Abstract
Polyphosphate kinase (PPK), responsible for the processive synthesis of inorganic polyphosphate (polyP) from ATP in Escherichia coli, can transfer in reverse the terminal phosphate residue of polyP to ADP to yield ATP. PolyP also serves as a donor in a polyP:AMP phosphotransferase (PAP) activity observed in extracts of Acinetobacter johnsonii and Myxococcus xanthus. We have found that overexpression of the gene encoding PPK results in a large enhancement of PAP activity in E. coli. The PAP activity requires both PPK and adenylate kinase in equimolar amounts. PPK and adenylate kinase form a complex in the presence of polyphosphate. We discuss a phosphotransfer mechanism that involves both enzymes and enables polyP to be a phospho-donor to AMP.
Inorganic linear polyphosphates (polyPs) are widespread in nature, having been found in all living organisms from bacteria to higher eukaryotes (1). Several enzymes using polyP have been reported, such as polyphosphate kinase (PPK) (2–4), exopolyphosphatase (5, 6), polyphosphate:AMP phosphotransferase (PAP) (7, 8), and polyphosphate glucokinase (9). Among these enzymes, PPK, which is responsible for the processive synthesis of polyP in bacteria, has been intensively studied, and PPK-encoding (ppk) genes have been identified in several species (10–13). PPK transfers the terminal phosphate residue of polyP to ADP to yield ATP in a freely reversible reaction (2–4). PPK of Escherichia coli has been shown to undergo ATP-dependent autophosphorylation at the site of a well-conserved His-461; the resulting phosphoprotein has been shown to be an intermediate of the phosphotransfer reaction to polyP (14).
Marked PAP activity was found in Acinetobacter johnsonii (7) and Myxococcus xanthus (8), where it phosphorylates AMP with polyP to give ADP. Weak PAP activity was also detected in E. coli (8). We have found that overexpression of the ppk gene in E. coli results in a drastic increase in PAP activity. In this paper, we discuss PAP activity as a phosphotransfer mechanism.
Materials and Methods
Overproduction of PPK and Adenylate Kinase (ADK).
The plasmid harboring the ppk gene, pTrc-PPK, was used for overproduction of PPK (15). The adk gene was amplified by PCR with primers 5′-ATGGATCCCCGTTTCAGCCCCAGGTGCC-3′ and 5′-ATAAGCTTGGCCTGAGATTGCTGATAAG-3′. The PCR product was cloned into pUC18 to give pUC-ADK. The adk gene was placed under the control of the lac promoter. E. coli JM109 (16) harboring pTrc-PPK or pUC-ADK was cultured in 2× YT broth (17) with 50 μg/ml of ampicillin at 30°C until the midlogarithmic growth phase. Then, isopropyl β-d-thiogalactopyranoside was added to 1 mM, and cultivation was continued for 5 h. Five grams of the harvested cells overproducing PPK was suspended in 50 ml of buffer P (25 mM Hepes-KOH, pH 7.5/0.1 mM EDTA/1 mM 2-mercaptoethanol/15% glycerol). Buffer A (50 mM Tris⋅HCl, pH 7.5/0.5 mM EDTA/1 mM 2-mercaptoethanol) was used to suspend the cells overproducing ADK. Each cell suspension was subjected to sonication and centrifuged at 30,000 × g for 30 min to prepare the cell lysate.
Purification of PPK and ADK.
PPK was purified as follows. The cell lysate containing overproduced PPK was put onto a DEAE-Toyopearl 650 M column (2.2 × 32 cm; Toso, Tokyo, Japan) equilibrated with buffer P, followed by elution with buffer P containing 300 mM NaCl. Proteins were eluted with 1 liter of linear NaCl gradient from 300 to 500 mM. The PPK-containing fractions were eluted at about 350 mM NaCl. This sample was further applied to a HiTrap Q column (5 ml; Amersham Pharmacia). Proteins were eluted with 20 ml of 500 mM KCl. The resultant PPK fraction was dialyzed against Hepes-KOH buffer (25 mM, pH 7.5) containing 500 mM KCl, 2 mM 2-mercaptoethanol, and 50% glycerol. ADK was purified as follows. Solid ammonium sulfate was added to the cell lysate containing overproduced ADK to 60% saturation. Ammonium sulfate was further added to the supernatant of the suspension to a final saturation of 80%. Precipitates were recovered and dissolved in 10 ml of buffer A. The sample was put onto a DEAE-Toyopearl 650 M column (2.2 × 32 cm) equilibrated with buffer A. Proteins were eluted with 1 liter of linear NaCl gradient from 0 to 100 mM. The ADK fractions were further applied to a HiTrap Q column (5 ml). Proteins were eluted with 10 ml of 150 mM KCl. Both proteins were monitored by means of SDS/PAGE.
