Abstract
enod40 is a plant gene that participates in the regulation of symbiotic interaction between leguminous plants and bacteria or fungi. Furthermore, it has been suggested to play a general role in non-symbiotic plant development. Although enod40 seems to have multiple functions, being present in many land plants, the molecular mechanisms of its activity are unclear; they may be determined though, by short peptides and/or RNA structures encoded in the enod40 genes. We utilized conserved RNA structures in enod40 sequences to search nucleotide sequence databases and identified a number of new enod40 homologues in plant species that belong to known, but also, to yet unknown enod40-containing plant families. RNA secondary structure predictions and comparative sequence analysis of enod40 RNAs allowed us to determine the most conserved structural features, present in all known enod40 genes. Remarkably, the topology and evolution of one of the conserved structural domains are similar to those of the expansion segments found in structural RNAs such as rRNAs, RNase P and SRP RNAs. Surprisingly, the enod40 RNA structural elements are much more stronger conserved than the encoded peptides. This finding suggests that some general functions of enod40 gene could be determined by the encoded RNA structure, whereas short peptides may be responsible for more diverse functions found only in certain plant families.
INTRODUCTION
While a majority of land plants are able to enter an endosymbiotic programme with mycorrhizal fungi (1–3), root nodule symbiosis is almost strictly confined to legumes and a few non-legumes that interact with rhizobia and other nitrogen-fixing bacteria (4,5). In both cases, specific signalling pathways activate, establish and maintain the symbiotic plant–microbe programme (6–9). The soyabean enod40 gene was initially identified as one of the plant genes that are expressed during the early stages of the formation of nitrogen-fixing root nodules in the symbiotic association of legumes with soil rhizobial bacteria (10,11). It is also activated in roots colonized by fungi forming phosphate-acquiring arbuscular mycorrhizae (12).The enod40 gene is present in all legumes studied so far, and is also found in many non-legume plants [reviewed in (13)].
In both legumes and non-legumes, various experiments have demonstrated enod40 expression to be important in nodule organogenesis and development [e.g. (14–25)]. The data accumulated so far on the biological effects of enod40 suggest that this gene may have multiple functions that are not restricted to the regulation of symbiosis. However, the molecular mechanisms of its activity are unclear. The enod40 genes lack long open reading frames (ORFs), but encode for short conserved peptides which were shown to be functional (26,27). The soyabean Enod40 peptides bind to sucrose synthase, suggesting a role in the regulation of sucrose utilization in nodules (27). The analysis of enod40 sequences and RNA secondary structures from various plants also depicts a role for enod40 as a regulatory RNA (14,26,28–30). This role is supported by experiments in alfalfa roots which showed that deletion of RNA structural elements in a mutated enod40 gene, while retaining proper translation, decreased enod40 activity with respect to stimulation of cortical cell division (26). Furthermore, an alfalfa enod40 RNA-binding protein MtRBP1 was isolated and found to co-localize with enod40 RNA in cytoplasmic granules during nodulation (31). MtRBP1 and its homologues possess an RNA recognition motif (RRM), but the binding sites in enod40 RNA have not yet been identified (31).
A comparative analysis of possible enod40 RNA structures (29) suggests that the presence of some structural domains correlates with the plant's ability to form nitrogen-fixing root nodules. While part of the enod40 structure seems to be well conserved in several plant families, certain domains are typical for legumes, the only group able to develop root nodule symbioses with rhizobia. Furthermore, a structured domain conserved in enod40 RNA of leguminous plants forming indeterminate nodules is completely eliminated in plants forming determinate nodules. In general, the non-legume enod40 RNAs seem to be less structured as compared to those of legumes (29).
The presence of strongly conserved RNA structural elements may be used to increase the efficiency of database mining for un-annotated enod40 homologues. The nucleotide sequence similarity in distantly related species is rather low, and only two high sequence similarity regions (named region I and region II) have been revealed [e.g. (13,17,32)]. While the most conserved short ORF I is encoded by region I, the highest conservation at the nucleotide level is observed in the short region II, where no conserved peptides can be proposed (13). On the other hand, the core of region II is flanked by previously identified (29) conserved RNA secondary structure elements. In this work, we have used this feature to search for unidentified enod40 orthologs in nucleotide sequence databases, in particular, in the GenBank database of expressed sequence tags (ESTs). This allowed us to extend considerably the number of known enod40-possessing non-legume families and species. Furthermore, the analysis of possible RNA secondary structures reveals structural elements that are conserved in enod40 RNAs across the plant kingdom. A comparison of the predicted structures suggests that the evolution of one of the conserved domains resembles that of expansion segments that are found in some structural RNAs.
RESULTS
The sequential application of sequence similarity searches and RNA structure predictions (see Materials and methods section) allowed us to identify a number of enod40-like sequences in various angiosperm species. The list of non-leguminous enod40 homologues found in GenBank at the time of writing is given in Table 1, together with nucleotide positions of conserved structural domains and ORFs. In the case of multiple enod40 EST sequences with minor variations, produced by large-scale sequencing projects for some species, we selected only one representative. In addition to the recent enod40 sequence compilation (13), we have found 22 new enod40 homologues. In particular, we discovered putative enod40 genes in another five plant families: Myrtales, Malvales, Brassicales, Apiales and Gentianales.
Table 1.
