Abstract
Metabolism represents a complex collection of enzymatic reactions and transport processes that convert metabolites into molecules capable of supporting cellular life. Here we explore the origins and evolution of modern metabolism. Using phylogenomic information linked to the structure of metabolic enzymes, we sort out recruitment processes and discover that most enzymatic activities were associated with the nine most ancient and widely distributed protein fold architectures. An analysis of newly discovered functions showed enzymatic diversification occurred early, during the onset of the modern protein world. Most importantly, phylogenetic reconstruction exercises and other evidence suggest strongly that metabolism originated in enzymes with the P-loop hydrolase fold in nucleotide metabolism, probably in pathways linked to the purine metabolic subnetwork. Consequently, the first enzymatic takeover of an ancient biochemistry or prebiotic chemistry was related to the synthesis of nucleotides for the RNA world.
Keywords: enzyme activity, evolution, metabolism, nucleotide metabolism
There is current interest in the processes underlying the biology of network because these offer insight into the organization and evolution of life (1). Cellular metabolism, one of the greatest achievements of science, is clearly the best-studied biological network. It represents a complex collection of enzymatic reactions and transport processes that convert metabolites into molecules capable of supporting cells and organisms. However, our knowledge of how modern metabolism originated and evolved is limited (2). One widely accepted hypothesis is that promiscuous catalytic activities in proteins provide a selective advantage and are recruited to perform new metabolic functions (3, 4). Considerable evidence supports a patchwork recruitment scenario in which recruited homologous enzymes are scattered over diverse pathways (2). For example, enzymes with α/β barrel fold structure that catalyze similar reactions occur across metabolic subnetworks (5, 6) and a small set of structural families dominates the small-molecule metabolism in Escherichia coli (7–10). The recruitment hypothesis assumes there is already an active enzymatic core with multifunctional enzymes from which proteins are drawn for metabolic innovation. Because history restricts the interplay between structure and function of metabolic enzymes, we here use evolutionary patterns in protein structure advantageously to study recruitment processes and metabolic network evolution.
The protein world has a hierarchical and redundant organization specified in terms of evolutionary units of molecular structure, the protein domains (11). Domains are generally unified into a comparatively small set of folding architectures, protein superfamilies, and these are further grouped into protein folds (12). Domain structure is generally maintained for long periods of evolutionary time. Consequently, the discovery of an architectural design constitutes an important and rare event in evolutionary history. The repertoire of architectures in proteomes can therefore be regarded as a collection of historical imprints or molecular fossils that carry considerable phylogenetic history. Using a genomic census of architecture, we recently generated phylogenies that describe the evolution of the protein world at different hierarchical levels of structural organization (13–15). These genomic-based phylogenies (phylogenomic trees) were used to classify proteins (mostly globular), define structural transformations, and uncover evolutionary patterns in structure. Interestingly, the same data were also used to build reasonable universal trees of life capable of describing the history of major organismal lineages satisfactorily. Because structural history limits recruitment, we also painted the relative ages (ancestries) of enzymes derived from rooted phylogenomic trees directly onto >100 metabolic subnetworks defined by the Kyoto Encyclopedia of Genes and Genomes (KEGG) (16), linked metabolic enzymes to fold architectures with hidden Markov models (HMMs) in almost 1 million genomic sequences, and used this information to build the molecular ancestry network (MANET) database (17). Evolutionarily painted subnetworks revealed a patchy distribution of ancestries [a literal evolutionary mosaic (8)] in metabolism that is indicative of widespread enzyme recruitment. This is illustrated in the metabolic diagrams of MANET [supporting information (SI) Fig. 5].
