Abstract
The human gut is home to trillions of microbes, thousands of bacterial phylotypes, as well as hydrogen-consuming methanogenic archaea. Studies in gnotobiotic mice indicate that Methanobrevibacter smithii, the dominant archaeon in the human gut ecosystem, affects the specificity and efficiency of bacterial digestion of dietary polysaccharides, thereby influencing host calorie harvest and adiposity. Metagenomic studies of the gut microbial communities of genetically obese mice and their lean littermates have shown that the former contain an enhanced representation of genes involved in polysaccharide degradation, possess more archaea, and exhibit a greater capacity to promote adiposity when transplanted into germ-free recipients. These findings have led to the hypothesis that M. smithii may be a therapeutic target for reducing energy harvest in obese humans. To explore this possibility, we have sequenced its 1,853,160-bp genome and compared it to other human gut-associated M. smithii strains and other Archaea. We have also examined M. smithii's transcriptome and metabolome in gnotobiotic mice that do or do not harbor Bacteroides thetaiotaomicron, a prominent saccharolytic bacterial member of our gut microbiota. Our results indicate that M. smithii is well equipped to persist in the distal intestine through (i) production of surface glycans resembling those found in the gut mucosa, (ii) regulated expression of adhesin-like proteins, (iii) consumption of a variety of fermentation products produced by saccharolytic bacteria, and (iv) effective competition for nitrogenous nutrient pools. These findings provide a framework for designing strategies to change the representation and/or properties of M. smithii in the human gut microbiota.
Keywords: archaeal-bacterial mutualism, comparative microbial genomics, functional genomics and metabolomics, gnotobiotic mice, human gut microbiota
The human gut microbiota is dominated by two divisions of bacteria, the Bacteroidetes and the Firmicutes, which together encompass >90% of all phylogenetic types (phylotypes). Archaea are also represented, most prominently by a methanogenic Euryarchaeote, Methanobrevibacter smithii, which comprises up to 10% of all anaerobes in the colons of healthy adults (1, 2).
Complex dietary polysaccharides (fiber) and proteins are digested by enzymes encoded by genes in the microbial community's collective genome (microbiome), but not in our human genome (3, 4). Bacterial fermentation of polysaccharides yields short chain fatty acids (SCFAs) (principally acetate, propionate, and butyrate), other organic acids (e.g., formate), alcohols (e.g., methanol and ethanol), and gases [e.g., hydrogen (H2) and carbon dioxide (CO2)]. Host absorption of SCFAs provides up to 10% of daily caloric intake, although this value varies depending on the glycan content of the diet (5). Archaeal methanogenesis improves the efficiency of polysaccharide fermentation in animal gut “bioreactors” by preventing the buildup of H2 and other reaction end products (6).
Several recent observations underscore the importance of delineating the genomic and metabolic underpinnings of M. smithii's contributions to energy balance and, if it is to be a therapeutic target for manipulation, the nature of its adaptations to the gut ecosystem. First, comparative metagenomic analysis of the gut microbiomes of genetically obese ob/ob mice and their lean ob/+ or +/+ littermates revealed that the obese community exhibits increased representation of archaea, division-wide increase in the proportion of Firmicutes relative to Bacteroidetes, and an accompanying enrichment of microbial genes involved in polysaccharide degradation (7). Moreover, transplantation of the gut microbiota from ob/ob or +/+ donors into lean +/+ germ-free (GF) mouse recipients disclosed that this increased energy harvesting capacity is transmissible: i.e., recipients of an obese donor's gut microbiota gained more body fat than did recipients of a lean donor's microbiota (7). Second, colonization of adult GF mice with M. smithii and/or Bacteroides thetaiotaomicron, a prominent sequenced human gut symbiont equipped with a large arsenal of glycoside hydrolases (GHs) not represented in our human proteome (8), revealed that the methanogen increased the efficiency, and changed the specificity of bacterial digestion of dietary glycans (6). Moreover, cocolonized mice exhibited a significantly greater increase in adiposity compared with mice colonized with either organism alone (6). Third, metagenomic studies of the colonic microbiomes of two healthy adults confirmed that M. smithii is a prominent component of this community and that enzymes involved in methanogenesis are well represented (3).
With these observations in mind, we have sequenced the genome of the M. smithii type strain PS, compared it to other M. smithii strains isolated from the human gut and to other archaeons, and have performed functional genomic and biochemical analyses of its properties in gnotobiotic mice that do or do not harbor B. thetaiotaomicron. The results provide insights about M. smithii's niche (profession), its evolved adaptations to its gut habitat, and strategies for identifying targets for development of antiarchaeal agents.
Results and Discussion
The 1,853,160-bp genome of the M. smithii-type strain PS contains 1,795 predicted protein coding genes [supporting information (SI) Tables 1–4 and Results in SI Text], 34 tRNAs, and two rRNA clusters. We compared its proteome with the proteomes of (i) Methanosphaera stadtmanae, a methanogenic Euryarchaeote that is a minor and inconsistent member of the human gut microbiota (1); (ii) nine “nongut methanogens” recovered from microbial communities in the environment; and (iii) these nongut methanogens plus an additional 17 sequenced Archaea (“all archaea”) (SI Table 5).