Nucleotide positions of conserved secondary structures and sORF I in the non-legume enod40 homologues
| Species | Family | sORF I | Domain 2 (size) | Domain 3 | Accession | enod40 annotationa |
|---|---|---|---|---|---|---|
| Prunus armeniaca | Rosales | 80–127 | 179–263 (85) | 287–302 | CV047471 | (13) |
| Malus x domestica | (<1)–24 | 79–188 (110) | 211–228 | CN917334 | This work | |
| Casuarina glauca | Fagales | 61–210 | 179–256 (78) | 281–299 | AJ487686 | (33) |
| Betula pendula | 14–163 | 124–196 (73) | 219–236 | CD271081 | This work | |
| Populus tremula | Malpighiales | 54–98 | 155–256 (102) | 279–296 | BU883953 | (13) |
| Euphorbia tirucalli | ns | 8–117 (110) | 142–157 | BP953888 | This work | |
| Bruguiera gymnorrhiza | 77–115 | 181–251 (71) | 277–294 | BP941035 | This work | |
| Manihot esculenta | 53–91 | 144–221 (78) | 244–262 | CK643649 | This work | |
| Eucalyptus gunnii | Myrtales | 38–76 | 144–248 (105) | 272–294 | CT987303 | This work |
| Gossypium hirsutum | Malvales | 83–115 | 185–288 (104) | 314–335 | DN804042 | This work |
| Citrus sinensis | Sapindales | 14–52 | 114–209 (96) | 232–250 | BQ624698 | (13) |
| Citrus unshiu | 35–73 | 138–233 (96) | 256–274 | C95533 | (13) | |
| Arabidopsis thaliana | Brassicales | – | 144–185 (42) | 208–226 | AK220907 | This work |
| Thlaspi caerulescens | – | 81–152 (72) | 175–198 | DN923678 | This work | |
| Brassica napus | – | 120–195 (76) | 218–236 | CX190651 | This work | |
| Daucus carotab | Apiales | – | 127–265 (139) | 286–305 | BI452209 | This work |
| Helianthus annuus | Asterales | – | 132–174 (43) | 198–215 | CD856145 | This work |
| Senecio aethnensis | – | 131–175 (45) | 199–216 | DY662668 | This work | |
| Lactuca sativa | – | 137–187 (51) | 211–228 | DW143889 | (13) | |
| Taraxacum officinale | – | 136–188 (53) | 212–229 | DY838401 | This work | |
| Lycopersicon esculentum | Solanales | 3282–3314 | 3485–3539 (55) | 3565–3583 | AY388519 | (21) |
| Solanum tuberosum | 98–130 | 305–385 (81) | 411–429 | CV503956 | (13) | |
| Nicotiana tabacum | 69–101 | 171–290 (120) | 313–331 | X98716 | (34) | |
| N. langsdorfii × N. sanderae | 72–104 | 170–264 (95) | ns | EB694790 | This work | |
| Antirrhinum majus | Lamiales | (<1)–15 | 80–186 (107) | 209–225 | AJ559999 | (13) |
| Plantago major | 147–179 | 232–288 (57) | 311–326 | AM156924 | This work | |
| Hedyotis terminalis | Gentianales | (<1?)–65 | 134–259 (126) | 281–299 | CB080316 | This work |
| Oryza sativa (1) | Poales | 2256–2294 | 2361–2405 (45) | 2431–2455 | AB024054 | (17) |
| Oryza sativa (2) | 29–67 | 151–225 (75) | 248–271 | AU101849 | (13) | |
| Oryza branchyantha | 2687–2725 | 2796–2837 (42) | 2864–2887 | AB024055 | (17) | |
| Zea mays (1) | 58–96 | 152–207 (56) | 229–252 | CD990776 | (13,35) | |
| Zea mays (2) | 82–123 | 221–314 (94) | 337–360 | DN209550 | (13,35) | |
| Sorghum bicolor | 95–136 | 229–308 (80) | 331–354 | BE362667 | (13) | |
| Saccharum officinarum | 54–92 | 149–202 (54) | 225–248 | CA155599 | This work | |
| Lolium perenne | 93–131 | 115–234 (120) | 267–290 | AF538350 | (36) | |
| Festuca arundinacea | (<1)–37 | 21–140 (120) | 173–196 | DT701589 | This work | |
| Hordeum vulgare | 13–51 | 37–156 (120) | 196–219 | AF542513 | (36) | |
| Leymus chinensis | ns | ns | 38–61 | CN465797 | This work | |
| Triticum aestivum | 41–79 | 63–167 (105) | 200–223 | BJ278615 | (13,35) | |
| Avena sativa | ns | ns | 63–86 | CN815024 | This work | |
| Brachypodium distachyon | 17–55 | 39–158 (120) | 191–214 | DV479239 | This work |
ns, not sequenced; ?, sequence uncertainty.
aenod40 annotation references. Domain nomenclature is from (29). Previously unknown enod40-containing families are shown in bold font.
bEST from the extra-radical mycelium of fungus Glomus intraradices during arbuscular mycorrhizal symbiosis with Daucus carota.
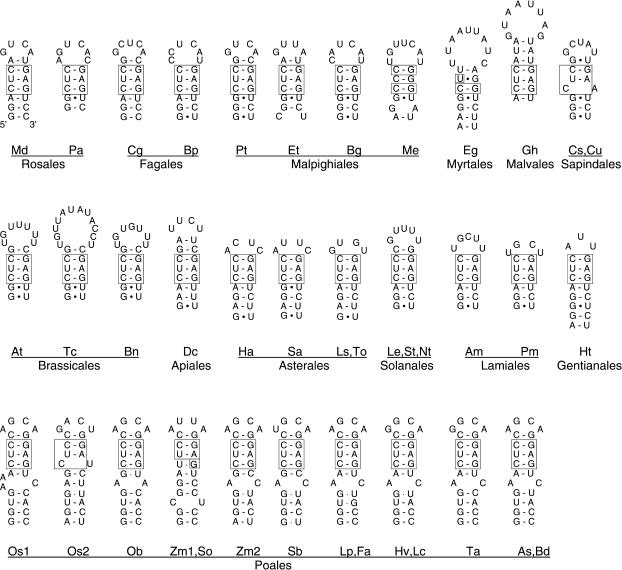
Despite the relatively low global sequence similarity in some cases, the suggested enod40 assignments are strongly supported by the presence of similar secondary structures. The deduced global enod40 structure is shown schematically in Figure 1. The most conserved structure is domain 3 [nomenclature of (29)], represented by a relatively small, though stable, hairpin found in all (putative) enod40 RNAs. Figure 2 shows these hairpins in non-legume sequences [legume analogues are described in (29)], with the nucleotide positions given in Table 1. The hairpins consist of 5–9 bp, sometimes interrupted by a mismatch. All hairpins are located in the 3′-proximal part of region II, where many base covariations are observed and sequence diversity is increased as compared to the 5′-end of the region. The hairpins are located at similar positions downstream of the conserved region II core (13) and are easily found by eye inspection of the alignment in this region, even without using an RNA folding program. The only sequence motif, present in almost all hairpins, is a 3-bp unit [CUC/GAG] in the middle of the stem. With some deviations, the motif is found in all hairpins. A similar pattern is observed in homologous legume structures (29).
