), and domains shortened at the C-terminus by 11 and 18 residues, respectively. The only interacting residues were found in the C-terminal 30-residue segment of τ, most of which is structurally disordered in free τC16. Since the N- and C-termini of the structured core of τC16 are located close to each other, this limits the possible distance between α and the pentameric δτ2γδ′ clamp–loader complex and, hence, between the two α subunits involved in leading- and lagging-strand DNA synthesis. Analysis of an N-terminally extended construct (τC22) showed that τC14 presents the only part of Domains IVa and V of τ which comprises a globular fold in the absence of other interaction partners.
INTRODUCTION
DNA polymerase III (Pol III) is the central enzyme of the Escherichia coli replisome. The holoenzyme is composed of several individual proteins, including the subunits α, β, γ, δ, δ′, ε, χ, ψ, τ and θ. 3D structures have been determined for a large fragment of α (1), β (2), the structured parts of ε (3), θ (4) and δ′ (5) and for multimeric complexes including a pentameric γ3δδ′ complex (6,7), and the βδ (8), χψ (9) and εθ (10) complexes. To date, no 3D structure of full-length α or τ has been published, though the complete α subunit could presumably be modeled on the corresponding structure from Thermus aquaticus (11).
The full-length τ subunit is a 71 kDa protein of 643 amino acid residues. The N-terminal 430 residues of τ (Domains I–III) are identical to γ (12,13) and can substitute for γ in the pentameric complexes with δ and δ′ (14,15). The 213 additional residues at the C-terminus of τ (Domains IVa + V, residues 430–643) have been shown to bind to α with nanomolar affinity (16,17) and to bind to the DnaB helicase, albeit much more weakly (18–20). In addition, a construct comprising Domains IVa and V has been shown to bind to primed DNA (21). The interaction site with α has been mapped to the 147 C-terminal residues of τ (Domain V, residues 499–643; Figure 1) (22) and the binding to DnaB was assigned to residues 430–496 (Domain IVa) (23). Weak DNA-binding activity has also been mapped to Domain IVa (24). The replisome contains two coupled α subunits, one for leading- and one for lagging-strand DNA synthesis (16,25). These two subunits are each linked to the same pentameric clamp–loader complex composed of γ, δ, δ′ and two τ subunits (14). The interaction of τ with α thus presents a critical structural link in the replisome, tying two α subunits to the clamp–loader complex. Here we report the 3D solution structure of the C-terminal Domain V of τ from E. coli. In addition, the structure of Domain IVa was probed in a construct comprising Domain IVa and the globular part of Domain V, and the interaction site of α on τ was mapped by NMR experiments using three different constructs of Domain V of τ. The results place restraints on the distances in the replisome between the two α subunits, their proximity to the DnaB helicase, and the flexibility of the complex of τ, α and DnaB.
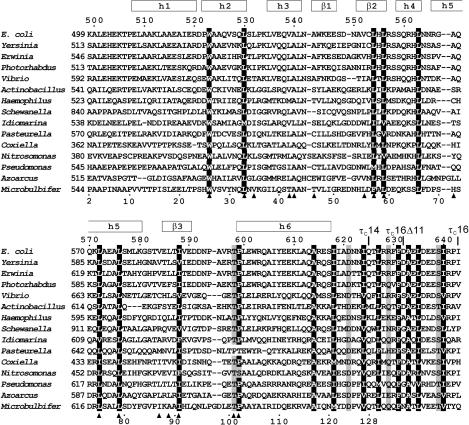
Figure 1.
Sequence alignment of τC16 with τ proteins from different bacteria. The sequence of τC16 comprises the 145 C-terminal residues of the τ subunit of E. coli DNA Pol III holoenzyme (residues 499–643 which constitute Domain V). Its sequence numbering is shown at the top. The constructs τC14, τC16Δ11 and τC16 share the same N-terminus. Their C-termini are indicated above the sequence. The sequence numbering shown below the alignment is that of the NMR sample of τC14 included for reference with previous work (41). The regular secondary structure elements of τC14 determined in this work are shown at the top. Positions with conserved hydrophobic residues are indicated by white characters on a black background. Conserved hydrophilic residues are shaded gray. Filled triangles below the alignment identify residues with side chains buried in the τC14 structure (<5% solvent accessibility). The following sequences from τ proteins are shown (abbreviation, organism, GenBank number): E. coli, Escherichia coli, 43320; Yersinia, Yersinia pseudotuberculosis, 51595342; Erwinia, Erwinia carotovora subsp. Atroseptica, 49610641; Photorhabdus, Photorhabdus lummines subsp. Laumondii tto1, 37527701; Vibrio, Vibrio vulnificus cmcp6, 27361492; Actinobacillus, Actinobacillus pleuropneumoniae serovar 1 STR, 32035419; Haemophilus, Haemophilus somnus, 32029123; Shewanella, Shewanella oneidensis MR-1, 24373576; Idiomarina, Idiomarina loihiensis L2TR, 56460949; Pasteurella, Pasteurella multocida subsp. multocida str. Pm70, 12720609; Coxiella, Coxiella burnetii RSA 493, 29541263; Nitrosomonsa, Nitrosomonas europaea ATCC 19718, 30138336; Pseudomonas, Pseudomonas fluorescens PfO-1, 48731928; Azoarcus, Azoarcus sp. EbN1, 56476074; Microbulbifer, Microbulbifer degradans 2-40, 48862900.