Compared with nongut methanogens and/or all archaea, M. smithii and M. stadtmanae are significantly enriched (binomial test, P < 0.01) for genes assigned to gene ontology (GO) categories involved in surface variation (e.g., cell wall organization and biogenesis; see below), defense (e.g., multidrug efflux/transport), and processing of bacteria-derived metabolites (SI Tables 6 and 7).
The M. smithii and M. stadtmanae genomes exhibit limited global synteny (SI Fig. 4) but share 968 proteins with mutual best BLAST hit E values ≤10−20 (46% of all M. smithii proteins; SI Table 8). A predicted interaction network of M. smithii clusters of orthologous groups (COGs) constructed based on STRING (9) shows that it contains more COGs for persistence, improved metabolic versatility, and machinery for genomic evolution compared with M. stadtmanae (SI Fig. 5 and SI Table 9).
Cell Surface Variation.
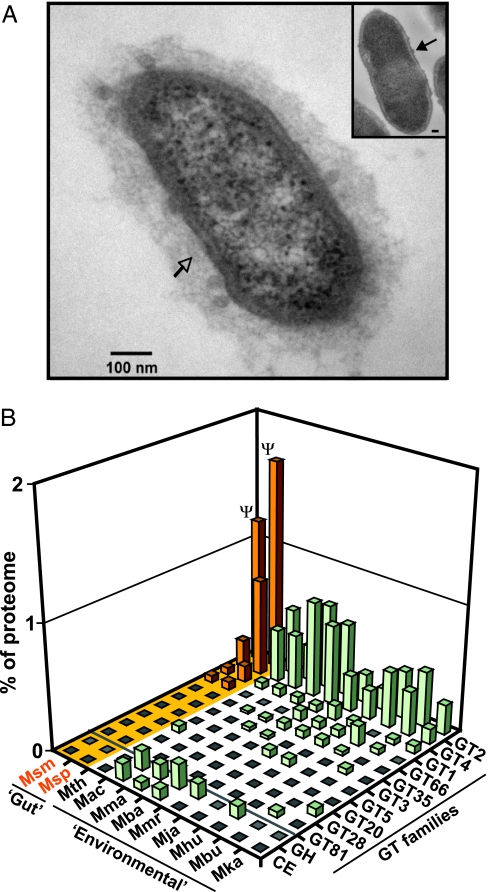
The ability to vary capsular polysaccharide surface structures in vivo by altering expression of glycosyltransferases (GTs) is a feature shared among sequenced bacterial species that are prominent in the distal human gut microbiota (4, 10–12). Transmission EM studies of M. smithii harvested from gnotobiotic mice after a 14-day colonization revealed that it too has a prominent capsule (Fig. 1A). The proteomes of both human gut methanogens also contain an arsenal of GTs [26 in M. smithii and 31 in M. stadtmanae; see SI Table 10 for a complete list organized on the basis of the Carbohydrate Active enZYme (CAZy) classification scheme (www.cazy.org); ref. 13]. Unlike the sequenced Bacteroidetes, which possess large repertoires of GH and carbohydrate esterases (CE) not represented in the human “glycobiome,” neither gut methanogen has any detectable GH or CE family members (Fig. 1B). Both M. smithii and M. stadtmanae dedicate a significantly larger proportion of their “glycobiome” to GT2 family glycosyltransferases than any of the sequenced nongut associated methanogens (binomial test; P < 0.00005; Fig. 1B). These GT2 family enzymes have diverse predicted activities, including synthesis of hyaluronan, a component of human glycosaminoglycans in the mucosal layer.
Fig. 1.
M. smithii decorates its cell surface to mimic the host glycan landscape. (A) Transmission electron microscopy (TEM) of M. smithii harvested from the ceca of adult germ-free (GF) mice after a 14-d colonization. (Inset) A comparable study of stationary phase M. smithii recovered from a batch fermentor containing Methanobrevibacter complex medium (MBC). Note that the size of the capsule is greater in cells recovered from the cecum (open vs. closed arrow). (Scale bar: 100 nm.) (B) Comparison of GT, GH, and CE families (defined in CAZy; SI Table 10) represented in the genomes of the following sequenced methanogens (see SI Table 5): M. smithii (Msm); M. stadtmanae (Msp); Methanothermobacter thermoautotrophicus (Mth); Methanosarcina acetivorans (Mac); Methanosarcina barkeri (Mba); Methanosarcina mazei (Mma); Methanococcus maripaludis (Mmr); Methanococcus jannaschii (Mja); Methanospirillum hungatei (Mhu); Methanococcoides burtonii (Mbu); and Methanopyrus kandleri (Mka). Gut methanogens (highlighted in orange) have no GH or CE family members but have a larger proportion of family 2 GTs (Ψ, P < 0.00005 on the basis of binomial test for enrichment vs. non-gut-associated methanogens).