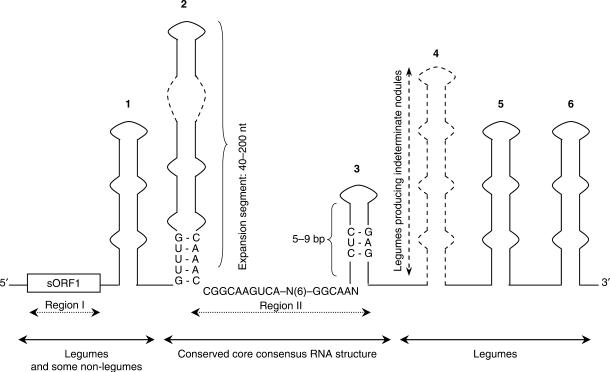
Figure 1.
Conserved structures and sequence motifs in enod40 RNAs. Domains 1–6 are shown schematically and not in scale, domain numbering is from (29). The locations of sORF1 and regions I and II (dashed double arrows) are also shown. Double arrows indicate the extent of conservation of particular structures. Some deviations from the shown consensus sequences are possible.
Figure 2.
The hairpins of the enod40 domain 3 [nomenclature from (29)] from various enod40 homologues. Nucleotide positions are given in Table 1. The sequences correponding to the CUC/GAG motif are boxed. For A. thaliana, a genomic sequence is given—it differs from GenBank entry AK220907 by one substitution in the loop of hairpin 3. Species names are abbreviated by the first two characters, for complete names see Table 1.
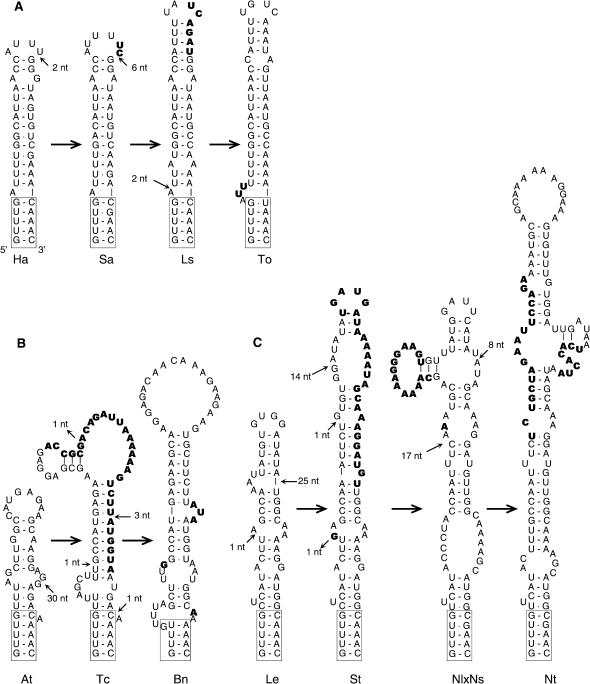
Domain 2 is more variable compared to domain 3. While the 3′-ends of the predicted domain 2 are all located at similar positions just upstream of the region II, the size of this structure varies in a broad range of 40–140 nt (Table 1). Nevertheless, the shape of the domain is strongly conserved in all putative enod40 sequences: it is an extended stem-loop structure, sometimes (in larger domains) with branching in the interior (Figure 3). Similar structures can also be folded in legume enod40 RNAs, typically with a size of 120–135 nt, which are sometimes extended in paralogous genes (29). Bearing in mind a very high diversity of enod40 sequences in this region, such a conserved shape is remarkable. Interestingly, the majority of structures contain rather similar paired sequences GUUUG and CAAAC, or their minor variations (examples are shown in Figure 3), preserving the pairing at the very ends, while interior sequences from different families are not similar and difficult to align.
Figure 3.
Examples of enod40 domain 2 evolution in families Asterales (A), Brassicales (B) and Solanales (C). Nucleotide positions are given in Table 1. The conserved closing stem is boxed. The insertion locations are indicated by small arrows, inserted nucleotides are in different letter font. Large arrows indicate the transitions between structures of various species determined by insertions (but they should not always correspond to real evolutionary events that may occur in reverse order or include branching). Species names are abbreviated by first two characters, for complete names see Table 1.
Within plant families, conservation of terminal sequences and structures is extended further inside domain 2, whereas the interior parts are more variable (multiple alignment of enod40 sequences is shown in Figure S1 of Supplementary Data). This variation originates from frequent insertions that occur predominantly in the loops. Examples of such domain 2 evolution within a family are shown in Figure 3 for Asterales, Brassicales and Solanales. For instance, comparison of four Solanales enod40 sequences (tomato, potato and two tobacco species) shows a gradual increase of the domain size from 55 to 120 nt while the sequence at the lowest part of the structure remains almost unchanged. Similar types of insertions are found in enod40 RNAs from other families as well (not shown). Interestingly, the same type of domain 2 extension occurs in some paralogous enod40 genes of legumes. For instance, in Lotus japonicus, domain 2 is extended from 132 nt in enod40-1 gene to 176 nt in the enod40-2 (29). A comparison of alignments for Trifolium repens enod40 sequences with structural predictions (20,29) indicates an insertion of 70 nt in the interior of enod40-3 domain 2 as compared to the homologous structure in enod40-1, resulting in a domain expansion from 135 to 205 nt. Such a remarkable pattern of domain 2 extensions, observed in enod40 RNA from various plant families, is similar to the evolution of expansion segments in ribosomal RNAs, SRP and RNase P RNAs (37–39).