In this paper, we uncover evolutionary patterns embedded in modern metabolism. This exploration assumes metabolism is a palimpsest that recapitulates earlier biochemistries (18) and prebiotic chemistries (19), and that protein architecture has preserved ancient structural designs as fossils of ancient biochemistries. We first discover that metabolism is ancient and arose very early in the history of the protein world. Folds appearing early in evolution were widely shared not only by proteomes in all organisms that have been fully sequenced but also by many metabolic subnetworks. We then survey the presence (abundance and occurrence) of folds in metabolism, reconstruct phylogenetic trees describing the evolution of subnetworks, and sort out patterns of enzyme recruitment and origin. This allows identification of ancient subnetworks and putative enzymatic activities as sites of origins of metabolism. The result of these analyses is surprising and provides further support for the existence of an ancient RNA world.
Results and Discussion
Ancient Fold Architectures Distribute Widely Throughout Metabolism.
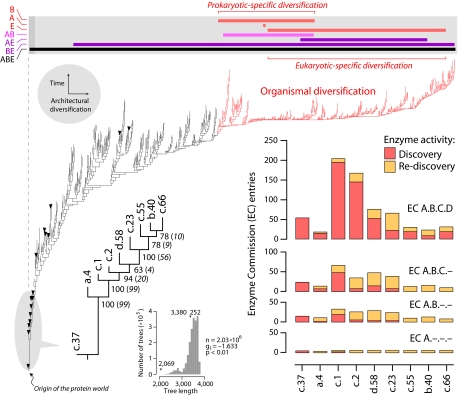
A phylogenomic tree (15) describing the evolution of 776 folds defined by the Structural Classification of Proteins (SCOP) (12) shows that folds appearing early in evolution were widely shared by proteomes in all organisms that have been fully sequenced (Fig. 1). Details on the evolutionary model used in phylogenetic analysis and the validity of rooting of our phylogenomic trees (summarized in Materials and Methods) have been described, together with limitations and biases of the reconstruction method (13–15). There were only 16 omnipresent folds, nine of which appeared at the base of the tree. Twelve of the omnipresent folds, including the nine most ancient and basal folds, contained omnipresent superfamilies that also appeared at the base of trees of superfamilies (15). These nine ancient folds represent architectures of fundamental importance (SI Table 1) undisputedly encoded in a genetic core that can be traced back to the universal ancestor of the three superkingdoms of life (20). These architectures are widespread in metabolism and are present even in parasitic organisms with highly reduced genomes and proteome complements. Phylogenomic reconstruction of evolutionary relationships between these ancestral folds showed that the P-loop-containing nucleoside triphosphate hydrolase fold (c.37) was the most ancient architecture, followed by the DNA/RNA-binding three-helical bundle fold (a.4), and then by the two most multifunctional and widely shared folds in metabolism, the TIM βα-barrel (c.1) and the NAD(P)-binding Rossmann (c.2) folds (Fig. 1). The P-loop hydrolase fold represents a single superfamily that was also basal in trees of superfamilies (15). Phylogenetic relationships in the tree of nine ancient folds were congruent with those in the global tree of architectures (Fig. 1). All of these omnipresent architectures were also widely distributed throughout metabolism. Using MANET, we identified metabolic enzymes with one or more domains having structures that match the nine ancient folds in 105 of 133 subnetworks, present in 11 mesonetworks defining core metabolism in KEGG (see SI Table 2 for data and nomenclature). The structural associations were also functional when the main enzymatic activities were linked directly to the ancient folds. These enzymes had highly diverse functions (Fig. 1), with 3–6, 8–33, 10–67, and 18–205 enzymatic activities defined at the first (class), second (subclass), third (subsubclass), and fourth (enzyme specificity) levels of Enzyme Commission (EC) classification, respectively (SI Table 3).
Fig. 1.