Sialic acids are a family of nine-carbon sugars that are abundantly represented in human mucus- and epithelial cell surface-associated glycans (14). N-acetylneuraminic acid (Neu5Ac) is the predominant type of sialic acid found in our species. Unique among sequenced archaea, M. smithii has a cluster of genes (MSM1535-1540) that encode all enzymes necessary for de novo synthesis of sialic acid from UDP-N-acetylglucosamine (SI Fig. 6A). Quantitative RT-PCR (qRT-PCR) assays of RNAs prepared from the cecal contents of 12-week-old gnotobiotic mice that had been colonized for 14 d with the archaeon alone or with B. thetaiotaomicron for 14 d followed by the addition of M. smithii for 14 d (n = 5–6 mice per treatment group) revealed that this cluster of genes is expressed in vivo at equivalent levels in mono- and cocolonized mice (SI Table 11). Biochemical analysis of extracts prepared from cultured M. smithii, plus histochemical staining of the microbe with the sialic acid-specific lectin, Sambucus nigra agglutinin (SNA), confirmed the presence of Neu5Ac (SI Fig. 6 B and C). Taken together, our findings indicate that M. smithii has developed mechanisms to decorate its surface with carbohydrate moieties that mimic those encountered in the glycan landscape of its intestinal habitat.
The genomes of both human gut methanogens also encode a class of predicted surface proteins that have features similar to bacterial adhesins (48 members in M. smithii and 37 members in M. stadtmanae). A phylogenetic analysis (see Materials and Methods in SI Text) indicated that each methanogen has a specific clade of these adhesin-like proteins (ALPs) (SI Fig. 7A). A subset of M. smithii ALPs has homology to pectin esterases (GO 0030599): this GO family, which is significantly enriched in this compared with other Archaea on the basis of the binomial test (P < 0.0005; SI Table 6) is associated with binding of chondroitin, a major component of mucosal glycosaminoglycans. Several other M. smithii ALPs have domains predicted to bind other sugar moieties (e.g., galactose-containing glycans; SI Fig. 7A). Both methanogens also have ALPs with peptidase-like domains (see SI Table 12 for a complete list of InterPro domains).
We conducted qRT-PCR assays of cecal RNAs from the mono- and cocolonized gnotobiotic mice described above. The results revealed one “sugar-binding” ALP (MSM1305) that was significantly up-regulated in the presence of B. thetaiotaomicron, four that were suppressed (including one with a GAG binding domain), and two that exhibited no statistically significant alterations (SI Fig. 7B). Regulated expression of distinct subsets of ALPs may direct this methanogen to specific intestinal microhabitats where close association with saccharolytic bacterial partners could promote establishment and maintenance of syntrophic relationships.
Methanogenic and Nonmethanogenic Removal of Bacterial End Products of Fermentation.
Compared with other sequenced non-gut-associated methanogens, M. smithii has significant enrichment of genes involved in utilization of CO2, H2 and formate for methanogenesis (GO 0015948; SI Table 6). They include genes that encode proteins involved in synthesis of vitamin cofactors used by enzymes in the methanogenesis pathway [methyl group carriers (F430 and corrinoids), riboflavin (precursor for F430 biosynthesis), and coenzyme M synthase (involved in the terminal step of methanogenesis)] (see SI Table 7 for a gene list and Fig. 2A for the metabolic pathways). M. smithii also has an intact pathway for molybdopterin biosynthesis to allow for CO2 utilization (SI Fig. 8). qRT-PCR assays demonstrated that although key central methanogenesis enzymes are constitutively expressed in the presence or absence of B. thetaiotaomicron [Fwd (tungsten formylmethanofuran dehydrogenase), Hmd (methylene-H4MPT dehydrogenase), and Mcr (methyl-CoM reductase)], ribofuranosylaminobenzene 5′-phosphate (RFA-P)-synthase (RfaS, MSM0848), an essential gene involved in methanopterin biosynthesis is significantly up-regulated with cocolonization (see Fig. 2A and SI Table 11 for qRT-PCR results). M. smithii also up-regulates a formate utilization gene cluster (FdhCAB, MSM1403–5) for methanogenic consumption of this B. thetaiotaomicron-produced metabolite (6).
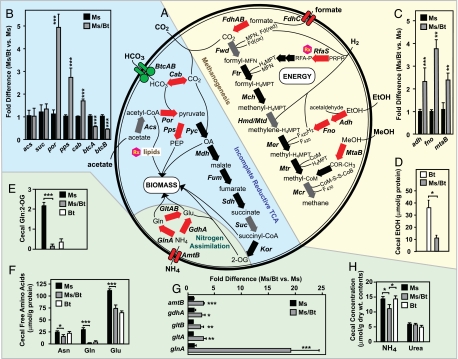
Fig. 2.