Domain 1, located upstream of the domain 2, has been previously predicted by various algorithms in a number of enod40 RNAs (26,28,29). It is conserved in legumes and represented by a stem-loop structure of variable length, usually with a purine-rich 5′-half and a pyrimidine-rich 3′-half, resulting in possible ‘flipping’ of base pairs. While we could putatively locate this structure in the majority of enod40 genes (not shown), in some of them the presence of alternative structures and poor conservation of sequences hampered accurate assignment of the domain borders. For instance, in the absence of sufficient sequence data, the previous analysis of enod40 RNA structures (29) apparently misinterpreted partial predictions for domain 2 in Hordeum vulgare and Lolium perenne sequences as putative domain 1 structures. Furthermore, in some of the RNAs, this part seems to be located in EST regions that are not reliably determined or not sequenced at all.
Upstream of domain 1, we could not reliably predict any conserved secondary structure. This region corresponds to the high-similarity region I containing translatable sORFs (17,18,21,26,27,32) and apparently evolves without strong secondary structure constraints. Also, similar to our previous conclusions (29), we could not detect any structure downstream of domain 3, that might be conserved across both leguminous and non-leguminous plants.
DISCUSSION
We have reliably identified the presence of two conserved secondary structure domains, common to both leguminous and non-leguminous plants in a rather large number of enod40 homologues from various angiosperm plant families. Named according to the previously used nomenclature (29) as domains 2 and 3, these two structures flank the core of region II which has the highest level of sequence similarity shared by enod40 genes (13,17,32). The sequences within the secondary structure domains are more diverse than the conserved spacer between them (Figure 1). Despite this diversity, the structural features of domains 2 and 3 are absolutely conserved in all currently found enod40 homologues.
The most frequently recurring motifs in double-stranded regions, both GUUUG/CAAAC at the bottom of domain 2 and CUC/GAG in the stem of the domain 3 hairpin (Figures 1–3), allow some deviations. Occurrence of the domain 2 motif seems to be different in eudicots and monocots, the latter represented by Poales. The motif is very conserved in eudicots, albeit sometimes with substitutions disrupting 2 bp at most, with a maximum of 3 nt changed, in Euphorbia tirucalli (UUUUG/CGGAC) and Casuarina glauca (CAUUG/CAAAU). In Poales, the motif, with some variations, is present in only four of the enod40 homologues found (two Zea mays genes, Sorghum bicolor and Saccharum officinarum) and the perfect combination is in one of the rice genes, enod40-2 (AU101849). In other Poales species, the motif seems to be lost.
The topology and evolution of the enod40 domain 2 resemble those of expansion segments, also called divergent or D-domains, which are well known for eukaryotic ribosomal RNAs (37,40–42) and have recently been discovered in signal recognition particle (SRP) RNA and RNase P (38,39). Similar to these structures, the domain 2 is an extended stem-loop structure of variable length with more diverse distal sequences as compared to the stems closing the domain. However, some differences either in an expansion mechanism or constraints imposed by secondary structure may exist. In rRNA expansion segments, domain elongation was suggested to proceed mostly via a compensatory slippage mechanism, with sequential duplications of short low complexity sequences (e.g. nucleotide repeats) on one of the helix strands during DNA replication, accompanied by similar compensatory changes on the opposite strand to restore helix symmetry (43–45). Such a mechanism explains the frequent occurrence of insertion ‘indels’ on both strands in the middle of a stem-loop structure (45–47). We did not find any examples of such internal insertions of stem modules in enod40 domain 2: when large insertions occurred in the middle of one of the strands, they were compensated simply by point substitutions on the other, frequently accompanied by opening of some stems and building new ones to preserve the shape, like in Brassicales and Solanales (Figure 3).
In addition to the replication slippage mechanism, other mechanisms of the length increase of expansion segments are possible as well. Particularly, the insertion of large sequences in some rRNA expansion segments was suggested to originate from the (quasi)palindromic character of sequences leading to the formation of stem-loop structures in one of the DNA strands during replication and hence to incorrect copying of the template strand (47,48). Such models may explain large insertions observed in the enod40 expansion domains (e.g. Figure 3).
In some of the rRNA expansion segments (46) and leguminous enod40 domains (29), a relatively frequent occurrence of U-rich bulges and internal loops has been observed. In rRNA, this has been associated with a slippage-like mechanism of helix-length increase leading to frequent ‘leftover’ bulged Us, in particular, in sequences with biased nucleotide composition (46). In leguminous enod40, such bulges and loops seem to play a functional role, because their positions in domain 2 are conserved and there is an additional domain with conserved U-containing loops in molecules from species forming a specific type of nodules, namely indeterminate nodules (29). Although enod40 domain 2 of non-legumes seem to expand in the same way as legume structures, U-containing loops are less frequent and their positions are variable. We did not notice any other statistically significant bias in nucleotide composition of loops in non-legume enod40 domain 2.
The function of expansion segments in structural RNAs is not clear. One of the hallmarks of their secondary structure––conserved terminal pairings embracing self-contained internal structure––apparently allows their hypervariability to be compatible with conserved functional cores of RNA molecules. This has led to the suggestion that in rRNA they do not have any function and are only tolerated because their elongation does not disrupt any functional domain (49). On the other hand, some of their structural features seem to be important for the biogenesis and stability of rRNA (50–52). Correlations between sequences and sizes of various rRNA expansion segments indicate possible functional relationships between them (53,54). Size correlations are also observed for RNase P variable domains (39). The size of enod40 expansion domain seems to weakly correlate with the plant's ability to form nitrogen-fixing root nodules: in legume, the domain is typically 123–135 nt (29) while in non-legumes, with some exceptions, it is usually smaller (Table 1). Similar to rRNA expansion segments, possible functions of the enod40 domain 2 may include stabilization of RNA structure and/or interactions with other enod40 domains or molecules.