Enzyme Assays.
The PPK assay mixture (100 μl) contained 50 mM Tris⋅HCl (pH 8.0), 50 mM ammonium sulfate, 10 mM MgCl2, 5 mM ADP, polyP (150 mM in terms of phosphate; chain lengths, 15–20; purchased from Sigma), and an enzyme sample. The PAP assay mixture (100 μl) contained 50 mM Tris⋅HCl (pH 8.0), 50 mM ammonium sulfate, 10 mM MgCl2, 5 mM AMP, polyP (75 mM in terms of phosphate; chain lengths, 15–20) and enzyme samples. The ADK assay mixture (100 μl) contained 50 mM Tris⋅HCl (pH 8.0), 50 mM ammonium sulfate, 10 mM MgCl2, 10 mM ATP, 5 mM AMP, and an enzyme sample. The ATP or ADP was quantified by HPLC. One unit of an enzyme was defined as the amount required to form 1 μmol of ATP in the PPK assay in a polyP-dependent manner, 1 μmol of the sum of ADP and ATP in the PAP assay, and 2 μmol of ADP in the ADK assay per min at 30°C. The PAP assay with [32P]polyP was performed as follows. [32P]PolyP was synthesized and isolated by essentially the same method as described by Wurst and Kornberg (6). Each assay mixture (20 μl) contained 50 mM Tris⋅HCl (pH 8.0), 50 mM ammonium sulfate, 10 mM MgCl2, [32P]polyP (500 μM in terms of phosphate), 5 mM AMP, and enzyme samples. After incubation for 30 min at 30°C, 1 μl of each product was applied in a line along the bottom of a polyethyleneimine-cellulose TLC (PEI-TLC) plate and developed with 1 M HCOOH and 0.6 M LiCl followed by autoradiography. The polyP-forming assay was performed as follows. Each reaction mixture (10 μl) containing 50 mM Hepes-NaOH (pH 7.4), 40 mM ammonium sulfate, 4 mM magnesium acetate, 5 mM phosphocreatine, 0.4 μg creatine kinase, 1 mM [γ-32P]ATP (20 cpm/pmol), and 5 μl of an enzyme sample was incubated at 37°C for 60 min, and 1 μl of each product was applied in a PEI-TLC plate and developed with 1 M HCOOH and 2 M LiCl followed by autoradiography.
Results
PAP Activity Increased in E. coli Cells Overproducing PPK.
E. coli JM109 possesses a very low level of intrinsic PAP activity that catalyzes the synthesis of ADP from AMP and polyP (Table 1). When PPK was overproduced in E. coli JM109 cells, not only PPK activity, but also PAP activity was increased by over 600-fold (Table 1).
Table 1.
PPK and PAP activity of the lysate containing overproduced PPK
| Lysate | PPK activity (units/mg protein × 10−2) | PAP activity (units/mg protein × 10−2) |
|---|---|---|
| Control | <0.01 | 0.0197 |
| PPK++ | 3.00 | 12.3 |
PPK Alone Fails to Catalyze PAP Activity.
The cell lysate containing overproduced PPK was subjected to anion-exchange column chromatography. Proteins were fractionated by NaCl elution in a stepwise manner between 0 and 500 mM. PPK was eluted at 350–450 mM NaCl in fractions VIII-X (Table 2). No fraction showed PAP activity. These results indicate that PPK alone does not catalyze the phosphorylation of AMP with polyP, and that an additional factor separated by fractionation is needed for the PAP activity observed in the cell lysate.