Functional genomic and biochemical assays of M. smithii metabolism in the ceca of gnotobiotic mice. (A) In silico metabolic reconstruction of M. smithii pathways involved in (i) methanogenesis from formate, H2/CO2, and alcohols, (ii) carbon assimilation from acetate and bicarbonate, and (iii) nitrogen assimilation from ammonium. Acs, acetyl-CoA synthase; Adh, alcohol dehydrogenase; AmtB, ammonium transporter; BtcA/B, bicarbonate (HCO3) ABC transporter; Cab, carbonic anhydrase; CH3, methyl; CoB, coenzyme B; CoM, coenzyme M; COR, corrinoid; F420, cofactor F420; F430, cofactor F430; Fd, ferredoxin (ox, oxidized; red, reduced); FdhAB, formate dehydrogenase subunits; FdhC, formate transporter; Fno, F420-dependent NADP oxidoreductase; Ftr, formylmethanofuran: tetrahydromethanopterin (H4MPT) formyltransferase; Fum, fumarate hydratase; Fwd, tungsten formylmethanofuran dehydrogenase; GdhA, glutamate dehydrogenase; GlnA, glutamine synthetase; GltA/B, glutamate synthase subunits A and B; Hmd, H2-forming methylene-H4MPT dehydrogenase; Kor, 2-oxoglutarate synthase; Mch, methenyl-H4MPT cyclohydrolase; Mcr, methyl-CoM reductase; Mdh, malate dehydrogenase; MeOH, methanol; Mer, methylene-H4MPT reductase; MFN, methanofuran; MtaB, methanol:cobalamin methyltransferase; Mtd, F420-dependent methylene-H4MPT dehydrogenase; Mtr, methyl-H4MPT:CoM methyltransferase; NH4, ammonium; OA, oxaloacetate; PEP, phosphoenol pyruvate; Por, pyruvate:ferredoxin oxidoreductase; Pps, phosphoenolpyruvate synthase; PRPP, 5-phospho-a-d-ribosyl-1-pyrophosphate; Pyc, pyruvate carboxylase; RfaS, ribofuranosylaminobenzene 5′-phosphate (RFA-P) synthase; Sdh, succinate dehydrogenase; Suc, succinyl-CoA synthetase. Potential drug targets are denoted by “Rx.” (B, C, and G) qRT-PCR assays of the expression of key M. smithii (Ms) genes in gnotobiotic mice that do or do not harbor B. thetaiotaomicron (Bt) (n = 5–6 animals per group; each sample assayed in triplicate; mean values ± SEM plotted; see SI Table 11 for full list of analyses). Results are summarized in A by using the following color codes: red, up-regulated; green, down-regulated; gray, assayed but no significant change; black arrows, transcript not assayed. (D) Ethanol (EtOH) levels in the ceca of mice colonized with B. thetaiotaomicron ± M. smithii (n = 5–7 animals per group representing two independent experiments; each sample assayed in duplicate; mean values ± SEM plotted). (E) Ratio of cecal concentrations of glutamine (Gln) and 2-oxoglutarate (2-OG) (n = 5 animals per group; samples assayed in duplicate; mean values ± SEM). (F) Cecal levels of free Gln, Glu (glutamate), and Asn (asparagine) (n = 5 animals per group; samples assayed in duplicate; mean values ± SEM). (H) Cecal ammonium and urea levels (n = 5–15 mice per group; three independent experiments). ∗, P < 0.05; ∗∗, P < 0.01; ∗∗∗, P < 0.005, according to Student's t test.
Our previous qRT-PCR and mass spectrometry studies revealed that cocolonization increased B. thetaiotaomicron acetate production [acetate kinase (BT3963) is 9-fold up-regulated vs. B. thetaiotaomicron monoassociated controls; P < 0.0005; n = 4–5 animals per group; ref. 6]. Although acetate is not converted to methane by M. smithii (15), we found that its proteome contains an “incomplete reductive tricarboxylic acid (TCA) cycle” that would allow it to assimilate acetate [Acs (acetyl-CoA synthetase, MSM0330), Por (pyruvate:ferredoxin oxidoreductase, MSM0557–60), Pyc (pyruvate carboxylase, MSM0765), Mdh (malate dehydrogenase, MSM1040), Fum (fumarate hydratase, MSM0447, MSM0563, MSM0769, MSM0929), Sdh (succinate dehydrogenase, MSM1258), Suc (succinyl-CoA synthetase, MSM0228, MSM0924), and Kor (2-oxoglutarate synthase, MSM0925–8) in Fig. 2A]. qRT-PCR assays disclosed that cocolonization up-regulated two important M. smithii genes associated with this pathway that participate in acetate assimilation: Por and Cab (carbonic anhydrase, MSM0654, MSM1223), which converts CO2 to bicarbonate, a substrate for Por (Fig. 2B).