Metabolism and the protein world. Reconstruction of a phylogenomic tree of protein fold architecture using data from a domain census in 185 fully sequenced genomes representing the three superkingdoms of life (15). One optimal most-parsimonious tree [85,644 steps; consistency index (CI) = 0.043; retention index (RI) = 0.770; length skewness (g1) = −0.136; permutation tail probability (PTP) test, P = 0.01] was recovered after a heuristic search with tree-bisection-reconnection branch swapping and 100 replicates of random addition sequence. Phylogenetically uninformative characters were excluded from the analysis. To decrease search times during branch swapping of suboptimal trees, no more than one tree was saved in each replicate. The tree depicted evolutionary relationships of 776 SCOP folds, was well resolved, had strong cladistic structure (P < 0.01), and was consistent with phylogenies generated from a set of 32 proteomes using a similar approach (13). Bullets identify 16 folds shared by the genomes analyzed (c.37, a.4, c.1, c.2, d.58. c.23, c.55, b.40, c.66, c.47, d.15, a.2, d.142, b.34, a.5, and c.120, from ancestral to derived; see SI Fig. 6 for fold names). All other terminal leaves are unlabeled because they would not be legible. A phylogenomic tree of the nine most ancient and widely shared folds identified in the global tree is described separately. An exhaustive maximum parsimony search resulted in one tree of 2,069 steps (CI = 0.687, RI = 0.728) that was well supported by bootstrap support (BS) values (shown below nodes) and decay indices (in parentheses) and measures of skewness in tree distribution (see Inset; PTP test, P = 0.01). Enzymatic activities associated with these nine ancestral folds were retrieved from MANET. These activities describe variability in reaction chemistry, indicating number of EC entries defined at the four different levels of classifications: class (A, one of six general enzyme categories), subclass (B, denoting type of chemical compound or group involved in the reaction), subsubclass (C, describing the type of reaction), and serial identifier (D, identification of individual enzymes). Discovered and rediscovered enzymatic activities are plotted in bar diagrams. The bar diagram above the universal tree shows range of distribution of folds unique to Archaea (A), Bacteria (B), and Eukarya (E) in the tree (red bars), those folds shared by prokaryotes (pink bar) and by other superkingdoms. The upper bound for organismal diversification is shown by coloring tree branches in red.
Most Enzymatic Functions Were Discovered at the Start of the Protein World.
The accumulation of newly discovered enzymatic activities along the entire phylogenomic tree of protein architecture (Fig. 2) showed that most activities defined at different levels of EC classification were clearly associated with the first nine, and to a lesser degree, with the first 24, folds (SI Fig. 6). These trends suggest that, during evolution of ancient architectures, there was a burst of enzymatic innovation starting in primordial metabolic networks and extending throughout modern metabolism. In fact, we found noticeable patterns of innovation, such as the existence of a burst of enzymes transferring phosphorus-containing groups with an alcohol group as acceptor (EC 2.7.1) associated with the ancient c.37 fold, a subsequent burst of enzymatic diversification associated with the c.1 fold involving discovery and diversification of isomerases (EC 5), discovery of glycosidases (EC 3.2.1), and diversification of lyases (EC 4), and episodes of diversification of dehydrogenases (EC 1.1.1) and of lyases associated with the c.2 fold. Functions associated with the nine ancestral folds are described in SI Text. Remarkably, the EC 2.7.1 transferase burst of enzymes harboring the c.37 fold appeared ancient, involved 11 subnetworks, and originated in the purine metabolism subnetwork (see below). Evidently, enzymatic diversification occurred very early, ≈300 folds away from folds delimiting episodes of prokaryotic and eukaryotic-specific protein diversification and defining upper bounds for organismal diversification (Fig. 1). Indeed, at the time of appearance of superkingdom-specific folds, most enzymatic activities had been already discovered at all levels of EC classification (Fig. 2). Consequently, the common ancestor of diversified life probably had a complete metabolic toolkit.
Fig. 2.
Discovery of enzymatic functions. The accumulation of newly discovered enzymatic activities along the phylogenomic tree of protein architecture was given as a function of distance in nodes from a hypothetical ancestral fold (nd) normalized to a 0–1 scale. The 9 and 24 most ancestral folds defined relative time frames (shaded area) in which newly discovered activities reached 80% and 100% of total EC entries analyzed at subclass (EC A.B) level, respectively. The dashed line delimits the upper bound for organismal diversification, at which time 100%, 100%, 98.2%, and 95.7% of enzymatic activities had been already discovered at first, second, third, and fourth levels of EC classification, respectively. Computational implementations are in SI Text.