MATERIALS AND METHODS
Proteins
The α subunit of E. coli DNA Pol III was prepared as described earlier (26). The proteins τC16 (N-terminal Met followed by C-terminal residues 499–643 of τ, comprising all of Domain V), τC16Δ11 (τC16 with the C-terminal 11 residues removed), τC14 (τC16 with the C-terminal 18 residues removed) and τC22 (N-terminal Met followed by residues 430–626 of τ, corresponding t Domain IVa and τC14) were isolated as described in the accompanying paper (24). Stable-isotope enriched samples were produced by expression on a minimal medium (M9) containing 15NH4Cl as the sole nitrogen source or on a 13C/15N-enriched medium (Silantes, Germany). In the case of τC14 the respective yields of purified protein were 9 and 21 mg/l of medium. In the case of τC22, the yield was 15 mg of purified 15N-labeled protein per liter of medium.
NMR sample preparation
All samples for NMR measurements were prepared in a buffer containing 10 mM sodium phosphate (pH 6.8), 100 mM NaCl, 1 mM dithiothreitol and 0.1 mM NaN3, with protein concentrations of up to 1.9 mM. Samples were prepared in 90% H2O/10% D2O or 100% D2O.
NMR measurements
All NMR measurements for the structure determination of τC14 were carried out at 30°C, using Varian Inova 600 and Bruker AV 800 NMR spectrometers. The backbone resonance assignment was obtained from the analysis of 3D HNCACB and CBCA(CO)NH together with NOESY-15N-HSQC and 2D NOESY spectra. The side-chain resonances were assigned through the analysis of 3D CC(CO)NH, HNHA, (H)CCH-TOCSY and TOCSY-15N-HSQC and HNCO spectra, together with NOESY-15N-HSQC, 13C-HSQC-NOESY and 2D NOESY and TOCSY spectra. The protons of aromatic groups were assigned with 2D DQF-COSY, NOESY and TOCSY spectra. A few resonances could be assigned only with the help of NOEs after initial structure calculations. ϕ and Ψ angle restraints were generated by the program TALOS (27) using the chemical shifts of CA, CB, CO, HA and N, and checked against the intensities of intraresidual and sequential NOEs between backbone protons.
In total, 2500 NOE cross-peaks were assigned and integrated, resulting in 1891 meaningful distance restraints. Most of these restraints came from a 2D NOESY spectrum of an unlabeled sample recorded with 40-ms mixing time, using t1max = 78 ms, t2max = 213 ms and ∼3 days of recording time. Stereo-specific resonance assignments were obtained with the program GLOMSA (28), using 3J(Hα,Hβ) coupling constant information derived from the DQF-COSY spectrum. Stereo-specific assignments of the isopropyl CH3-groups of valine and leucine residues were achieved by recording constant-time 13C-HSQC spectra of a 10% biosynthetically fractionally 13C-labeled sample (29). Pairs of methyl groups from the same isopropyl group were identified by methyl–methyl NOEs observed in a diagonal-suppressed constant-time 13C-HSQC spectrum with NOE relay (30) recorded of uniformly 13C-labeled protein.
15N-relaxation data of τC22 were recorded at 25°C using standard experiments (31). The relaxation delays used were 9, 17, 26, 43, 60, 78, 95 and 112 ms in the R2 experiment and 3, 30, 100, 200, 350, 600, 900 and 1300 ms in the R1 experiment. The complete set of R1 and R2 relaxation data was recorded in about 18 h per protein sample, using t1max = 50 ms and t2max = 100 ms for each spectrum.
Structure calculations
The cross-peaks in the NOESY spectra of τC14 were assigned and integrated using the program XEASY (32). The NMR structure was calculated using the program DYANA (28), starting from 200 random conformers that were annealed in 40 000 steps using torsion-angle dynamics. The 20 conformers with the lowest residual restraint violations were energy minimized in a shell of water using the program OPAL with standard parameters (33).
Table 1 shows an overview of the restraints used and structural statistics. The Ramachandran plot was analyzed using PROCHECK-NMR (34). No residue was consistently in a forbidden region in all 20 conformers.
Table 1.
Structural statistics for the NMR conformers of τC14
| Parameter | Value |
|---|---|
| Number of assigned NOE cross-peaks | 2500 |
| Number of non-redundant NOE upper-distance limits | 1891 |
| Number of scalar coupling constants (Hα–Hβ)a | 64 |
| Number of dihedral-angle restraints | 241 |
| Intra-protein AMBER energy (kcal/mol) | −5388 ± 321 |
| Maximum NOE-restraint violations (Å) | 0.09 ± 0.00 |
| Maximum dihedral-angle restraint violations (°) | 2.0 ± 0.5 |
| r.m.s.d. to the mean for N, Cα and C′ (Å)b | 0.55 ± 0.14 |
| r.m.s.d. to the mean for all heavy atoms (Å)b | 0.99 ± 0.11 |
| Ramachandran plot appearancec | |
| Most favored regions (%) | 90.2 |
| Additionally allowed regions (%) | 9.5 |
| Generously allowed regions (%) | 0.3 |
| Disallowed regions (%) | 0.0 |
aStereo-specific resonance assignments were obtained for 49 pairs of CβH2, one pair of CγH2 and two pairs of CδH2 protons. bFor residues 509–609. cFrom PROCHECK-NMR (34).