M. smithii also possesses enzymes that in other methanogens facilitate utilization of two other products of bacterial fermentation, methanol and ethanol (16, 17). qRT-PCR assays showed that cocolonization significantly increased expression of a methanol:cobalamin methyltransferase (MtaB, MSM0515), an NADP-dependent alcohol dehydrogenase (Adh, MSM1381), and an F420-dependent NADP oxidoreductase (Fno, MSM0049) (2.4 ± 0.3, 2.3 ± 0.4, and 3.7 ± 0.4 fold vs. monoassociated controls, respectively; P < 0.01; see Fig. 2A for pathway information, and Fig. 2C for qRT-PCR results]. Follow-up biochemical studies confirmed a significant decrease in ethanol levels in the ceca of cocolonized mice (11 ± 2 μmol/g total protein in cecal contents vs. 35 ± 6 μmol/g in mice with B. thetaiotaomicron alone; n = 5–7 animals per group; P < 0.05; Fig. 2D). Moreover, levels of ethanol in M. smithii monoassociated controls were not significantly different from background levels defined in germ-free controls (n = 5–7 animals per group; P > 0.05; data not shown). Expression of B. thetaiotaomicron's alcohol dehydrogenases (BT4512 and BT0535) was not altered by cocolonization (6), indicating that the reduction in cecal ethanol levels observed in cocolonized mice is not due to diminished bacterial production but rather to increased archaeal consumption.
Collectively, these findings indicate that M. smithii supports methanogenic and nonmethanogenic removal of diverse bacterial end products of fermentation: this capacity may endow it with greater flexibility to form syntrophic relationships with a broad range of bacterial members of the distal human gut microbiota.
M. smithii Utilization of Ammonium as a Primary Nitrogen Source.
Host metabolism of amino acids by glutaminases associated with the intestinal mucosa (18) or deamination of amino acids during bacterial degradation of dietary proteins yields ammonium (19). The M. smithii proteome contains a transporter for ammonium (AmtB; MSM0234) plus two routes for its assimilation: (i) the ATP-dependent glutamine synthetase–glutamate synthase pathway, which has a high affinity for ammonium and thus is advantageous under nitrogen-limited conditions, and (ii) the ATP-independent glutamate dehydrogenase pathway, which has a lower affinity for ammonium (20).
Microanalytic biochemical assays revealed that the ratio of glutamine to 2-oxoglutarate was 32-fold lower in the ceca of cocolonized gnotobiotic mice compared with animals colonized with M. smithii alone and that was 5-fold lower compared with B. thetaiotaomicron monoassociated hosts (P < 0.0001; Fig. 2E; n = 5 mice per group). In addition, levels of several polar amino acids were also significantly reduced in mice containing the saccharolytic bacterium and methanogen (Fig. 2F; n = 5 mice per group), providing additional evidence for a more nitrogen-limited gut environment. qRT-PCR analysis established that many of the key M. smithii genes involved in ammonium assimilation are up-regulated with cocolonization, particularly those in the high-affinity glutamine synthetase–glutamate synthase pathway [GlnA (glutamine synthetase, MSM1418); GltA/GltB (two subunits of glutamate synthase, MSM0027, MSM0368); Fig. 2 A and G]. GeneChip analysis of the transcriptional responses of B. thetaiotaomicron to cocolonization with M. smithii indicated that it also up-regulates a high-affinity glutamine synthetase (BT4339; 2.4-fold vs. B. thetaiotaomicron monoassociated mice; n = 4–5 mice per group; P < 0.001; ref. 6). This prioritization of ammonium assimilation by B. thetaiotaomicron and M. smithii is accompanied by a decrease in cecal ammonium levels in cocolonized hosts (11.1 ± 1.3 μmol/g dry weight of cecal contents vs. 14.4 ± 0.6 and 14.3 ± 0.9 in M. smithii- and B. thetaiotaomicron-monoassociated animals, respectively; n = 5–15 per group; P < 0.05; Fig. 2H). Together, these studies indicate that ammonium provides a key source of nitrogen for M. smithii when it exists in isolation in the gut of gnotobiotic mice, and that it must compete with B. thetaiotaomicron for this nutrient resource.
Considering Targets for Development of Anti-M. smithii Agents.
As noted in the Introduction, manipulation of the representation of M. smithii in our gut microbiota could provide a means for treating obesity. When considering how to manipulate the representation of M. smithii, several obvious questions arise: (i) is the targeted M. smithii gene/pathway expressed in vivo and is its expression affected by the presence of actively fermenting bacteria, and (ii) are the therapeutic targets being considered conserved among different M. smithii strains? We addressed these questions in a final set of experiments.
Functional genomics studies in gnotobiotic mice illustrate one way to approach issue i. For example, inhibitors exist for several M. smithii enzymes. A class of N-substituted derivatives of para-aminobenzoic acid (pABA) interfere with methanogenesis by competitively inhibiting ribofuranosylaminobenzene 5′-phosphate synthase (RfaS; MSM0848; ref. 21). As noted above, this enzyme, which participates in the first committed step in synthesis of methanopterin, is up-regulated with cocolonization (4.6 ± 0.9 fold vs. monoassociated controls; P < 0.01; Fig. 2A).
