Abstract
Aberrant expression of the TCL1 oncoprotein promotes malignant transformation of germinal center (GC) B cells. Repression of TCL1 in GC B cells facilitates FAS-mediated apoptosis and prevents lymphoma formation. However, the mechanism for this repression is unknown. Here we show that the CREB coactivator TORC2 directly regulates TCL1 expression independent of CREB Ser-133 phosphorylation and CBP/p300 recruitment. GC signaling through CD40 or the BCR, which activates pCREB-dependent genes, caused TORC2 phosphorylation, cytosolic emigration, and TCL1 repression. Signaling via cAMP-inducible pathways inhibited TCL1 repression and reduced apoptosis, consistent with a prosurvival role for TCL1 before GC selection and supporting an initiating role for aberrant TCL1 expression during GC lymphomagenesis. Our data indicate that a novel CREB/TORC2 regulatory mode controls the normal program of GC gene activation and repression that promotes B cell development and circumvents oncogenic progression. Our results also reconcile a paradox in which signals that activate pCREB/CBP/p300 genes concurrently repress TCL1 to initiate its silencing.
Keywords: gene regulation, lymphomagenesis, signal transduction
The germinal center (GC) is home to T cell-dependent antigen-driven B cell maturation, memory B and plasma cell production, and the site of origin for most human B cell lymphomas (1–3). GC B cells undergo critical changes in gene expression that are required for proper development and protection from oncogenesis (4, 5). The generation of an appropriate humoral response is insured by negative selection, primarily mediated by FAS-induced apoptosis (6–8). Escape from apoptosis results in autoimmunity and B cell transformation (8).
TCL1 functions as a coactivator of the cell survival kinase AKT (9, 10). In mature T cell tumors, TCL1 expression is aberrantly elevated by rearrangements into T cell receptor loci (11). Physiologic TCL1 expression is largely limited to B lineage cells and is robust from pre-B cells through peripheral naïve B cells, followed by a critical 40–60% repression in GC B cells and complete silencing in post-GC memory B and plasma cells (12, 13). Most B cell lymphomas that arise by transformation of GC-experienced B cells exhibit elevated TCL1 expression by escape from GC mechanism(s) of TCL1 repression (12, 14, 15). Interestingly, transgenic mice with TCL1 expression levels stabilized in GC lymphocytes develop cancers that resemble a spectrum of human GC B cell lymphomas (16).
Unrepressed TCL1 expression inhibits FAS-induced B cell apoptosis independent of activation by BCR survival signaling (17, 18). Impaired apoptosis from failed TCL1 repression in GC B cells connects TCL1 dysregulation in patient lymphomas (12, 14) to a mechanism for increased transformation (16, 18). Factors so far suggested to regulate TCL1, including Nur77 (19, 20), miRNA-29 and miRNA-181 (21), Sp1 (22), and EBV infection (23), fail to adequately explain TCL1 repression in GC B cells and its continued aberrant expression in lymphomas (11). Its role in promoting B cell development and preventing lymphomagenesis makes TCL1 a highly important direct target of the regulatory program that controls a battery of GC B cell genes, although the regulatory program controlling TCL1 is unknown (24). Therefore, we focused on the mechanism(s) of TCL1 regulation, especially during GC B cell processes, as a strategy for identifying new regulatory programs that control key GC B cell genes and for discovering key sites of TCL1 dysregulation in B cells that could promote malignant transformation.
Results
CREB Controls TCL1 Expression.