Secondary structure elements and root mean square deviation (r.m.s.d.) values were calculated using the program MOLMOL (35). Side-chain solvent accessibilities were measured with a spherical probe of 1.4 Å radius and calculated in percent of the accessibilities measured for a fully extended side chain of residue X in a helical Gly-X-Gly peptide (36). The values obtained were averaged over the 20 NMR conformers.
The chemical shifts and coordinates of the structure have been deposited in the BioMagResBank (accession code 6869) and PDB (accession code 2AYA).
RESULTS
NMR resonance assignments
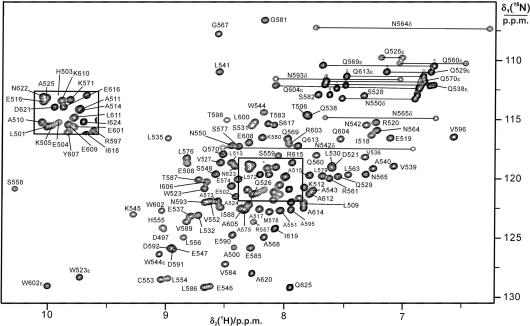
The 15N-HSQC spectrum of τC14 presented well-dispersed resonances characteristic of a folded protein, with line widths consistent with it being monomeric (Figure 2). Virtually complete resonance assignments were obtained for the polypeptide backbone between residues 500 and 625, except that the amide cross-peak of His562 was missing and the amide cross-peaks of Lys534 and Arg566 were very broad, so that no NOEs could be observed with these HN resonances. 15N-relaxation measurements of τC16Δ11 indicated that the loop region with Lys534 was affected by chemical exchange (37), while the line broadening of Arg566 may have been due to fast amide proton exchange. A constant-time 13C-HSQC spectrum with NOE relay and suppression of the diagonal peaks proved particularly useful for the identification of the 13C–1H methyl cross-peaks belonging to the same isopropyl group, since constant-time 13C-HSQC spectra can be recorded with much higher resolution than HCCH-TOCSY experiments.
Figure 2.
15N-HSQC spectrum of τC14. The spectrum was recorded at 30°C with a 1.8 mM solution of uniformly 15N/13C-labeled τC14 in 90% H2O/10% D2O at pH 6.8 in phosphate buffer (10 mM phosphate, 100 mM NaCl, 1.0 mM dithiothreitol and 0.1 mM NaN3). The cross-peaks are assigned using one-letter amino acid symbols and residue numbers. Side-chain resonances are identified by Greek characters. Cross-peaks from side-chain amides are connected by horizontal lines.
Structure of τC14 and comparison with the KH fold
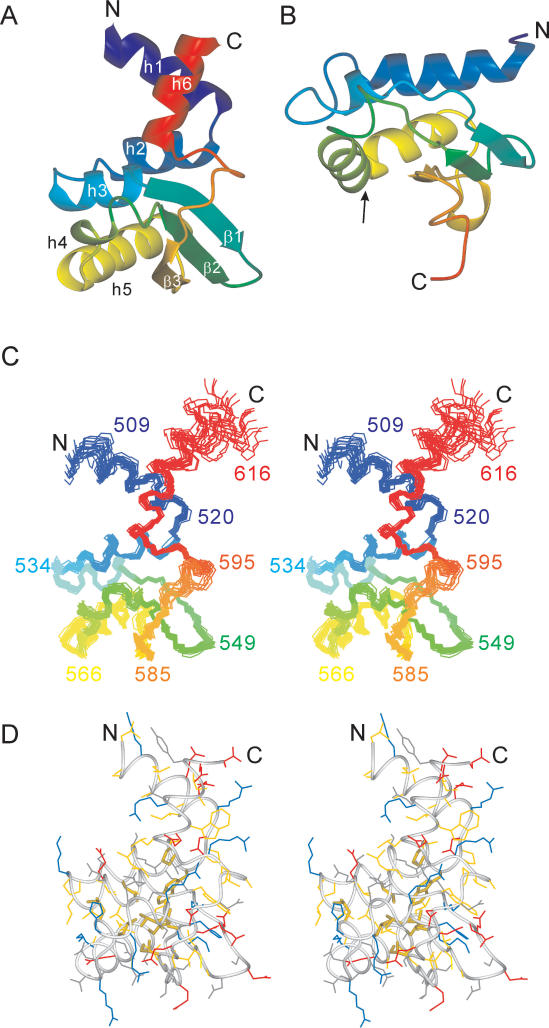
The structure of τC14 comprises six helices and a three-stranded mixed parallel/antiparallel β-sheet (Figures 1 and 3). The amino- and carboxyl-terminal helices point away from the structural core of the protein (Figure 3). Their orientation towards the terminal ends becomes increasingly uncertain, as the NMR structure of this part of the protein is based exclusively on short-range distance restraints.
Figure 3.
Solution structure of τC14 and comparison with the KH domain of protein MPN156. (A) Ribbon representation of τC14, including residues 509–609. The secondary structure elements are labeled as in Figure 1. (B) Ribbon representation of MPN156 from Mycoplasma pneumoniae (PDB code: 1PA4). The arrow identifies the RNA-binding helix-loop-helix motif of the KH-domain type II fold. (C) Stereo view of a superposition of the backbone atoms of 20 NMR conformers, including residues 507–617. (D) Stereo view of a heavy-atom representation of the conformer closest to the mean structure of τC14 (only residues 509–609 are shown). The side-chains are color coded in blue (Lys, Arg, His), red (Asp, Glu), yellow (Ala, Cys, Ile, Leu, Met, Phe, Pro, Trp, Val) and gray (Asn, Gln, Ser, Thr, Tyr). The side chains of the strictly conserved hydrophobic residues Trp523, Leu530, Leu554, Leu556, Leu563, Leu572, Leu576, Ile588 and Pro599 are highlighted with broader lines. This figure was prepared using the program MOLMOL (35).