Archaeal membrane lipids, unlike bacterial lipids, contain ether linkages. A key enzyme in the biosynthesis of archaeal lipids is hydroxymethylglutaryl (HMG)-CoA reductase (MSM0227), which catalyzes the formation of mevalonate, a precursor for membrane (isoprenoid) biosynthesis (22). Some HMG-CoA reductase inhibitors (statins) have been reported to inhibit growth of Methanobrevibacter species in vitro (22). qRT-PCR revealed that MSM0227 is expressed at high levels in vivo in the presence or absence of B. thetaiotaomicron (P > 0.05; SI Table 11). Although statins are commonly used for treating hypercholesterolemia in humans, we are not aware of any studies that report a causal association between their consumption and weight loss. However, the efficacy of statins as antiarchaeal agents will depend on factors such as their activity against the M. smithii enzyme, their capacity to enter the archaeal cell, and their concentration and stability within the distal human gut.
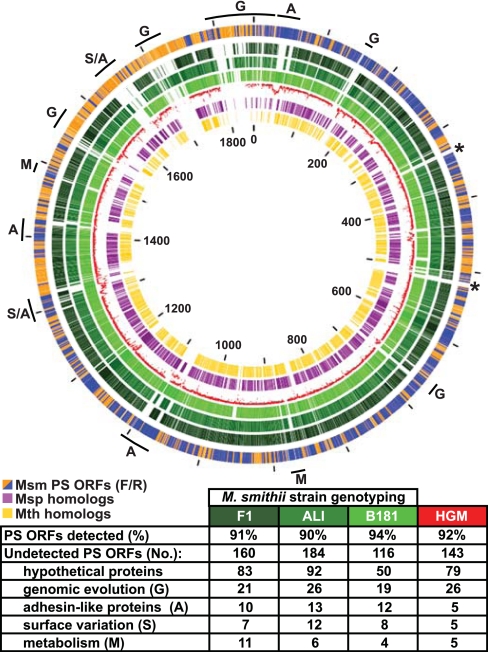
To address issue ii, we designed a custom GeneChip containing probe sets directed against 99.1% of M. smithii's 1,795 predicted protein-coding genes (see SI Table 13 for details). This GeneChip was used to perform whole-genome genotyping of M. smithii PS (control) plus three other strains recovered from the feces of healthy humans: F1 (DSMZ 2374), ALI (DSMZ 2375), and B181 (DSMZ 11975). Replicate hybridizations indicated that 100% of the ORFs represented on the GeneChip were detected in M. smithii PS, whereas 90–94% were detected in the other strains, including the potential drug targets mentioned above (SI Table 2 and Fig. 3). Approximately 50% of the undetectable ORFs in each strain encode hypothetical proteins. The other undetectable genes are involved in genome evolution [e.g., recombinases, transposases, insertion sequence (IS) elements, and type II restriction modification (R-M) systems], are components of a putative archaeal prophage in strain PS (SI Table 14) or are related to surface variation, including several adhesin-like proteins (ALPs) (e.g., MSM0057 and MSM1585–7, MSM1590; SI Fig. 7). Strains F1 and ALI also appear to lack redundant gene clusters encoding subunits of formate dehydrogenase (MSM1462–3) and methyl-CoM reductase (MSM0902–4) that are found in the PS strain (the latter cluster is also undetectable in strain B181). In addition, the only methanol utilization cluster present in the PS strain (MSM0515–8) was not detectable in strain F1 (SI Table 2).
Fig. 3.
Analysis of the M. smithii pan-genome. Schematic depiction of the conservation of M. smithii PS genes [depicted in the outermost circle where the color code is orange for forward strand ORFs (F) and blue for reverse strand ORFs (R)] in (i) other M. smithii strains (GeneChip-based genotyping of strains F1, ALI, and B181; circles in increasingly lighter shades of green, respectively; see SI Table 2 for details), (ii) the fecal microbiomes of two healthy individuals [human gut microbiome (HGM), shown as the red plot in the fifth innermost circle with nucleotide identity plotted from 80% (closest to the purple circle) to 100% (closest to lightest green ring); see also SI Fig. 9 for details], and (iii) two other members of the Methanobacteriales division [M. stadtmanae (Msp) (purple circle), another human gut methanogen, and M. thermoautotrophicus (Mth) (yellow circle), an environmental thermophile; mutual best BLASTP hits (E value <10−20)]. Tick marks indicate nucleotide number in kilobases. Asterisks denote the positions of ribosomal rRNA operons. Letters highlight distinguishing features among M. smithii genomes: the table below the figure summarizes differences in M. smithii gene content between strains F1, ALI, and B181, as well as the two human fecal metagenomic data sets.