DNase I footprint and MatInspector revealed a CREB response element (CRE)-like half-site (GACGT) within the TCL1 promoter [supporting information (SI) Fig. 7A] (25). Electrophoretic mobility shift assays using B cell nuclear extracts, CREB-specific antiserum, and purified CREB-1 protein demonstrated that CREB bound this CRE half-site (SI Fig. 7 B–E). To provide physiologic relevance, ChIP studies demonstrated CREB and pCREB associated with the endogenous TCL1 promoter in TCL1 expressing Nalm-6 pre-B and Ramos B cells (SI Fig. 8). The effect of CREB on TCL1 promoter activity was determined in HEK293T cells using 424 bp of the TCL1 promoter (−424luc), as done before (SI Fig. 9) (22). TCL1 is not expressed in HEK293T cells, although its promoter is highly active in transient reporter assays (22). CREB transactivation is mediated by increased cAMP, which results in protein kinase A or PKC activation and phosphorylation of CREB on Ser-133 (pCREB-133) (26–28). pCREB-133 recruits CBP (CREB-binding protein) or its paralogue, p300, to activate CREB-responsive promoters (27). Incubation with dibutyryl cAMP (dbcAMP) resulted in a 3-fold induction, whereas mutation of the CRE half-site (GACGT→GATCT) to block CREB binding abolished cAMP responsiveness. However, a TCL1 promoter response to cAMP could still be CREB-independent (27). Cotransfection of a CREB expression plasmid with the −424luc reporter resulted in a 2.5-fold induction, whereas cotransfection with the −m424luc reporter failed to activate TCL1, indicating that cAMP and CREB transactivation was direct at the CRE half-site. CREB shRNA validated a role for CREB in regulating TCL1, with knockdown to 25% of native levels resulting in robust repression of TCL1 promoter activity. Overall, the data indicate that CREB along with Sp1 supports robust basal activity of the TCL1 promoter (22).
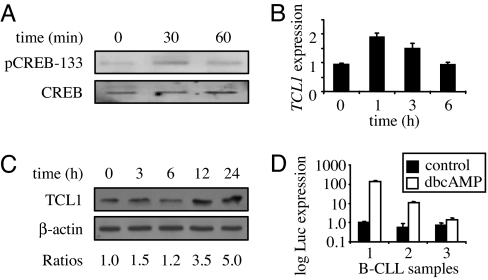
We next examined a role for cAMP in regulating endogenous TCL1 in B cells. Ramos B cells treated with dbcAMP showed a modest increase in pCREB-133 at 30 min that returned to baseline after 1 h (Fig. 1A), paralleling a 2-fold increase in TCL1 mRNA (Fig. 1B). After 6 h of dbcAMP, TCL1 expression returned to baseline, duplicating the induction by dbcAMP in transient reporter assays (SI Fig. 9B and data not shown). Significantly, TCL1 protein increased between 6 and 24 h (Fig. 1C), with an ≈5-fold increase at 24 h. dbcAMP treatment of Jurkat T cells, which do not express TCL1, failed to induce TCL1 (data not shown), likely because dbcAMP cannot overcome epigenetic silencing at the TCL1 locus (22). In contrast, B-chronic lymphohocytic leukemia (B-CLL) cells likely require TCL1 for survival (12, 14, 29, 30), and the −424luc reporter was activated by dbcAMP in at least two of three patient samples between 14- and 140-fold over untreated levels (Fig. 1D). Combined, these data indicate that cAMP induces TCL1 in GC-derived B cell lines and primary B-CLL samples, supporting physiologic and pathologic roles for TCL1 regulation in vivo.
Fig. 1.
cAMP increases TCL1 expression in B cells and patient B-CLL samples. (A) Ramos B cells were incubated with 0.1 mM dbcAMP for the times indicated, and whole-cell lysates were immunoblotted with CREB or pCREB-133 Abs. (B) TCL1 mRNA levels in Ramos cells responding to dbcAMP for the indicated times were measured by real-time quantitative RT-PCR (SYBRgreen), with expression normalized to a 36B4 gene control. (C) Immunoblot for TCL1 and β-actin in Ramos cells responding to dbcAMP for the indicated times. Band ratios were determined with densitometry. (D) −424luc reporter activity in three different patient B-CLL samples stimulated with 0.1 mM dbcAMP for 72 h. Data are representative of two independent experiments.
TORC2 Controls TCL1 Promoter Activity.