Table 2.
Distribution of PPK and PAP activity
| Fraction* | Protein (mg) | PPK Activity (units) | PAP Activity (units) |
|---|---|---|---|
| Before fractionation | 87.5 | 5.34 | 2.63 |
| I | 2.85 | <0.05 | <0.005 |
| II | 0.363 | <0.05 | <0.005 |
| III | 0.638 | <0.05 | <0.005 |
| IV | 17.0 | <0.05 | <0.005 |
| V | 25.0 | <0.05 | <0.005 |
| VI | 21.0 | <0.05 | <0.005 |
| VII | 9.50 | <0.05 | <0.005 |
| VIII | 11.3 | 0.930 | <0.005 |
| IX | 7.13 | 0.990 | <0.005 |
| X | 5.63 | 1.07 | <0.005 |
| XI | 0.538 | <0.05 | <0.005 |
The cell lysate containing overproduced PPK (87.5 mg of proteins) was put onto a DEAE-Toyopearl 650M column (1.6 × 12.5 cm; Toso) equilibrated with buffer P, followed by stepwise elution with buffer P containing 0, 50, 100, 150, 200, 250, 300, 350, 400, 450, and 500 mM NaCl. The eluents from each step were defined as fractions I to XI, respectively. Each fraction was subjected to the PPK and the PAP assays.
PPK Requires Another Factor for PAP Activity.
The PAP activity of each fraction (Table 2) was determined in the presence of a fixed volume of the PPK-containing fraction IX was analyzed (Table 3). When fractions III or IV, relatively low-salt eluents, were incubated with the PPK fraction IX, significant PAP activity was observed. These results indicate that a factor in fractions III or IV combine with PPK to catalyze the phosphorylation of AMP in a polyP-dependent manner. ADK was a plausible candidate because of its capacity to phosphorylate AMP with ATP in a freely reversible reaction (18). The distribution of ADK activity in the eluted fractions precisely overlapped the PAP activity (Table 3).
Table 3.
Reconstitution of the PAP activity
PPK and ADK Together Constitute the PAP Activity.
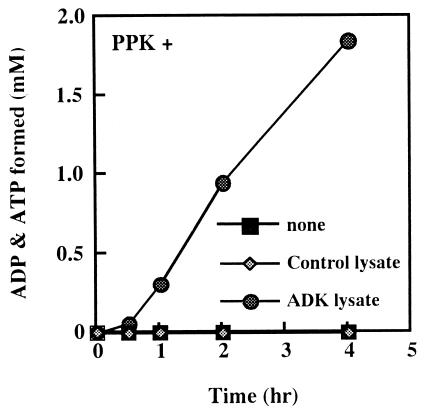
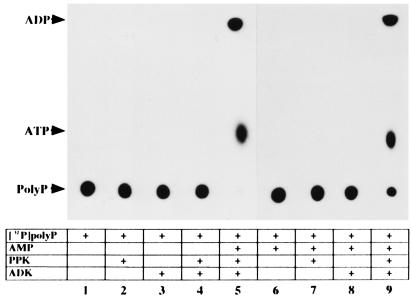
To examine the constitution of PAP activity by PPK and ADK, we overexpressed the adk gene encoding ADK in E. coli JM109 to obtain an ADK-enriched lysate which shows about 6-fold higher specific activity of ADK (42.0 units/mg protein) than control lysate (7.58 units/mg protein), and purified PPK homogeneity (0.235 units/mg protein). When the purified PPK and a small amount of a lysate of E. coli JM109 harboring a control vector were incubated together, very low PAP activity was expressed (Fig. 1). However, incubation of both the purified PPK and the ADK-enriched lysate resulted in a marked increase in PAP activity (Fig. 1). We further confirmed PAP activity to be the combined actions of PPK and ADK. ADK was purified to homogeneity (192 units/mg protein). [32P]PolyP synthesized from [γ-32P]ATP by PPK (6) was free [γ-32P]ATP (Fig. 2, lane 1). Only when [32P]polyP was incubated with AMP, purified PPK, and ADK, were ADP and ATP detected (Fig. 2, lanes 5 and 9). No ADP or ATP appeared when any one of the three components was omitted. Once ADP is formed by PAP activity, ATP can be readily synthesized by PPK or ADK (Fig. 2, lanes 5 and 9).