To further assess the degree of nucleotide sequence divergence among M. smithii strains, we compared the sequenced PS type strain to a 78-Mb metagenomic data set generated from the aggregate fecal microbiome of two healthy humans (3). Their sequenced microbiomes contained 92% of the ORFs in the type strain (Fig. 3 and SI Table 2), including the potential drug targets described above. Several restriction modification (R-M) system gene clusters (MSM0157–8, MSM1742–8), a number of transposases, a DNA repair gene cluster (MSM0690–95), and all ORFs in the prophage were not evident in the two microbiomes. Sequence divergence was also observed in 33 of the 48 ALP genes plus two “surface variation” gene clusters (MSM1288–1313 and MSM1590–1616) that encode 11 glycosyltransferases and 9 proteins involved in pseudomurein cell wall biosynthesis (SI Fig. 9). A redundant methyl-CoM reductase cluster (MSM0902–5), an F420-dependent NADP oxidoreductase (MSM0049) involved in consumption of bacteria-derived ethanol, and two subunits of the bicarbonate ABC transporter (MSM0990–1; carbon utilization) exhibited heterogeneity in the M. smithii populations present in the gut microbiota of these two adults (SI Table 2 and SI Fig. 9).
Prospectus.
Ongoing comparisons of the genomes of multiple strains of a bacterial “species” have revealed remarkable variations in gene content. This has led to the concept that understanding the nature of a microbial “species” requires that we consider the sum of all genes present in all strains (i.e., its pan-genome; ref. 23). Our results provide an initial glimpse of the genetic diversity of a gut archaeon and the operations of one strain's transcriptome and metabolome in vivo. Although there is overall conservation of gene content among different strains of M. smithii, there are differences in metabolic capacities and surface properties represented in the organism's pan-genome. These differences may influence its partitioning within and adaptations to its gut habitat, as well as its relationship with members of the bacterial community.
Tractable genetic systems for manipulating the M. smithii genome are not available at present. However, further characterization of the M. smithii pan-genome by sequencing isolates obtained from related and unrelated individuals represents an opportunity to evaluate genome conservation and evolution in an archaeon that coexists with bacteria in an incredibly dense microbial community. In addition, gnotobiotic mice colonized with different strains of M. smithii together with sequenced representatives of human gut-derived Bacteroidetes and Firmicutes, should help guide drug discovery programs aimed at identifying antiarchaeal therapeutic agents with broad efficacy.
Materials and Methods
Genome Sequencing and Annotation.
M. smithii strain PS (ATCC 35061) was grown as described in Materials and Methods in SI Text for 6 d at 37°C. DNA was recovered from harvested cell pellets by using the Qiagen (Valencia, CA) Genomic DNA Isolation kit with mutanolysin (1 unit/mg wet-weight cell pellet; Sigma, St. Louis, MO) added to facilitate lysis of the microbe. An ABI 3730xl instrument (Applied Biosystems, Foster City, CA) was used for paired end-sequencing of inserts in a plasmid library (average insert size 5 kb; 42,823 reads; 11.6-fold coverage), and a fosmid library (average insert size of 40 kb; 7,913 reads; 0.6-fold coverage). Phrap and PCAP (24) were used to assemble the reads. A primer-walking approach was used to fill-in sequence gaps. Physical gaps and regions of poor quality (as defined by Consed; ref. 25) were resolved by PCR-based resequencing. The assembly's integrity and accuracy was verified by clone constraints. Regions containing insufficient coverage or ambiguous assemblies were resolved by sequencing spanning fosmids. Sequence inversions were identified on the basis of inconsistency of constraints for a fraction of read pairs in those regions. The final assembly consisted of 12.6× sequence coverage with a Phred base quality value ≥40. ORFs were identified and annotated as described in Materials and Methods in SI Text.
qRT-PCR Analyses.
All mouse experiments were performed using protocols approved by the animal studies committee of Washington University. Gnotobiotic male mice belonging to the NMRI inbred strain (n = 5–6 mice per group per experiment) were colonized with either M. smithii (14 d) or B. thetaiotaomicron (28 d) alone or first with B. thetaiotaomicron for 14 d followed by cocolonization with M. smithii for 14 d. All mice were killed at 12 weeks of age. Cecal contents from each mouse were flash frozen and stored at −80°C. RNA was extracted from an aliquot of the harvested cecal contents (100–300 mg) and used to generate cDNA for qRT-PCR assays (see Materials and Methods in SI Text). qRT-PCR data were normalized to 16S rRNA (ΔΔCT method) before comparing treatment groups. PCR primers are listed in SI Table 15. All amplicons were 100–150 bp.
Biochemical Assays.
Perchloric and hydrochloric acid extracts and alkali extracts of freeze-dried cecal contents were prepared, and established pyridine nucleotide-linked microanalytic assays (26) were used to measure metabolites (see Materials and Methods in SI Text for details).
Supplementary Material
Acknowledgments
We thank W. B. Whitman (University of Georgia, Athens, GA), F. Rohwer, and R. Edwards (both at San Diego State University, San Diego, CA), and our colleagues J. Amend, L. Angenent, M. Mahowald, R. Ley, E. Martens, J. Sonnenburg, J. McCutcheon, T. Jones, M. Bjursell, M. Giannakis, and J. Oh (all at Washington University) for many useful suggestions. We also thank L. Fulton, S. Clifton, and J. Xu for their help and guidance with genome sequencing; S. Wagoner, M. Karlsson, and D. O'Donnell for assistance with husbandry of gnotobiotic mice; H. Wynder for transmission EM studies; and the University of California, San Diego, Glycotechnology Core Facility for sialic acid analysis. This work was funded by National Institutes of Health Grants DK30292 and DK70977 and by the W. M. Keck Foundation. B.S.S. is a recipient of National Science Foundation Graduate Research Fellowship DGE-0202737.