In contrast to the expansion segments of rRNAs, RNase P and SRP RNAs, the enod40 expansion domain 2 does not seem to be inserted into a conserved structural core, but is located upstream of conserved sequence motif. Whatever the function of the enod40 RNA structure, the overall configuration of secondary structure elements in enod40 RNA (Figure 1) is more conserved than the encoded sORFs. The most conserved sORF is located in region I (sORF I) and encodes a short peptide of 10–15 amino acids shown to be translated in several species (17,18,21,26,27,32). Although the homologous ORF is found in almost all enod40 genes where the appropriate region is sequenced, there are a number of exceptions (Table 1). The first is a deletion of one nucleotide in both Fagales enod40 RNAs (C. glauca and Betula pendula), leading to far longer encoded peptides due to a frameshift. Nevertheless, a major part of the characteristic peptide motif is present. The second, which is more puzzling, is the complete absence of this motif in all enod40 homologues from Brassicales, Apiales and Asterales families. Among all possible reading frames in these sequences, we could not identify any that would be similar to known enod40 sORF I sequences. On the other hand, the suggested enod40 assignments are supported by the presence of typical enod40 RNA structures (Figures 2 and 3). In case of the Arabidopsis thaliana enod40 homologue, the cDNA sequence (AK220907) is also validated by genomic BLAST comparison showing only two substitutions in the transcript, which are neutral for the proposed structure model.
Probably, for some of the enod40 functions the secondary structure of domains 2 and 3 is absolutely required, while conserved peptide sequences are needed for other purposes and the constraints to preserve them may be released in some species. For instance, the absence of conserved sORFs in Brassicales enod40 sequences (A. thaliana, Thlaspi caerulescens and Brassica napus) could be related to the fact that, in contrast to the majority of angiosperms, these species in natural environment do not form effective symbiotic mycorrhizal associations with fungi (3,55). Mycorrhizal symbioses are probable evolutionary predecessors of nitrogen-fixing nodule symbioses (7,8), and enod40 seems to be involved in both (12,19). There is a precedent of lower similarity of an A. thaliana homologue of multifunctional protein required for symbiotic nodule development: the A. thaliana calcium/calmodulin-dependent protein kinase is different from related proteins, presumably because of the nonmycotrophic character of Arabidopsis (56). On the other hand, it is more difficult to explain our failure to find any trace of the conserved enod40 sORFs in sequences from Apiales and Asterales (Table 1): these plants can form arbuscular mycorrhiza (3). Moreover, according to the database annotation, the EST-encoding putative Daucus carota enod40 (BI452209) was isolated from a fungus extra-radical mycelium during arbuscular mycorrhizal symbiosis with the plant. Of course, it is also possible that in some of available ESTs, the 5′-proximal sORFI-encoding enod40 sequences are missing.
Apparently, multifunctionality of enod40 is determined by a complex combination of functions of both encoded peptide(s) and RNA structure(s). sORF I is less conserved than the topology of domains 2 and 3, but more conserved than domains 1, 4, 5 and 6, predicted in some species (29). While domain 1 is probably present in many species, domains 4–6 seem to be specific for legumes only. Thus, the enod40 RNA 5′-proximal region has properties of a peptide-encoding mRNA, while the core of enod40 RNA (Figure 3) has hallmarks of structural RNAs, namely a strongly conserved secondary structure topology despite very high sequence diversity. The non-coding character of the enod40 core is further emphasized by the presence of an expansion segment reminiscent of highly ordered RNAs such as rRNAs, RNase P and SRP RNAs. Furthermore, the conserved RNA structural domains 2 and 3 seem to determine some general enod40 functions whereas enod40-encoded peptides may be responsible for more diverse specific roles.
MATERIALS AND METHODS
Sequence database search
Due to the high sequence diversity of enod40, a straightforward BLAST search (57) using complete enod40 sequences as queries was not very efficient to retrieve distant enod40 homologies. Therefore, we restricted sequence similarity searches by conserved regions only and complemented it with RNA secondary structure analyses. The most conserved region in both legume and non-legume enod40 genes is the so-called region II (13,17,32). Thus, the most conserved region II core sequences (∼30 nt, the location is indicated in Figure S1 of the Supplementary Data) were used as queries in BLAST searches ‘for short, nearly exact matches’. This BLAST option is more suitable for retrieving distant short similarities due to shorter ‘word’ size (7 nt) used in the initial search for matches, as compared to that of standard BLAST search (11 nt). The relatively significant (E < 1) non-redundant sequence hits were further analysed for the presence of conserved secondary structure elements, so-called domains 2 and 3, located near the potential region II (29). The searches were done in GenBank including EST sequences. We started from the recent compilation of enod40 sequences (13), and the region II of newly found enod40 genes were subsequently used as queries for similar searches as well.
In order to distinguish putative enod40 homologues from BLAST hits produced by chance, two criteria, derived from known enod40 gene features, were used. First, the potential for coding amino acid sequences homologous to known enod40 sORF1 was explored. The second, independent, criterion required the possibility of folding of characteristic structural domains 2 and 3, flanking the conserved core of the potential region II, described (with small deviations) as the consensus sequence CGGCAAGUCA-N(6)-GGCAAN (Figure 1). Both domains should be located at 1–3 nt upstream or downstream of the consensus core, domain 3 being a stable hairpin and domain 2 being an extended structure flanked by typical sequences GUUUG and CAAAC or their variations. The sequences satisfying one or two of the described criteria were considered as enod40 homologues.