Figure 1.
Time course of polyP-dependent AMP phosphorylation. The reaction mixtures (100 μl) containing 50 mM Tris⋅HCl (pH 8.0), 50 mM ammonium sulfate, 10 mM MgCl2, 5 mM AMP, polyP (75 mM of phosphate, chain length, 15–20), 4 μg purified PPK, and a cell lysate (0.8 μg of proteins) was incubated at 30°C. The ADP and ATP formed were monitored and plotted as the sum of both levels. ■, No lysate; ⧫, the cell lysate of JM109 harboring the control vector pTrc99A; ●, the cell lysate of JM109 overproducing ADK.
Figure 2.
Autoradiogram showing phosphotransfer from [32P]polyP to AMP by the combined action of both purified enzymes. Each or both of 4 μg purified PPK and ADK were used for enzyme samples in the PAP assay with [32P]polyP. Each sample was analyzed by PEI-TLC followed by autoradiography: lanes 1 and 6, reaction compounds with no enzyme; lanes 2 and 7, reaction compounds containing PPK; lanes 3 and 8, reaction compounds containing ADK; lanes 4, 5, and 9, reaction compounds containing PPK and ADK; lanes 1–4, AMP was omitted from the reaction mixtures. Other details are given in Materials and Methods.
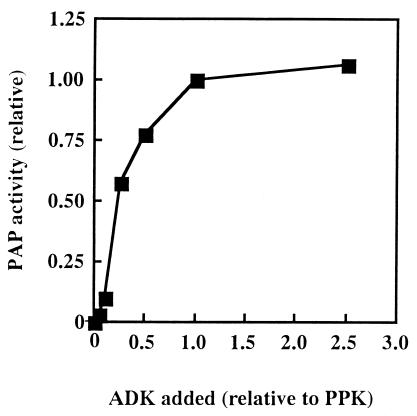
PAP Requires Equimolar Concentrations of ADK and PPK for Optimal Activity.
With ADK varied in amount from 0 to 2.5 times the molar amounts of PPK, optimal PAP activity was observed when they were equimolar (Fig. 3). With ADK in excess, PAP activity increased only slightly.
Figure 3.
A stoichiometry of PPK and ADK required for the PAP activity. The purified PPK (25 pmol) and various doses of the purified ADK (1.3, 2.5, 6.3, 13, 25, and 63 pmol) were used for enzyme samples in the PAP assay. The relative activity to the value observed for 25 pmol of both enzymes was plotted against the dose of ADK added, the value of which was expressed relative to the amount of PPK.
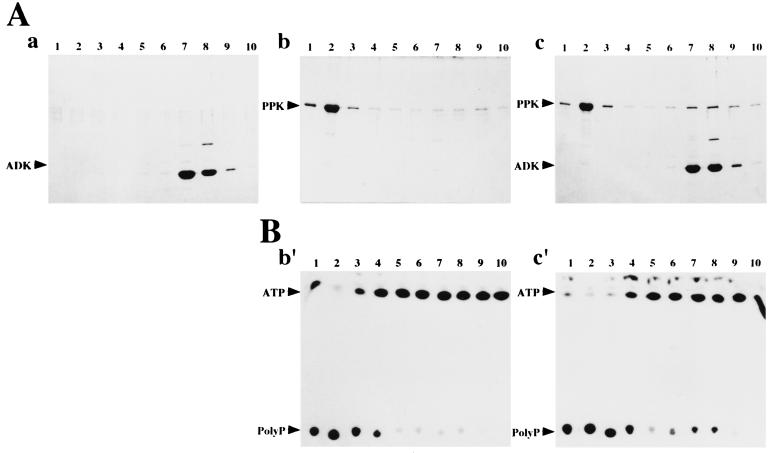
PPK and ADK Form a Complex in the Presence of PolyP.