Abbreviations
- ALP
adhesin-like protein
- CE
carbohydrate esterase
- GH
glycoside hydrolase
- GO
gene ontology
- GT
glycosyltransferase
- qRT-PCR
quantitative RT-PCR.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequence reported in this paper for the M. smithii PS genome has been deposited in the GenBank database (accession no. CP000678).
This article contains supporting information online at www.pnas.org/cgi/content/full/0704189104/DC1.
References
- 1.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miller TL, Wolin MJ. Syst Appl Microbiol. 1986;7:223–229. [Google Scholar]
- 3.Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, Gordon JI, Relman DA, Fraser-Liggett CM, Nelson KE. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sonnenburg JL, Xu J, Leip DD, Chen CH, Westover BP, Weatherford J, Buhler JD, Gordon JI. Science. 2005;307:1955–1959. doi: 10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- 5.McNeil NI. Am J Clin Nutr. 1984;39:338–342. doi: 10.1093/ajcn/39.2.338. [DOI] [PubMed] [Google Scholar]
- 6.Samuel BS, Gordon JI. Proc Natl Acad Sci USA. 2006;103:10011–10016. doi: 10.1073/pnas.0602187103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 8.Xu J, Bjursell MK, Himrod J, Deng S, Carmichael LK, Chiang HC, Hooper LV, Gordon JI. Science. 2003;299:2074–2076. doi: 10.1126/science.1080029. [DOI] [PubMed] [Google Scholar]
- 9.von Mering C, Huynen M, Jaeggi D, Schmidt S, Bork P, Snel B. Nucleic Acids Res. 2003;31:258–261. doi: 10.1093/nar/gkg034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sonnenburg ED, Sonnenburg JL, Manchester JK, Hansen EE, Chiang HC, Gordon JI. Proc Natl Acad Sci USA. 2006;103:8834–8839. doi: 10.1073/pnas.0603249103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. Cell. 2005;122:107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 12.Coyne MJ, Reinap B, Lee MM, Comstock LE. Science. 2005;307:1778–1781. doi: 10.1126/science.1106469. [DOI] [PubMed] [Google Scholar]
- 13.Coutinho PM, Henrissat B. In: Recent Advances in Carbohydrate Bioengineering. Gilbert HJ, Davies G, Henrissat B, Svensson B, editors. Cambridge, UK: R Soc Chem; 1999. pp. 3–12. [Google Scholar]
- 14.Vimr ER, Kalivoda KA, Deszo EL, Steenbergen SM. Microbiol Mol Biol Rev. 2004;68:132–153. doi: 10.1128/MMBR.68.1.132-153.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller TL, Wolin MJ, de Macario EC, Macario AJ. Appl Environ Microbiol. 1982;43:227–232. doi: 10.1128/aem.43.1.227-232.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fricke WF, Seedorf H, Henne A, Kruer M, Liesegang H, Hedderich R, Gottschalk G, Thauer RK. J Bacteriol. 2006;188:642–658. doi: 10.1128/JB.188.2.642-658.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berk H, Thauer RK. Arch Microbiol. 1997;168:396–402. doi: 10.1007/s002030050514. [DOI] [PubMed] [Google Scholar]
- 18.McCauley R, Kong SE, Heel K, Hall JC. Int J Biochem Cell Biol. 1999;31:405–413. doi: 10.1016/s1357-2725(98)00121-6. [DOI] [PubMed] [Google Scholar]
- 19.Wallace RJ. J Nutr. 1996;126:1326S–1334S. doi: 10.1093/jn/126.suppl_4.1326S. [DOI] [PubMed] [Google Scholar]
- 20.Cabello P, Roldan MD, Moreno-Vivian C. Microbiology. 2004;150:3527–3546. doi: 10.1099/mic.0.27303-0. [DOI] [PubMed] [Google Scholar]
- 21.Dumitru R, Palencia H, Schroeder SD, DeMontigny BA, Takacs JM, Rasche ME, Miner JL, Ragsdale SW. Appl Environ Microbiol. 2003;69:7236–7241. doi: 10.1128/AEM.69.12.7236-7241.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wolin MJ, Miller TL. Int Congr Ser. 2006;1293:131–137. [Google Scholar]
- 23.Medini D, Donati C, Tettelin H, Masignani V, Rappuoli R. Curr Opin Genet Dev. 2005;15:589–594. doi: 10.1016/j.gde.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 24.Huang X, Wang J, Aluru S, Yang SP, Hillier L. Genome Res. 2003;13:2164–2170. doi: 10.1101/gr.1390403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gordon D, Abajian C, Green P. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- 26.Passonneau JV, Lowry OH. Enzymatic Analysis: A Practical Guide. Totawa, NJ: Humana; 1993. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.