Phylogenetic Analysis of Structure Identifies Ancient Metabolic Subnetworks.
We then focused on the presence of the nine ancestral folds in metabolic subnetworks and devised a phylogenetic method to make inferences about the history of subnetworks. For this purpose, we introduced a previously undescribed phylogenetic feature (character), the abundance or occurrence of an ancient fold in a subnetwork (see assumptions in SI Text). The phylogenetic criterion of primary homology underlying the use of these characters was the sharing of ancient protein architectures by the subnetworks resulting from enzyme recruitment processes. Analysis of occurrence and abundance of folds in enzymes of the 133 subnetworks (SI Table 2) shows that 28 subnetworks did not contain any of the nine most ancient folds and should be considered evolutionarily derived (SI Table 4). They were removed from further analysis. Nine of these lacked structural assignments and were uninformative. These 28 subnetworks belonged to seven mesonetworks, one to metabolism of other amino acids (AA2), one to metabolism of cofactors and vitamins (COF), two to energy metabolism (NRG), four to glycan biosynthesis and metabolism (GLY), six to biosynthesis of polyketides and nonribosomal peptides (POL), nine to biosynthesis of secondary metabolites (SEC), and five to biodegradation of xenobiotics (XEN). Two derived energy-linked subnetworks stand out in the list, oxygenic mitochondrial ATP synthesis (NRG 00193) and oxygenic photosynthesis (NRG 00195), suggesting these important functions appeared late in evolution, well after discovery of most enzymatic activities. This is consistent with molecular and geological records that suggest life achieved considerable complexity before the appearance of oxygen in the atmosphere, and with enzyme distribution in aerobic pathways that suggests adaptation to oxygen occurred after major prokaryotic divergences in the tree of life (21). Subnetworks with many ancient folds belonging to the remaining mesonetworks, amino acid metabolism (AAC), carbohydrate metabolism (CAR), lipid metabolism (LIP) and nucleotide metabolism (NUC), were clearly ancestral and part of the early enzymatic burst.
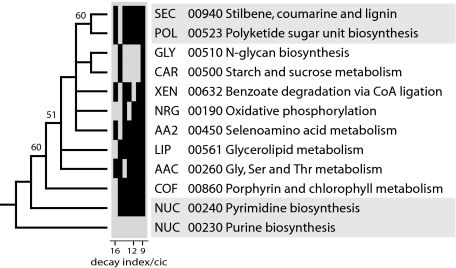
We used this phylogenetic method to generate rooted trees of subnetworks for each mesonetwork. We focused on mesonetworks because the global tree of subnetworks was poorly resolved. Trees reconstructed from fold abundance in subnetworks (SI Fig. 7) were generally congruent with those reconstructed from fold occurrence but carried more phylogenetic information (not shown). Clearcut subnetwork candidates of origin for each mesonetwork were identified at the base of individual trees, and these subnetworks were used to generate a tree of ancient subnetworks (Fig. 3). This tree was congruent with a tree describing the evolution of mesonetworks (SI Fig. 8), providing further confidence in statements of subnetwork evolution. In the tree of ancient subnetworks, the two subnetworks of the nucleotide metabolism mesonetwork, purine metabolism (NUC 00230) and pyrimidine metabolism (NUC 00240), were placed at its base. These subnetworks were followed by the porphyrin and chlorophyll metabolism subnetwork (COF 00860). This is noteworthy because nucleotides, and to a lesser extent selected cofactors in the COF mesonetwork, should be considered linked to RNA, conserved throughout life (18), and important components of an ancient RNA world (22). Two subnetworks were clearly derived, the polyketide sugar unit biosynthesis (POL 00523) and the stilbene, coumarine, and lignin (SEC 00940) subnetworks. These subnetworks belong to POL and XEN, mesonetworks that also harbor the largest number of subnetworks lacking ancestral folds. Other interesting evolutionary patterns were evident. For example, the citrate cycle (CAR 000200) subnetwork is derived in the CAR mesonetwork (SI Fig. 7), and CAR is quite derived within mesonetworks (Fig. 3 and SI Fig. 8). However, scenarios for the prebiotic evolution of metabolism suggest that the citric acid cycle was one of the first pathways to evolve (23, 24). Consequently, our results suggest prebiotic pathways evolved in a sequence unrelated to the pattern of subsequent enzymatic takeovers.