RNA secondary structure predictions
RNA secondary structure predictions were performed using the genetic algorithm of STAR package (58) and Mfold program (59).
SUPPLEMENTRY DATA
Supplementry Data are available at NAR Online.
Supplementary Material
ACKNOWLEDGEMENTS
We thank H. Spaink and R. Olsthoorn for helpful discussions and comments on the manuscript. The Open Access publication charge for this paper has been waivered by Oxford Journals.
Conflict of interest statement. None declared.
REFERENCES
- 1.Read D.J., Ducket J.G., Francis R., Ligron R., Russell A. Symbiotic fungal associations in ‘lower’ land plants. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2000;355:815–830. doi: 10.1098/rstb.2000.0617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brundrett M. Coevolution of roots and mycorrhizas of land plants. New Phytol. 2002;154:275–304. doi: 10.1046/j.1469-8137.2002.00397.x. [DOI] [PubMed] [Google Scholar]
- 3.Wang B., Qiu Y.-L. Phylogenetic distribution and evolution of mycorrhizas in land plants. Mycorrhiza. 2006;16:299–363. doi: 10.1007/s00572-005-0033-6. [DOI] [PubMed] [Google Scholar]
- 4.Doyle J.J. Phylogenetic perspectives on nodulation: evolving views of plants and symbiotic bacteria. Trends Plant Sci. 1998;3:473–478. [Google Scholar]
- 5.Pawlowski K., Bisseling T. Rhizobial and actinorhizal symbioses. What are the shared features? Plant Cell. 1996;8:1899–1913. doi: 10.1105/tpc.8.10.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bladergroen M.R., Spaink H.P. Genes and signal molecules involved in the rhizobia-Leguminoseae symbiosis. Curr. Opin. Plant Biol. 1998;1:353–359. doi: 10.1016/1369-5266(88)80059-1. [DOI] [PubMed] [Google Scholar]
- 7.Albrecht C., Geurts R., Bisseling T. Legume nodulation and mycorrhizae formation: two extremes in host specificity meet. EMBO J. 1999;18:281–288. doi: 10.1093/emboj/18.2.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kistner C., Parniske M. Evolution of signal transduction in intracellular symbiosis. Trends Plants Sci. 2002;7:511–518. doi: 10.1016/s1360-1385(02)02356-7. [DOI] [PubMed] [Google Scholar]
- 9.Limpens E., Bisseling T. Signaling in symbiosis. Curr. Opin. Plant Biol. 2003;6:343–350. doi: 10.1016/s1369-5266(03)00068-2. [DOI] [PubMed] [Google Scholar]
- 10.Yang W.-C., Katinakis P., Hendriks P., Smolders A., de Vries F., Spee J., van Kammen A., Bisseling T., Franssen H. Characterization of Gmenod40, a gene showing novel patterns of cell-specific expression during soybean nodule development. Plant J. 1993;3:573–585. doi: 10.1046/j.1365-313x.1993.03040573.x. [DOI] [PubMed] [Google Scholar]
- 11.Kouchi H., Hata S. Isolation and characterization of novel nodulin cDNAs representing genes expressed at early stages of soybean nodule development. Mol. Gen. Genet. 1993;238:106–119. doi: 10.1007/BF00279537. [DOI] [PubMed] [Google Scholar]
- 12.Van Rhijn P., Fang Y., Galili S., Shaul O., Atzmon N., Wininger S., Eshed Y., Lum M., Li Y., et al. Expression of early nodulin genes in alfalfa mycorrhizae indicates that signal transduction pathways used in forming arbuscular mycorrhizae and Rhizobium-induced nodules may be conserved. Proc. Natl. Acad. Sci. USA. 1997;94:5467–5472. doi: 10.1073/pnas.94.10.5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ruttink T. PhD Thesis. Wageningen University; 2003. ENOD40 affects phytohormone cross-talk. ISBN:9058089797. [Google Scholar]
- 14.Crespi M.D., Jurkevitch E., Poiret M., d’Auberton-Carafa Y., Petrovics G., Kondorosi E., Kondorosi A. Enod40, a gene expressed during nodule organogenesis, codes for a non-translatable RNA involved in plant growth. EMBO J. 1994;13:5099–5112. doi: 10.1002/j.1460-2075.1994.tb06839.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Charon C., Johansson C., Kondorosi E., Kondorosi A., Crespi M. enod40 induces dedifferentiation and division of root cortical cells in legumes. Proc. Natl. Acad. Sci. USA. 1997;94:8901–8906. doi: 10.1073/pnas.94.16.8901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Charon C., Sousa C., Crespi M., Kondorosi A. Alteration of enod40 expression modifies Medicago truncatula root nodule development induced by Sinorhizobium meliloti. Plant Cell. 1999;11:1953–1966. doi: 10.1105/tpc.11.10.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kouchi H., Takane K., So R.B., Ladha J.K., Reddy P.M. Rice ENOD40: isolation and expression analysis in rice and transgenic soybean root nodules. Plant J. 1999;18:121–129. doi: 10.1046/j.1365-313x.1999.00432.x. [DOI] [PubMed] [Google Scholar]
- 18.Flemetakis E., Kavroulakis N., Quaedvlieg N.E.M., Spaink H.P., Dimou M, Roussis A., Katinakis P. Lotus japonicus contains two distinct enod40 genes that are expressed in symbiotic, nonsymbiotic, and embryonic tissues. Mol. Plant - Microbe Interact. 2000;9:987–994. doi: 10.1094/MPMI.2000.13.9.987. [DOI] [PubMed] [Google Scholar]