A search of the protein data bank with the program DALI (38) revealed only remote structural homologues. The best match was with the ribosome-binding factor A from Mycoplasma pneumoniae (MNP156), with a r.m.s.d. of 2.5 Å for 66 superimposed residues. This protein, like the next best matches detected by DALI, assumes a KH type-II fold (39). The KH fold does not include helices corresponding to helices 1 and 6 in τC14. Furthermore, the KH fold includes a helix-turn-helix motif (identified by an arrow in Figure 3B) that is involved in RNA binding and contains a characteristic GxxG turn signature (39). The τC14 structure does not feature a similar motif. In particular, helix 4 is a short 310 helix with clear Hα(i) − HN(i + 2) NOEs rather than a long α-helix as in the KH fold. There is also no evidence for an extended, positively charged surface in this or any other region of the protein which could favor binding to the phosphodiester backbone of RNA or DNA.
The fold of τC14 includes N- and C-terminal helices that protrude from the globular core of the structure into the solvent (Figure 3). These helices appear to be more than mere extensions of the KH fold since they are attached to the rest of the protein in a very specific manner: helix 6 partly extends the helix dipole of helix 3 and helices 1 and 6 form intimate contacts that define their relative orientation. For example, ring currents from the side chain of Trp602 in helix 6 cause significant up-field shifts of the resonances of Arg520 CβH2 in helix 1.
Helices 1 and 6 border flexible polypeptide segments near the N- and C-termini, respectively. The virtual absence of medium-range NOEs for the residues preceding Pro507 indicated that these residues are structurally disordered. Also the C-terminal end of helix 6 is conformationally unstable, as the mobility of the polypeptide chain gradually increases towards the C-terminus, as evidenced by the observation of exchange peaks between the water resonance and the backbone amide protons for Ala612 and the following residues in NOESY and TOCSY experiments. This conclusion was further supported by the observation of increasing signal intensities for Ala612 and subsequent residues in experiments with long periods of T2 relaxation. Helix 6 seems to end at residue Ser617, as the chemical shifts of Ile618 were indicative of an extended backbone conformation and the chemical shifts and NOEs of the following residues were characteristic of random coil conformations.
Conservation of the domain fold
A search for homologous proteins using BLAST (40) exclusively revealed τ domains from other bacterial species (Figure 1). Our data show that all these domains are structurally conserved and closely related to the structure of E. coli τC14. First, any insertions and deletions occur exclusively in the loop regions between regular secondary structure elements. Second, almost all of the strictly conserved hydrophobic residues (highlighted in Figure 3D for residues between Leu509 and Glu609) are buried in the 3D structure with very low solvent accessibility, as expected for residues that are critical to maintain the fold of the protein. The only exception is Leu564, the side chain of which is somewhat solvent exposed (about 20%). Finally, residues with buried side chains in the structure of τC14 are consistently uncharged in the sequence alignment of Figure 1. The only exception is Arg925 in the Shewanella protein located at the position of Val584 in E. coli τC14. Since the side chain of arginine is longer than that of valine, its charge is quite possibly solvent exposed in the Shewanella protein.
Structure of τC16 (Domain V)
Many conserved residues are located after residue 613 (Figure 1). All those present in the NMR structure of E. coli τC14 are highly solvent exposed, indicating that their conservation is of functional importance. Samples comprising the entire Domain V of τ, τC16, were highly sensitive to proteolytic cleavage (24). The 15N-HSQC spectra of 15N-labeled τC16 samples that had been freshly prepared in a cell-free protein synthesis system (41) showed increasingly narrow and more intense signals for the segment following Ser617, indicating substantially increased mobility. In particular, there was no evidence for any ordered association of this unstructured segment with the folded part of τC14 (41).
Interaction with the α subunit and relaxation data analysis
The interaction of Domain V of τ with the α subunit of Pol III was probed by monitoring changes observed in the 15N-HSQC spectra of 15N-labeled τC14, τC16 and τC16Δ11 upon addition of purified α. No spectral changes were observed at all when α was added to τC14, indicating no binding (data not shown). This is consistent with gel filtration and surface plasmon resonance (SPR) data that also showed that τC14 does not interact with α (24). Upon addition of increasing amounts of α, the 15N-HSQC spectrum of τC16 became weaker without changing its appearance (data not shown). This indicated tight binding, where only the spectrum of free τC16 is observed because the signals of bound τC16 are broadened beyond detection. Therefore, the last 18 residues of τ present in τC16 but not τC14 are critical for the α–τ interaction. The results are in accord with the gel filtration and SPR data (24).
In contrast, the previous study showed that removal of eleven residues from the C-terminus of τC16 produced a protein (τC16Δ11) that could no longer be observed by SPR to interact with α, indicating KD > 10 μM for the α–τC16Δ11 complex (24). However, at the higher concentrations used for NMR experiments, τC16Δ11 could be observed to bind to α with a KD of about 0.9 mM, resulting in an observable NMR spectrum of τC16Δ11 even in the presence of an excess of α (37). This indicates the existence of fast exchange between free and bound τC16Δ11. Averaging of the NMR signals between free and bound τC16Δ11 was further indicated by the fact that the signals shifted continuously for different ratios of τC16Δ11 to α.