Fig. 3.
Evolution of ancient subnetworks in mesonetworks. Two optimal most-parsimonious trees of 119 steps (CI = 0.580, RI = 0.587; g1 = −0.538; PTP test, P = 0.01) describing the origins of mesonetworks were recovered after a branch-and-bound search. The tree shown represents a strict consensus of the two trees. Branches with BS values >50% are shown above nodes. Vertical bars in the bar diagram describe the identity of terminal taxa joined by individual reduced cladistic consensus (RCC) support trees derived from double decay (DD) analysis. Within the seven RCC topologies, total decay ranged from 112 to 223 steps, and cladistic information content (cic) values ranged from 6.7 to 21.0. RCC topologies are presented in order, starting with the most informative (i.e., with higher decay-to-cic values), and support the phylogenetic statement.
Metabolism Originated in Nucleotide Metabolism Subnetworks.
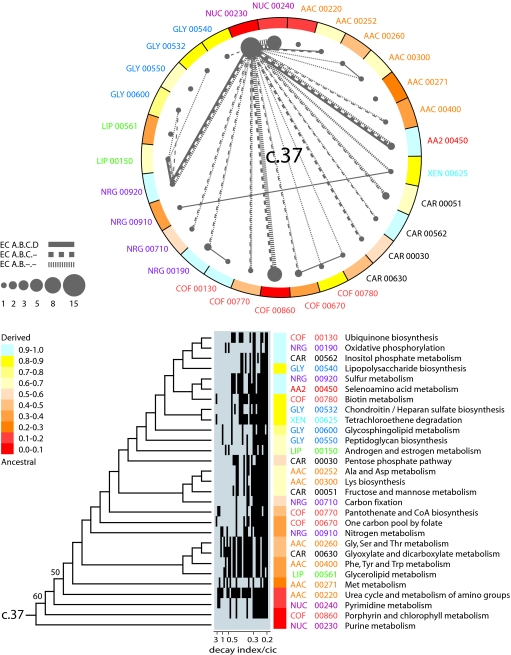
Because recruitment erases historical patterns of enzymes in networks, we used “subnetwork wheels” to reveal patterns of origin and evolution in metabolism. For each fold, these graphs represent subnetworks as vertices (nodes) and sharing of enzymatic activities (EC numbers at different levels of classification) as edges (lines connecting nodes). We assume that in network evolution, enzymes take over ancient or prebiotic reactions. In this process, a copy of a protein domain used in one metabolic context (donor site) begins functioning in a new context (host site), performing that function de novo or taking it over from the previous catalyst at the host site. This process overlaps with the invention of new architectures, beginning with the most ancient one, each new one contributing novel functions and new opportunities for recruitment. Although extant donor and host domains may differ, we assume successful recruitment results in evolutionary lockin at a structural level [structural canalization (25)] necessary to guarantee the maintenance of the fold architecture. Similarly, we consider that change is costly, and that takeovers are more plausible among sublevels within each EC classification level. Given these assumptions, four criteria were used to reveal evolutionary patterns of recruitment between subnetworks: (i) the abundance of the fold in each subnetwork, (ii) the ancestry of each subnetwork derived from trees of subnetworks, (iii) the sharing of enzymatic activities by subnetworks at different levels of EC classification, and (iv) phylogenomic superfamily relationships of the shared enzymes. These criteria provided weights to the vertices and edges of the subnetwork wheels that helped establish direction of enzyme recruitment.