- 19.Staehelin C., Charon C., Boller T., Crespi M., Kondorosi A. Medicago truncatula plants overexpressing the early nodulin gene enod40 exhibit accelerated mycorrhizal colonization and enhanced formation of arbuscules. Proc. Natl. Acad. Sci. USA. 2001;98:15366–15371. doi: 10.1073/pnas.251491698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Varkonyi-Gasic E., White D.W.R. The white clover enod40 gene family. Expression patterns of two types of genes indicate a role in vascular function. Plant Physiol. 2002;129:1107–1118. doi: 10.1104/pp.010916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vleghels I., Hontelez J., Ribeiro A., Fransz P., Bisseling T., Franssen H. Expression of ENOD40 during tomato plant development. Planta. 2003;218:42–49. doi: 10.1007/s00425-003-1081-9. [DOI] [PubMed] [Google Scholar]
- 22.Grönlund M., Roussis A., Flemetakis E., Quaedvlieg N.E.M., Schlaman H.R.M., Umehara Y., Katinakis P., Stougaard J., Spaink H.P. Analysis of promoter activity of the early nodulin Enod40 in Lotus japonicus. Mol. Plant Microbe Interact. 2005;18:414–427. doi: 10.1094/MPMI-18-0414. [DOI] [PubMed] [Google Scholar]
- 23.Takeda N., Okamoto S., Hayashi M., Murooka Y. Expression of LjENOD40 genes in response to symbiotic and non-symbiotic signals: LJENOD40-1 and LjENOD40-2 are differentially regulated in Lotus japonicus. Plant Cell Physiol. 2005;46:1291–1298. doi: 10.1093/pcp/pci138. [DOI] [PubMed] [Google Scholar]
- 24.Kumagai H., Kinoshita E., Ridge R.W., Kouchi H. RNAi knock-down of ENOD40s leads to significant suppression of nodule formation in Lotus japonicus. Plant Cell Physiol. 2006;47:1102–1111. doi: 10.1093/pcp/pcj081. [DOI] [PubMed] [Google Scholar]
- 25.Ruttink T., Boot K., Kijne J., Bisseling T., Franssen H. ENOD40 affects elongation growth in tobacco Bright Yellow-2 cells by alteration of ethylene biosynthesis kinetics. J. Exp. Bot. 2006;57:3271–3282. doi: 10.1093/jxb/erl089. [DOI] [PubMed] [Google Scholar]
- 26.Sousa C., Johansson C., Charon C., Manyani H., Sautter C., Kondorosi A., Crespi M. Translational and structural requirements of the early nodulin gene enod40, a short-open reading frame-containing RNA, for elicitation of a cell-specific growth response in the alfalfa root cortex. Mol. Cell. Biol. 2001;21:354–366. doi: 10.1128/MCB.21.1.354-366.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rohrig H., Schmidt J., Miklashevichs E., Schell J., John M. Soybean ENOD40 encodes two peptides that bind to sucrose synthase. Proc. Natl. Acad. Sci. USA. 2002;99:1915–1920. doi: 10.1073/pnas.022664799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hofacker I.L., Fekete M., Stadler P.F. Secondary structure prediction for aligned RNA sequences. J. Mol. Biol. 2002;319:1059–1066. doi: 10.1016/S0022-2836(02)00308-X. [DOI] [PubMed] [Google Scholar]
- 29.Girard G., Roussis A., Gultyaev A.P., Pleij C.W.A., Spaink H.P. Structural motifs in the RNA encoded by the early nodulation gene enod40 of soybean. Nucleic Acids Res. 2003;31:5003–5015. doi: 10.1093/nar/gkg721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Caetano-Anolles G. Grass evolution inferred from chromosomal rearrangements and geometrical and statistical features in RNA structure. J. Mol. Evol. 2005;60:635–652. doi: 10.1007/s00239-004-0244-z. [DOI] [PubMed] [Google Scholar]
- 31.Campalans A., Kondorosi A., Crespi M. Enod40, a short open reading frame-containing mRNA, induces cytoplasmic localization of a nuclear RNA binding protein in Medicago truncatula. Plant Cell. 2004;16:1047–1059. doi: 10.1105/tpc.019406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vijn I., Yang W.-C., Pallisgard N., Jensen E.O., van Kammen A., Bisseling T. VsENOD5, VsENOD12 and VsENOD40 expression during Rhizobium-induced nodule formation in Vicia sativa roots. Plant Mol. Biol. 1995;28:1111–1119. doi: 10.1007/BF00032671. [DOI] [PubMed] [Google Scholar]
- 33.Santi C, von Groll U., Ribeiro A., Chiurazzi M, Auguy F., Bogusz D., Franche C., Pawlowski K. Comparison of nodule induction in legume and actinorhizal symbioses: the induction of actinorhizal nodules does not involve enod40. Mol. Plant Microbe Interact. 2003;16:808–816. doi: 10.1094/MPMI.2003.16.9.808. [DOI] [PubMed] [Google Scholar]
- 34.Matvienko M., van de Sande K., Pawlowski K., van Kammen A., Franssen H., Bisseling T. Nicotiana tabacum SR1 contains two ENOD40 homologs. In: Stacey G., Mullin B., Gresshoff P.M., editors. Biology of Plant-Microbe Interactions. International Society for Molecular Plant-Microbe Interactions, St. Paul, MN; 1996. pp. 387–391. [Google Scholar]
- 35.Compaan B., Ruttink T., Albrecht C., Meeley R., Bisseling T., Franssen H. Identification and characterization of a Zea mays line carrying a transposon-tagged ENOD40. Biochim. Biophys. Acta. 2003;1629:84–91. doi: 10.1016/j.bbaexp.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 36.Larsen K. Molecular cloning and characterization of a cDNA encoding a ryegrass (Lolium perenne) ENOD40 homologue. J. Plant Physiol. 2003;160:675–687. doi: 10.1078/0176-1617-00962. [DOI] [PubMed] [Google Scholar]
- 37.Michot B., Bachellerie J.-P. Comparisons of large subunit rRNAs reveal some eukaryote-specific elements of secondary structure. Biochimie. 1987;69:11–23. doi: 10.1016/0300-9084(87)90267-7. [DOI] [PubMed] [Google Scholar]