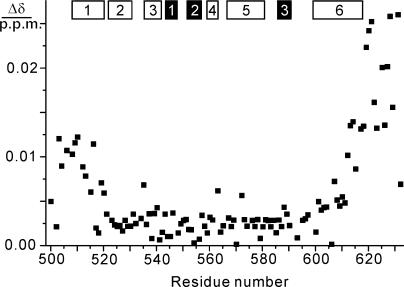
As expected for weak binding, only small chemical shift changes were observed, most of which were within the line width of the cross-peaks (Figure 4). These changes were predominantly observed for the C-terminal 20 residues of τC16Δ11 including part of helix 6, but also among the N-terminal residues including helix 1. Extensive line broadening was observed only for the residues following Gln613. Notably, Ala614 is one of the few totally conserved residues in τC16 in the alignment of Figure 1. These data confirm the importance of the conserved C-terminal residues for binding and suggest that the chemical shift changes observed in helix 1 may be caused by small structural adjustments in helix 6 (since the two helices are close together, as shown in Figure 3A) rather than by direct contacts with α.
Figure 4.
Changes in amide chemical shifts of 15N-labeled τC16Δ11 observed upon binding of α at pH 6.8 and 25°C. The data were recorded with concentrations of τC16Δ11 and α of 90 and 120 μM, respectively. The chemical shift change Δδ was calculated as ({[Δδ(1H)]2 + [0.1Δδ(15N)]2}/2)1/2, where Δδ(1H) and Δδ(15N) denote the chemical shift changes in p.p.m. observed in the 1H and 15N dimensions of the 15N-HSQC spectrum. The locations of the α-helices and β-strands in τC14 are indicated by open and filled bars, respectively.
Binding of τC16Δ11 to α was also manifested by exchange broadening of the 15N-HSQC cross-peaks of the residues following Ile619. In contrast, the presence of α caused no exchange broadening for the N-terminal residues of τC16Δ11. In summary, these data stress the importance of the C-terminal unstructured segment of τ and of the C-terminal residues of helix 6 of Domain V for binding to α, whereas no other parts of τC16 seem to contact α.
Structure of τC22 (Domain IVa + V)
The 69 additional residues in τC22 N-terminal of τC14, which represent Domain IVa, were found to assume no defined conformation. 15N-HSQC cross-peaks of τC14 superimposed perfectly with 15N-HSQC cross peaks of τC22 (Supplementary Data, Figure S1), showing that the additional residues present in τC22 did not affect the structure of τC14 or interact with it in any specific manner. In agreement with a random coil conformation of Domain IVa, the additional 15N-HSQC cross-peaks present in the spectrum of τC22 were observed at 1H chemical shifts characteristic of random coil conformations. Finally, uniformly large R1(15N) and small R2(15N) relaxation rates were measured for Domain IVa, and the averaged R2/R1 ratio was 5.4 for Domain IVa and 19.8 for the τC14 domain in the τC22 construct, indicating that all residues of Domain IVa were highly mobile (Supplementary Data, Figure S2).
DISCUSSION
Escherichia coli τC14 is the first example of a C-terminal domain from a τ subunit of Pol III holoenzyme from any organism for which a 3D structure has been determined. As demonstrated by amino acid sequence conservation of buried hydrophobic residues, the fold is representative of all known bacterial τC domains (Figure 1).
The core of the fold of Domain V of τ displays overall structural similarity with KH type-II domains. Yet, pronounced structural differences in the region of the helix-turn-helix motif that is required for RNA binding in KH domains indicate that the overall structural similarity does not extend to functional similarity. The fold of Domain V of τ can thus be considered to be a unique fold that delivers a fixed-angle relationship between an N-terminal and a C-terminal α-helix (Figure 3).
The apparent absence of significant amino acid conservation on the surface of the domain suggests that interactions between the folded part of the domain and other proteins are not conserved. In stark contrast, the highest sequence conservation is found among the thirty C-terminal residues of τ that are required for binding to the Pol III α subunit. Three of the six positions of complete sequence conservation are found among these thirty residues (Figure 1). Remarkably, this polypeptide segment is highly solvent-exposed in the free protein and most of it is highly mobile. The sequence conservation in this segment is thus entirely determined by its interaction with a site in α. The conserved amino acids may be optimal for maximum binding affinity across a relatively small protein–protein interface. Alternatively, the sequence conservation may indicate interactions with different proteins during the lifecycle of τ which would be less tolerant to compensating mutations emerging simultaneously in τ and α. At present, it is not known whether a second binding partner exists for this segment of τ.
The τC16 domain binds to α considerably more tightly than a peptide comprising the 32 C-terminal residues of τ (24). Yet, no evidence could be found that any other residues of τ interact directly with α. This apparent inconsistency could be explained if the pre-formed secondary structure of helix 6 were important for binding to α. A correctly pre-formed structure has been shown before to greatly enhance binding affinity (42). In addition, it is not clear how much the binding affinities are affected by the generation of charged N- and C-termini in the peptide and in τC16Δ11, respectively, which are not present in the τC16 domain.