Fig. 4 shows a subnetwork wheel for the most ancient architecture, the P-loop hydrolase fold. Twenty-nine subnetworks had enzymes that shared this fold, and a tree of these subnetworks again had purine metabolism, pyrimidine metabolism, and porphyrin and chlorophyll metabolism at its base. Fold abundance was also maximal in these three subnetworks. Purine metabolism appeared as the fundamental vertex of enzymatic sharing in the c.37 wheel, judged by the high degree of connectivity of this subnetwork at different levels of EC classification and the direction of enzyme recruitment. It is noteworthy that highly weighted connectivities were also established among these three most ancient subnetworks, especially at subclass level, most notably between the nucleotide metabolism subnetworks. There was also significant enzymatic sharing between purine metabolism and both sulfur (NRG 00920) and selenoamino acid metabolism (AA2 00450), but these two subnetworks had low fold abundance and were clearly derived in the set. We believe these instances of sharing represent late recruitment processes.
Fig. 4.
A metabolic subnetwork wheel for the P-loop hydrolase fold. The graph shows subnetworks containing the c.37 fold as vertices, with numerical properties of vertices describing fold abundance and ancestries of the subnetworks and sharing of EC number at different levels of classification as edges, with line values describing sharing frequency. Node area is proportional to fold abundance, and line width is proportional to sharing of enzymatic activities. A single optimal most-parsimonious tree (208 steps; CI = 0.380, RI = 0.590; g1 = −0.495; PTP test, P = 0.01) describing the evolution of subnetworks harboring the c.37 fold (shown below the wheel) was recovered after a heuristic search with tree-bisection-reconnection branch swapping and 10 replicates of random addition sequence. Branches with BS values >50% are shown above nodes. Despite low BS values, RCC support trees derived from double decay (DD) analysis (described by rows in the bar diagram; see Fig. 3) showed that the topology of the tree was reliable. Within the 34 RCC topologies, total decay ranged from 5 to 27 steps and cladistic information content (cic) values ranged from 1.6 to 116.9. Subnetwork ancestries derived from the tree of subnetworks are given as a function of distance in nodes from a hypothetical ancestral subnetwork and are color coded and used to paint the wheel of subnetworks.
The ancestral enzymes in nucleotide metabolism were probably phosphotransferases transferring P-containing groups with an alcohol (EC 2.7.1) or a phosphate group (EC 2.7.4) as acceptors, hydrolases acting on P-containing acid anhydrides (EC 3.6) and perhaps ligases forming C–N bonds (EC 6.3.4) (SI Tables 5 and 6). It is likely that these enzymes were not part of ancient purine and pyrimidine biosynthetic pathways. Instead, they were involved in nucleotide interconversion, distribution (storage and recycling) of chemical energy in acid-anhydride bonds of nucleotides, and terminal production of nucleotides and cofactors. In this regard, enzymatic activities shared between the purine metabolism and the porphyrin and chlorophyll metabolism subnetworks involved phosphotransferases (e.g., that phosphorylate adenosylcobinamide; EC 2.7.1) and ligases that form C–N bonds (EC 6.3).
Conclusions
Our results suggest strongly that modern metabolism originated in nucleotide metabolism, probably in pathways of purine metabolism. This is of great significance. The first enzymatic takeover of an ancient biochemistry or prebiotic chemistry involved processes related to the synthesis of nucleotides for a world in which RNA was the only genetically encoded catalyst (26). Although the RNA world has considerable explanatory power, explaining, for example, why RNA is at the core of translation (27), we know little of how this world transitioned into modern biochemistry (28). The origin of protein synthesis must have been the first step toward a ribonucleoprotein world, and the transition was probably driven by the superior catalytic ability of polypeptides and then proteins. Our findings suggest that modern metabolism developed early at the onset of protein discovery and had origins that benefited the formation of building blocks for the RNA world.