Each or both of PPK and ADK were incubated in the presence of polyP, and then subjected to size-exclusion chromatography. Proteins were fractionated by isocratic elution with the polyP-containing buffer. When PPK or ADK alone was incubated and fractionated, PPK was detected in fractions 1 and 2 by means of SDS/PAGE (Fig. 4Ab) and polyP-forming assay (Fig. 4Bb′), whereas ADK was in fractions 7 and 8 (Fig. 4Aa). When both enzymes were incubated together, a part of PPK was eluted in the ADK-containing fractions 7 and 8 (Fig. 4 Ac and Bc′). Coelution of PPK and ADK observed in this experiment indicates the complex formation between both enzymes in the presence of polyP. Although a mechanism underlying the delayed elution-behavior of PPK-ADK complex is not clear, we speculate that both enzymes might weakly associate and run in cycles to attach/detach and the association manner and/or the presence of polyP might cause the delayed behavior of the complex.
Figure 4.
Complex formation between PPK and ADK in the presence of polyP. The reaction mixture (1 ml) containing 0.5 mg of each or both of PPK and ADK, 50 mM Tris⋅HCl (pH 8.0), 50 mM ammonium sulfate, 10 mM MgCl2, 5 mM AMP and polyP (375 mM in terms of phosphate, chain lengths, 15–20) was incubated at 30°C for 30 min. The resultant mixtures were applied onto AKTA FPLC column system with HiLoad 16/60 Superdex 200pg (Amersham Pharmacia) and separated by isocratic elution with polyP-containing buffer [50 mM Tris⋅HCl, pH 8.0, 50 mM ammonium sulfate, 10 mM MgCl2 and polyP (15 mM in terms of phosphate, chain lengths, 15–20)]. After the void-volume (45 ml) of the eluent was wasted, each 5 ml of the eluent was fractionated and defined as fractions 1–10, respectively. Fraction 1 is the largest molecular weight fraction in this chromatography. (A) The results of SDS/PAGE analysis of fractions 1–10. Proteins in each fractions (200 μl) were precipitated by 5% trichloroacetic acid and analyzed. The fractions in the case that only ADK was incubated followed by the chromatography were analyzed in Aa, those in the case that only PPK was in Ab, and those in the case that PPK and ADK together were in Ac. Corresponding fraction numbers analyzed were designated at the top of each lane. (B) The result of polyP-forming assay of the fractions 1–10. The fractions in the case that only PPK was incubated followed by the chromatography were analyzed in Bb′, those in the case that PPK and ADK together were in Bc′. Corresponding fraction numbers analyzed were designated at the top of each lane.
Discussion
In this study, we demonstrated that PPK and ADK together constitute the PAP activity, where it phosphorylates AMP with polyP. The weak intrinsic activity observed in native E. coli cells may also be the consequence of the combined action of PPK and ADK originated from chromosomal copy of each gene.
Although massive PAP activity was found in A. johnsonii, the activity was associated with a 55-kDa protein and was free ADK (7). This indicates that the PAP activity in A. johnsonii might be expressed in a different manner from that in E. coli. We found prominent PAP activity also in extract of Pseudomonas aeruginosa strain PAO1, which could only be abolished by means of size-exclusion column chromatography. Incubation of PPK and ADK partially purified from the extract of P. aeruginosa PAO1 resulted in the reconstitution of the PAP activity, suggesting that the mechanism underlying the expression of the PAP activity clarified in E. coli is conserved in this organism (unpublished work).
PPK has been shown to undergo ATP-dependent autophosphorylation at the site of a well-conserved His-461, and the resulting phosphoprotein has been shown to be an intermediate of the phosphotransfer reaction to polyP in a forward reaction (14). We found that PPK also autophosphorylates itself in a [32P]polyP-dependent manner to give phospho-PPK, a potent phosphodonor (data not shown). Although ADK cannot use polyP as a phosphodonor directly, with the phosphoryl group of the intermediate instead of ATP might it phosphorylate AMP. We speculate that the complex formation between PPK and ADK in the presence of polyP might support the phosphotransfer mechanism.