Interestingly, most of the conserved amino acid residues between Ala614 and Phe630 are hydrophobic and spaced along the amino acid sequence in a manner such that they could lie on the same face of an amphipathic α-helix (Figure 1). This suggests that these residues form a helix upon binding to α (24). A recent report showed that the point mutations R557C and P599L in full-length τ affect the interaction with α (43). Yet, the side chains of these residues are buried with, respectively, 15 and 2% solvent accessibility in the solution structure of τC14. This suggests that these mutations affect the interaction with α indirectly by disrupting the structure of τ.
The close proximity of the N- and C-termini in τC14 presents a restraint for the maximum spatial separation between the two α subunits in the replisome that simultaneously replicate the leading and lagging DNA strands. First, the N-terminal domains of the τ subunits are located close together in the natural δτ2γδ′ clamp loader complex (44). Second, the monomeric protein formed by the C-terminal 213 residues of τ binds to DnaB with a dissociation constant greater than 1 μM (20), which is too weak to maintain the complex under physiological conditions. DnaB is, however, a hexamer and binding to tetrameric full-length τ was reported to be much tighter (23). Since the replisome contains two τ molecules but only a single DnaB hexamer (25), both τ proteins are available for binding to the same DnaB hexamer. In view of the results that (i) the DnaB-binding site has been mapped to Domain IVa (residues 430–496) of τ (23), (ii) the structured part of Domain V starts at residue Leu509, and (iii) Domain V is rigidly attached to α, the complex consisting of DnaB, two τ and two α subunits could be structurally quite well defined. This would bring the α subunits into close proximity, in particular as both the DnaB hexamer (310 kDa) and α (130 kDa) are large proteins. These proposals are further supported by an atomic force microscopy study of the DnaB–τ interaction in Bacillus, where two τ subunits seem to bind to two neighboring C-terminal hexamerization domains of DnaB (45).
While the residues of Domain IVa were all mobile in the construct comprising Domains IVa and V (τC22), significant parts of Domain IVa (which is only 69 residues long) would presumably be immobilized by the interaction with DnaB. Further immobilization may result from interactions with DNA which depend on the positively charged Domain IVa rather than the overall negatively charged Domain V (24). The E. coli τ segment connecting the C-terminus of Domain III (Pro368) with the N-terminus of Domain IVa (Lys430), however, comprises a linker which could be designed for flexibility. The amino acid sequence of this linker contains eleven proline residues between residues 370 and 410 and no regular secondary structure is predicted for this segment. Therefore, τ may connect the clamp–loader complex with DnaB via a long flexible linker, whereas the relative orientations of DnaB and the α subunits are more rigidly defined.
SUPPLEMENTARY DATA
Supplementary Data is available at NAR Online.
ACKNOWLEDGEMENTS
We thank Dr Kiyoshi Ozawa for initial sample characterization by selective 15N-labeling and TOCSY spectra. The project and the purchase of the 800 MHz NMR spectrometer were supported by grants from the Australian Research Council. GO acknowledges an ARC Federation Fellowship. Access to the NMR Facility at the Australian National University (ANU) is is gratefully acknowledged. Funding to pay the Open Access publication charge was provided by ANU.
Conflict of interest statement. None declared.
REFERENCES
- 1.Lamers M.H., Georgescu R.E., Lee S.G., O’Donnell M., Kuriyan J. Crystal structure of the catalytic α subunit of E. coli replicative DNA polymerase III. Cell. 2006;126:881–892. doi: 10.1016/j.cell.2006.07.028. [DOI] [PubMed] [Google Scholar]
- 2.Kong X.-P., Onrust R., O’Donnell M., Kuriyan J. Three-dimensional structure of the β subunit of E. coli DNA polymerase III holoenzyme: a sliding DNA clamp. Cell. 1992;69:425–437. doi: 10.1016/0092-8674(92)90445-i. [DOI] [PubMed] [Google Scholar]
- 3.Hamdan S., Carr P.D., Brown S.E., Ollis D.L., Dixon N.E. Structural basis for proofreading during replication of the Escherichia coli chromosome. Structure. 2002;10:535–546. doi: 10.1016/s0969-2126(02)00738-4. [DOI] [PubMed] [Google Scholar]
- 4.Mueller G.A., Kirby T.W., DeRose E.F., Li D., Schaaper R.M., London R.E. Nuclear magnetic resonance solution structure of the Escherichia coli DNA polymerase III θ subunit. J. Bacteriol. 2005;187:7081–7089. doi: 10.1128/JB.187.20.7081-7089.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guenther B., Onrust R., Sali A., O’Donnell M., Kuriyan J. Crystal structure of the δ′ subunit of the clamp–loader complex of E. coli DNA polymerase III. Cell. 1997;91:335–345. doi: 10.1016/s0092-8674(00)80417-1. [DOI] [PubMed] [Google Scholar]