Materials and Methods
Phylogenomic trees of protein architectures were derived from an HMM-driven genomic census of protein folds (defined by using SCOP 1.67) (15) in 19 archaeal, 129 bacterial, and 37 eukaryal fully sequenced genomes. Normalized fold abundance data were coded as polarized linearly ordered multistate phylogenetic characters and subjected to phylogenetic analysis using maximum parsimony as the optimality criterion in PAUP* (29). Trees were rooted without the need of external hypotheses (outgroups) by polarizing characters directly with an evolutionary model in which protein architectures that are more prevalent in nature (i.e., reused in many biological contexts) originate from innovations in structural design that occur earlier in evolutionary time (13). The ancestral condition for architectures in proteomes (popular but not necessarily widely shared) was specified by inclusion of a hypothetical ancestor in the search for optimal trees. Because folds or superfamilies are retained over long evolutionary time scales, their gain or loss constitutes an important evolutionary event that appears to be relatively independent of the vagaries of horizontal gene transfer and other convergent evolutionary processes (30). Additional details on character argumentation, absence of circularity in assumptions, and rarity of convergent evolutionary processes can be found in SI Text and elsewhere (13–15).
Metabolic networks were analyzed by using MANET release 1.0 and Perl scripts associated with the database (17). Enzymatic activities associated with ancient folds either directly (the fold harbored the active site) or indirectly (the fold provided structural or auxiliary functions) were identified and used to build a data matrix for phylogenetic analysis. Phylogenetic trees of KEGG subnetworks and mesonetworks were reconstructed by using maximum parsimony from polarized binary and linearly ordered characters describing the presence or abundance of the nine most ancient folds in the networks. Phylogenetic reliability was evaluated by bootstrap and double decay analyses. Metabolic subnetwork wheels were visualized by using PAJEK (31). See SI Text for a detailed description of assumptions and methods.
Supplementary Material
Acknowledgments
We thank Minglei Wang for phylogenomic reconstructions and Gloria Caetano-Anollés for continued encouragement. The research was supported in part with funds from the University of Illinois at Urbana–Champaign and by the Office of Naval Research (Grant TRECC A6538-A76, to G.C.-A.) and the National Science Foundation (Grant MCB-0343126, to G.C.-A.).
Abbreviations
- EC
Enzyme Commission
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- MANET
molecular ancestry network
- RCC
reduced cladistic consensus
- SCOP
Structural Classification of Proteins
- BS
bootstrap support
- CI
consistency index
- PTP
permutation tail probability.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0701214104/DC1.
References
- 1.Barabási AL, Oltvai ZN. Nat Rev Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 2.Schmidt S, Sunyaev S, Bork P, Dandekar T. Trends Biochem Sci. 2003;28:336–341. doi: 10.1016/S0968-0004(03)00114-2. [DOI] [PubMed] [Google Scholar]
- 3.Ycas M. J Theor Biol. 1974;44:145–160. doi: 10.1016/s0022-5193(74)80035-4. [DOI] [PubMed] [Google Scholar]
- 4.Jensen RA. Annu Rev Microbiol. 1976;30:409–425. doi: 10.1146/annurev.mi.30.100176.002205. [DOI] [PubMed] [Google Scholar]