ADK is responsible for the maintenance of the appropriate energy state of the cell, which is essential for cell growth (18). Moreover, ADK has been known to function as an NDP kinase to generate NTP from NDP with ATP (19). It is of particular interest that ADK can use polyP as an indirect phosphodonor by means of PPK. By the catalysis of PPK and ADK, polyP accumulated in E. coli cells possibly participates in the global maintenance of an intracellular nucleotide pool.
Acknowledgments
We thank Dr. Arthur Kornberg for helpful suggestions and comments on the manuscript.
Abbreviations
- polyP
polyphosphate
- ADK
adenylate kinase
- PAP
polyphosphate:AMP phosphotransferase
- PEI-TLC
polyethyleneimine-cellulose TLC
- PPK
polyphosphate kinase
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.011518098.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.011518098
References
- 1.Kulaev I S. The Biochemistry of Inorganic Polyphosphates. New York: Wiley; 1979. [DOI] [PubMed] [Google Scholar]
- 2.Ahn K, Kornberg A. J Biol Chem. 1990;265:11734–11739. [PubMed] [Google Scholar]
- 3.Kornberg A, Kornberg S R, Simms E S. Biochim Biophys Acta. 1956;20:215–227. doi: 10.1016/0006-3002(56)90280-3. [DOI] [PubMed] [Google Scholar]
- 4.Kornberg S R. Biochim Biophys Acta. 1957;26:294–300. doi: 10.1016/0006-3002(57)90008-2. [DOI] [PubMed] [Google Scholar]
- 5.Akiyama M, Crooke E, Kornberg A. J Biol Chem. 1993;268:633–639. [PubMed] [Google Scholar]
- 6.Wurst H, Kornberg A. J Biol Chem. 1994;269:10996–11001. [PubMed] [Google Scholar]
- 7.Bonting C F C, Kortstee J J, Zehnder A J B. J Bacteriol. 1991;173:6484–6488. doi: 10.1128/jb.173.20.6484-6488.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kornberg A. J Bacteriol. 1995;177:491–496. doi: 10.1128/jb.177.3.491-496.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hsieh P-C, Shenoy B C, Jentoft J E, Phillips N F B. Protein Expression Purif. 1993;4:76–84. doi: 10.1006/prep.1993.1012. [DOI] [PubMed] [Google Scholar]
- 10.Akiyama M, Crooke E, Kornberg A. J Biol Chem. 1992;267:22556–22561. [PubMed] [Google Scholar]
- 11.Kato J, Yamamoto T, Yamada K, Ohtake H. Gene. 1993;137:237–242. doi: 10.1016/0378-1119(93)90013-s. [DOI] [PubMed] [Google Scholar]
- 12.Tinsley C R, Gotschlich E C. Infect Immun. 1995;63:1624–1630. doi: 10.1128/iai.63.5.1624-1630.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ishige K, Kameda A, Noguchi T, Shiba T. DNA Res. 1998;5:157–182. doi: 10.1093/dnares/5.3.157. [DOI] [PubMed] [Google Scholar]
- 14.Kumble K D, Ahn K, Kornberg A. Proc Natl Acad Sci USA. 1996;93:14391–14395. doi: 10.1073/pnas.93.25.14391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Noguchi T, Shiba T. Biosci Biotechnol Biochem. 1998;62:1594–1596. doi: 10.1271/bbb.62.1594. [DOI] [PubMed] [Google Scholar]
- 16.Amann E, Ochs B, Abel K J. Gene. 1988;69:301–31510. doi: 10.1016/0378-1119(88)90440-4. [DOI] [PubMed] [Google Scholar]
- 17.Maniatis T, Fritsch E F, Sambrook J. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1982. [Google Scholar]
- 18.Noda L. In: The Enzymes. Boyer P D, editor. Vol. 8. New York: Academic; 1973. pp. 279–305. [Google Scholar]
- 19.Lu Q, Inoue M. Proc Natl Acad Sci USA. 1996;93:5720–5725. doi: 10.1073/pnas.93.12.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]