- 6.Jeruzalmi D., O’Donnell M., Kuriyan J. Crystal structure of the processivity clamp loader gamma (γ) complex of E. coli DNA polymerase III. Cell. 2001;106:429–441. doi: 10.1016/s0092-8674(01)00463-9. [DOI] [PubMed] [Google Scholar]
- 7.Kazmirski S.L., Podobnik M., Weitze T.F., O’Donnell M., Kuriyan J. Structural analysis of the inactive state of the Escherichia coli DNA polymerase clamp–loader complex. Proc. Natl. Acad. Sci. U.S.A. 2004;101:16750–16755. doi: 10.1073/pnas.0407904101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jeruzalmi D., Yurieva O., Zhao Y., Young M., Stewart J., Hingorani M., O’Donnell M., Kuriyan J. Mechanism of processivity clamp opening by the delta subunit wrench of the clamp loader complex of E. coli DNA polymerase III. Cell. 2001;106:417–428. [PubMed] [Google Scholar]
- 9.Gulbis J.M., Kazmirski S.L., Finkelstein J., Kelman Z., O’Donnell M., Kuriyan J. Crystal structure of the χ:Ψ subassembly of the Escherichia coli DNA polymerase clamp–loader complex. Eur. J. Biochem. 2004;271:439–449. doi: 10.1046/j.1432-1033.2003.03944.x. [DOI] [PubMed] [Google Scholar]
- 10.Pintacuda G., Park A.Y., Keniry M.A., Dixon N.E., Otting G. Lanthanide labeling offers fast NMR approach to 3D structure determinations of protein-protein complexes. J. Am. Chem. Soc. 2006;128:3696–3702. doi: 10.1021/ja057008z. [DOI] [PubMed] [Google Scholar]
- 11.Bailey S., Wing R.A., Steitz T.A. The structure of T. aquaticus DNA polymerase III is distinct from eukaryotic replicative DNA polymerases. Cell. 2006;126:893–904. doi: 10.1016/j.cell.2006.07.027. [DOI] [PubMed] [Google Scholar]
- 12.Flower A.M., McHenry C.S. The γ subunit of DNA polymerase III holoenzyme of Escherichia coli is produced by ribosomal frameshifting. Proc. Natl. Acad. Sci. U.S.A. 1990;87:3713–3717. doi: 10.1073/pnas.87.10.3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsuchihashi Z., Kornberg A. Translational frameshifting generates the γ subunit of DNA polymerase III holoenzyme. Proc. Natl. Acad. Sci. U.S.A. 1990;87:2516–2520. doi: 10.1073/pnas.87.7.2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao D., McHenry C.S. τ binds and organizes Escherichia coli replication proteins through distinct domains. Domain III, shared by γ and τ binds δδ′ and χΨ. J. Biol. Chem. 2001;276:4447–4453. doi: 10.1074/jbc.M009827200. [DOI] [PubMed] [Google Scholar]
- 15.Glover B.P., Pritchard A.E., McHenry C.S. τ binds and organizes Escherichia coli replication proteins through distinct domains. Domain III, shared by γ and τ, oligomerizes DnaX. J. Biol. Chem. 2001;276:35842–35846. doi: 10.1074/jbc.M103719200. [DOI] [PubMed] [Google Scholar]
- 16.Studwell-Vaughan P.S., O’Donnell M. Constitution of the twin polymerase of DNA polymerase III holoenzyme. J. Biol. Chem. 1991;266:19833–19841. [PubMed] [Google Scholar]
- 17.Onrust R., Finkelstein J., Naktinis V., Turner J., Fang L., O’Donnell M. Assembly of a chromosomal replication machine: two DNA polymerases, a clamp loader, and sliding clamps in one holoenzyme particle. I. Organization of the clamp loader. J. Biol. Chem. 1995;270:13348–13357. doi: 10.1074/jbc.270.22.13348. [DOI] [PubMed] [Google Scholar]
- 18.Kim S., Dallman H.G., McHenry C.S., Marians K.J. τ couples the leading- and lagging-strand polymerases at the Escherichia coli DNA replication fork. J. Biol. Chem. 1996;271:21406–21412. doi: 10.1074/jbc.271.35.21406. [DOI] [PubMed] [Google Scholar]
- 19.Yuzhakov A., Turner J., O’Donnell M. Replisome assembly reveals the basis for asymmetric function in leading and lagging strand replication. Cell. 1996;86:877–886. doi: 10.1016/s0092-8674(00)80163-4. [DOI] [PubMed] [Google Scholar]
- 20.Dallmann H.G., Kim S., Pritchard A.E., Marians K.J., McHenry C.S. Characterization of the unique C terminus of the Escherichia coli τ DnaX protein. Monomeric C-τ binds α and DnaB and can partially replace τ in reconstituted replication forks. J. Biol. Chem. 2000;275:15512–15519. doi: 10.1074/jbc.M909257199. [DOI] [PubMed] [Google Scholar]
- 21.Leu F.P., Georgescu R., O’Donnell M. Mechanism of the E. coli τ processivity switch during lagging-strand synthesis. Mol. Cell. 2003;11:315–327. doi: 10.1016/s1097-2765(03)00042-x. [DOI] [PubMed] [Google Scholar]
- 22.Gao D., McHenry C.S. τ binds and organizes Escherichia coli replication proteins through distinct domains. Partial proteolysis of terminally tagged τ to determine candidate domains and to assign domain V as the α binding domain. J. Biol. Chem. 2001;276:4433–4440. doi: 10.1074/jbc.M009828200. [DOI] [PubMed] [Google Scholar]
- 23.Gao D., McHenry C.S. τ binds and organizes Escherichia coli replication proteins through distinct domains. Domain IV, located within the unique C terminus of τ, binds the replication fork helicase, DnaB. J. Biol. Chem. 2001;276:4441–4446. doi: 10.1074/jbc.M009830200. [DOI] [PubMed] [Google Scholar]