- 5.Copley RR, Bork P. J Mol Biol. 2000;303:627–640. doi: 10.1006/jmbi.2000.4152. [DOI] [PubMed] [Google Scholar]
- 6.Nagano N, Orengo CA, Thornton JM. J Mol Biol. 2002;321:741–765. doi: 10.1016/s0022-2836(02)00649-6. [DOI] [PubMed] [Google Scholar]
- 7.Teichmann SA, Rison SC, Thornton JM, Riley M, Gough J, Cothia C. J Mol Biol. 2001;311:693–708. doi: 10.1006/jmbi.2001.4912. [DOI] [PubMed] [Google Scholar]
- 8.Teichmann SA, Rison SCG, Thornton JM, Riley M, Gough J, Chothia C. Trends Biotechnol. 2001;19:482–486. doi: 10.1016/s0167-7799(01)01813-3. [DOI] [PubMed] [Google Scholar]
- 9.Saqui MAS, Sternberg JE. J Mol Biol. 2001;313:1195–1206. doi: 10.1006/jmbi.2001.5107. [DOI] [PubMed] [Google Scholar]
- 10.Rison SCG, Teichmann SA, Thornton JM. J Mol Biol. 2002;318:911–932. doi: 10.1016/S0022-2836(02)00140-7. [DOI] [PubMed] [Google Scholar]
- 11.Chothia C, Gough J, Vogel C, Teichmann SA. (2003) Science. 2003;300:1701–1703. doi: 10.1126/science.1085371. [DOI] [PubMed] [Google Scholar]
- 12.Murzin AG, Brenner SE, Hubbard T, Chothia C. J Mol Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 13.Caetano-Anollés G, Caetano-Anollés D. Genome Res. 2003;13:1563–1571. doi: 10.1101/gr.1161903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caetano-Anollés G, Caetano-Anollés D. J Mol Evol. 2005;60:484–498. doi: 10.1007/s00239-004-0221-6. [DOI] [PubMed] [Google Scholar]
- 15.Wang M, Boca SM, Kalelkar R, Mittenthal JE, Caetano-Anollés GA. Complexity. 2006;12:27–40. [Google Scholar]
- 16.Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. Nucleic Acids Res. 2004;32:D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim HS, Mittenthal JE, Caetano-Anollés G. BMC Bioinformatics. 2006;7:351. doi: 10.1186/1471-2105-7-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Benner SA, Ellington AD, Tauer A. Proc Natl Acad Sci USA. 1989;86:7054–7058. doi: 10.1073/pnas.86.18.7054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morowitz HJ. Beginning of Cellular Life. New Haven, CT: Yale Univ Press; 1992. [Google Scholar]
- 20.Harris JK, Kelley ST, Spiegelman GB, Pace NR. Genome Res. 2003;13:407–412. doi: 10.1101/gr.652803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raymond J, Segrè D. Science. 2006;311:1764–1767. doi: 10.1126/science.1118439. [DOI] [PubMed] [Google Scholar]
- 22.White HB., Jr J Mol Evol. 1976;7:101–104. doi: 10.1007/BF01732468. [DOI] [PubMed] [Google Scholar]
- 23.Wächtershäuser G. Prog Biophys Mol Biol. 1992;58:85–201. doi: 10.1016/0079-6107(92)90022-x. [DOI] [PubMed] [Google Scholar]
- 24.Morowitz HJ, Kostelnik JD, Yang J, Ody GD. Proc Natl Acad Sci USA. 2000;97:7704–7708. doi: 10.1073/pnas.110153997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fontana W. BioEssays. 2002;24:1164–1177. doi: 10.1002/bies.10190. [DOI] [PubMed] [Google Scholar]
- 26.Gilbert W. Nature. 1989;319:618. [Google Scholar]
- 27.Orgel LE. Crit Rev Biochem Mol Biol. 2005;39:99–123. doi: 10.1080/10409230490460765. [DOI] [PubMed] [Google Scholar]
- 28.Penny D. Biol Phyl. 2005;20:633–671. [Google Scholar]
- 29.Swofford DL. Phylogenetic Analysis Using Parsimony and Other Programs (PAUP*) Sunderland, MA: Sinauer; 2002. Ver 4.0. [Google Scholar]
- 30.Gough J. Bioinformatics. 2005;21:1464–1471. doi: 10.1093/bioinformatics/bti204. [DOI] [PubMed] [Google Scholar]
- 31.Batagelj A, Mvar A. Connections. 1998;21:47–57. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.