- 38.van Nues R.W., Brown J.D. Saccharomyces SRP RNA secondary structures: a conserved S-domain and extended Alu-domain. RNA. 2004;10:75–89. doi: 10.1261/rna.5137904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marquez S.M., Harris J.K., Kelley S.T., Brown J.W., Dawson S.C., Roberts E.C., Pace N.R. Structural implications of novel diversity in eucaryal RNase P RNA. RNA. 2005;11:739–751. doi: 10.1261/rna.7211705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hadjiolov A.A., Georgiev O.I., Nosikov V.V., Yavachev L.P. Primary and secondary structure of rat 28 S ribosomal RNA. Nucleic Acids Res. 1984;12:3677–3693. doi: 10.1093/nar/12.8.3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Clark C.G., Tague B.W., Ware V.C., Gerbi S.A. Xenopus laevis 28S ribosomal RNA: a secondary structure model and its evolutionary and functional implications. Nucleic Acids Res. 1984;12:6197–6220. doi: 10.1093/nar/12.15.6197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hassouna N., Michot B., Bachellerie J.-P. The complete nucleotide sequence of mouse 28S rRNA gene. Implications for the process of size increase of the large subunit rRNA in higher eukaryotes. Nucleic Acids Res. 1984;12:3563–3583. doi: 10.1093/nar/12.8.3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hancock J.M., Dover G.A. “Compensatory slippage” in the evolution of ribosomal RNA genes. Nucleic Acids Res. 1990;18:5949–5954. doi: 10.1093/nar/18.20.5949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nunn G.B., Theisen B.F., Christensen B., Arctander P. Simplicity-correlated size growth of the nuclear 28S ribosomal RNA D3 expansion segment in the crustacean order Isopoda. J. Mol. Evol. 1996;42:211–223. doi: 10.1007/BF02198847. [DOI] [PubMed] [Google Scholar]
- 45.Hancock J.M., Vogler A.P. How slippage-derived sequences are incorporated into rRNA variable-region secondary structure: implications for phylogeny reconstruction. Mol. Phylogenet. Evol. 2000;14:366–374. doi: 10.1006/mpev.1999.0709. [DOI] [PubMed] [Google Scholar]
- 46.Crease T.J., Taylor D.J. The origin and evolution of variable-region helices in V4 and V7 of the small-subunit ribosomal RNA of branchiopod crustaceans. Mol. Biol. Evol. 1998;15:1430–1446. doi: 10.1093/oxfordjournals.molbev.a025871. [DOI] [PubMed] [Google Scholar]
- 47.McTaggart S.J., Crease T.J. Selection on the structural stability of a ribosomal RNA expansion segment in Daphnia obtusa. Mol. Biol. Evol. 2005;22:1309–1319. doi: 10.1093/molbev/msi119. [DOI] [PubMed] [Google Scholar]
- 48.Hinkle G., Leipe D.D., Nerad T.A., Sogin M.L. The unusually long small subunit ribosomal RNA of Phreatamoeba balamuthi. Nucleic Acids Res. 1994;22:465–469. doi: 10.1093/nar/22.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gerbi S.A. The evolution of eukaryotic ribosomal DNA. BioSystems. 1986;19:247–258. doi: 10.1016/0303-2647(86)90001-8. [DOI] [PubMed] [Google Scholar]
- 50.Sweeney R., Chen L., Yao M.-C. An rRNA variable region has an evolutionarily conserved essential role despite sequence divergence. Mol. Cell. Biol. 1994;14:4203–4215. doi: 10.1128/mcb.14.6.4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Nues R.W., Venema J., Planta R.J., Raue H.A. Variable region V1 of Saccharomyces cerevisiae 18S rRNA participates in biogenesis and function of the small ribosomal subunit. Chromosoma. 1997;105:523–531. doi: 10.1007/BF02510489. [DOI] [PubMed] [Google Scholar]
- 52.Jeeninga R.E., van Delft Y., de Graaff-Vincent M., Dirks-Mulder A., Venema J., Raue H.A. Variable regions V13 and V3 of Saccharomyces cerevisiae contain structural features essential for normal biogenesis and stability of 5.8S and 25S rRNA. RNA. 1997;3:476–488. [PMC free article] [PubMed] [Google Scholar]
- 53.Crease T.J., Colbourne J.K. The unusually long small-subunit ribosomal RNA of the Crustacean, Daphnia pulex: sequence and predicted secondary structure. J. Mol. Evol. 1998;46:307–313. doi: 10.1007/pl00006307. [DOI] [PubMed] [Google Scholar]
- 54.Alkemar G., Nygard O. A possible tertiary rRNA interaction between expansion segments ES3 and ES6 in eukaryotic 40S ribosomal subunits. RNA. 2003;9:20–24. doi: 10.1261/rna.2108203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Regvar M., Vogel K., Irgel N., Wraber T., Hildebrandt U., Wilde P., Bothe H. Colonization of pennycresses (Thlaspi spp.) of the Brassicaeae by arbuscular mycorrhizal fungi. J. Plant Physiol. 2003;160:615–626. doi: 10.1078/0176-1617-00988. [DOI] [PubMed] [Google Scholar]
- 56.Mitra R.M., Gleason C.A., Edwards A., Hadfield J., Downie J.A., Oldroyd G.E.D., Long S.R. A Ca2+/calmodulin-dependent protein kinase required for symbiotic nodule development: gene identification by transcript-based cloning. Proc. Natl. Acad. Sci. USA. 2004;101:4701–4705. doi: 10.1073/pnas.0400595101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. ‘Basic local alignment search tool.’. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 58.Gultyaev A.P., van Batenburg F.H.D., Pleij C.W.A. The computer simulation of RNA folding pathways using a genetic algorithm. J. Mol. Biol. 1995;250:37–51. doi: 10.1006/jmbi.1995.0356. [DOI] [PubMed] [Google Scholar]
- 59.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.