- 24.Jergic S, Ozawa K, Williams NK, Su X-C, Scott DD, Hamdan SM, Crowther JA, Otting G, Dixon NE. 2006. The unstructured C-terminus of the τ subunit of Escherichia coli DNA polymerase III holoenzyme is the site of interaction with the α subunit. Nucleic Acids Res. accompanying paper. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Benkovic S.J., Valentine A.M., Salinas F. Replisome-mediated DNA replication. Annu. Rev. Biochem. 2001;70:181–208. doi: 10.1146/annurev.biochem.70.1.181. [DOI] [PubMed] [Google Scholar]
- 26.Wijffels G., Dalrymple B.P., Prosselkov P., Kongsuwan K., Epa V.C., Lilley P.E., Jergic S., Buchardt J., Brown S.E., et al. Inhibition of protein interactions with the β2 sliding clamp of Escherichia coli DNA polymerase III by peptides from β2-binding proteins. Biochemistry. 2004;43:5661–5671. doi: 10.1021/bi036229j. [DOI] [PubMed] [Google Scholar]
- 27.Cornilescu G., Delaglio F., Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 28.Güntert P., Mumenthaler C., Wüthrich K. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 1997;273:283–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- 29.Neri D., Szyperski T., Otting G., Senn H., Wüthrich K. Stereospecific nuclear magnetic resonance assignments of the methyl groups of valine and leucine in the DNA-binding domain of the 434 repressor by biosynthetically directed fractional 13C labeling. Biochemistry. 1989;28:7510–7516. doi: 10.1021/bi00445a003. [DOI] [PubMed] [Google Scholar]
- 30.Wu J., Fan J., Pascal S.M., Yang D. General method of suppression of diagonal peaks in heteronuclear-edited NOESY spectroscopy. J. Am. Chem. Soc. 2004;126:15018–15019. doi: 10.1021/ja045300l. [DOI] [PubMed] [Google Scholar]
- 31.Farrow N.A., Muhandiram R., Singer A.U., Pascal S.M., Kay C.M., Gish G., Shoelson S.E., Pawson T., Forman-Kay J.D., et al. Backbone dynamics of a free and a phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry. 1994;33:5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- 32.Bartels C., Xia T., Güntert P., Billeter M., Wüthrich K. The program XEASY for computer-supported NMR spectral analysis. J. Biomol. NMR. 1995;5:1–10. doi: 10.1007/BF00417486. [DOI] [PubMed] [Google Scholar]
- 33.Luginbühl P., Güntert P., Billeter M., Wüthrich K. The new program OPAL for molecular dynamics simulations and energy refinements of biological macromolecules. J. Biomol. NMR. 1996;8:136–146. doi: 10.1007/BF00211160. [DOI] [PubMed] [Google Scholar]
- 34.Laskowski R.A., Rullmann J.A.C., MacArthur M.W., Kaptein R., Thornton J.M. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 35.Koradi R., Billeter M., Wüthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 36.Sevilla-Sierra P., Otting G., Wüthrich K. Determination of the nuclear magnetic resonance structure of the DNA-binding domain of the P22 c2 repressor (1 to 76) in solution and comparison with the DNA-binding domain of the 434 repressor. J. Mol. Biol. 1994;235:1003–1020. doi: 10.1006/jmbi.1994.1053. [DOI] [PubMed] [Google Scholar]
- 37.Su XC, Jergic S, Ozawa K, Burns ND, Dixon NE, Otting G. J. Biomol. NMR. 2007. Measurement of dissociation constants of high-molecular weight protein-protein complexes by transferred 15N-relaxation. In press. [DOI] [PubMed] [Google Scholar]
- 38.Holm L., Sander C. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 39.Grishin N.V. KH domain: one motif, two folds. Nucleic Acids Res. 2001;29:638–643. doi: 10.1093/nar/29.3.638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215: 403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 41.Wu P.S.C., Ozawa K., Jergic S., Su X.-C., Dixon N.E., Otting G. Amino acid type identification in 15N-HSQC spectra by combinatorial selective 15N-labelling. J. Biomol. NMR. 2006;34:13–21. doi: 10.1007/s10858-005-5021-9. [DOI] [PubMed] [Google Scholar]
- 42.Liepinsh E., Berndt K.D., Sillard R., Mutt V., Otting G. Solution structure and dynamics of PEC-60, a protein of the Kazal-type inhibitor family, determined by nuclear magnetic resonance. J. Mol. Biol. 1994;239:137–153. doi: 10.1006/jmbi.1994.1356. [DOI] [PubMed] [Google Scholar]
- 43.Pham P.T., Zhao W., Schaaper R.M. Mutator mutants of Escherichia coli carrying a defect in the DNA polymerase III τ subunit. Mol. Microbiol. 2006;59:1149–1161. doi: 10.1111/j.1365-2958.2005.05011.x. [DOI] [PubMed] [Google Scholar]
- 44.Bullard J.M., Pritchard A.E., Song M.-S., Glover B.P., Wieczorek A., Chen J., Janjic N., McHenry C.S. A three-domain structure for the δ subunit of the DNA polymerase III holoenzyme. δ domain III binds δ′ and assembles into the DnaX complex. J. Biol. Chem. 2002;277:13246–13256. doi: 10.1074/jbc.M108708200. [DOI] [PubMed] [Google Scholar]
- 45.Haroniti A., Anderson C., Doddridge Z., Gardiner L., Roberts C.J., Allen S., Soultanas P. The clamp–loader-helicase interaction in Bacillus. Atomic force microscopy reveals the structural organisation of the DnaB-τ complex in Bacillus. J. Mol. Biol. 2004;336:381–393. doi: 10.1016/j.jmb.2003.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.