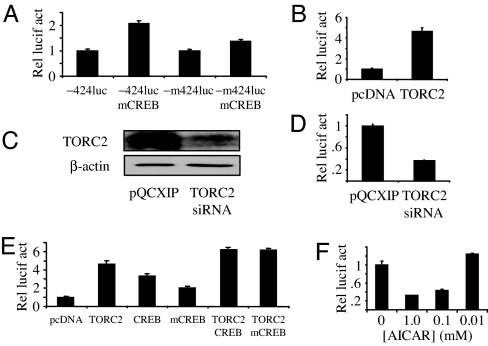
Our data predict that cAMP-mediated protein kinase A or PKC activation led to CREB Ser-133 phosphorylation, promoter binding, and CBP/p300 coactivator recruitment as a mechanism for TCL1 expression. To examine this idea further, a dominant-negative mutant CREB (mCREB) construct was cotransfected with −424luc or −m424luc reporters into HEK293T cells. mCREB contains a Ser-133→Ala-133 substitution, blocking CREB Ser-133 phosphorylation and activation of the canonical CREB transactivation pathway without blocking CREB binding to CRE-containing promoters (27). Because CREB knockdown repressed basal TCL1 promoter activity (SI Fig. 9 D and E), coexpression of mCREB was expected to also decrease TCL1 promoter activity (27). Unexpectedly, mCREB expression resulted in an ≈2-fold induction of −424luc reporter activity over robust unstimulated levels (Fig. 2A). mCREB did not appreciably affect −m424luc reporter activity (<2-fold change), supporting the specificity of mCREB for the CRE half-site in the TCL1 promoter. Unexpectedly, the data suggest that pCREB-133 is dispensable for CREB activation of the TCL1 promoter.
Fig. 2.
TORC2-dependent, pCREB-133-independent TCL1 activation. (A) −424luc or −m424luc reporter activity in HEK293T cells 48 h after transfection, with or without cotransfection of the mCREB (Ser-133→Ala-133) expression construct. (B) −424luc reporter activity in HEK293T cells 48 h after transfection, with or without cotransfection of the psportTORC2 (M. Montminy) expression construct. (C) Immunoblot for TORC2 repression in HEK293T cells 48 h after transfection with a TORC2 siRNA expression construct. (D) −424luc reporter activity in HEK293T cells at 48 h with or without cotransfection of a TORC2 siRNA expression construct. (E) −424luc reporter activity in HEK293T cells at 48 h with or without cotransfection of TORC2, CREB, or mCREB expression constructs. (F) −424luc reporter activity in HEK293T cells 48 h after transfection, with or without the indicated concentrations of AICAR added 6 h before assay harvest. For all experiments the data were normalized to a cotransfected Renilla luciferase (pRLCMV luciferase) reporter construct. Data represent the mean ± SD of two independent experiments, each done in triplicate.
An alternative mechanism for CREB control of TCL1 expression was next considered. A leading candidate for testing was the transducer of regulated CREB (TORC) protein. TORC proteins bind to CREB and activate target genes independent of pCREB-133 (31). TORC1 had no effect on the −424luc reporter (data not shown), consistent with the lack of TORC1 expression in human B cells (ref. 31 and data not shown), whereas TORC2 expression activated −424luc ≈5-fold (Fig. 2B). HEK293T cells, which express endogenous TORC2 (Fig. 2C), were cotransfected with TORC2 siRNA and the −424luc reporter, resulting in decreased TCL1 promoter activity to ≈30% of control levels (Fig. 2D), which was similar to the repression detected with CREB shRNA (SI Fig. 9E). When coexpressed, CREB and TORC2 increased TCL1 promoter activity additively by >6-fold (Fig. 2E). An identical induction of the −424luc reporter was also achieved by using mCREB with TORC2 cotransfection, further demonstrating the pCREB-133-independent, TORC2-dependent transactivation of TCL1.
Incubation with 5-aminoimidazole-4-carboxamide riboside (AICAR, an AMP analogue) results in TORC2 Ser-171 phosphorylation (pTORC2–171) and translocation out of nucleus into the cytoplasm, thereby repressing TORC2-dependent target genes (32). Because TCL1 promoter activity depended on TORC2 expression and not pCREB-133, we postulated that AICAR would repress TCL1. Consistent with this notion, AICAR caused a dose-dependent repression of the TCL1 promoter to ≈30% of untreated control cells (Fig. 2F). These data indicate that CREB and TORC2 transactivate TCL1 and that unstimulated, or robust basal, TCL1 expression depends on these interacting transfactors.
TORC2 Regulates TCL1 Expression in B Cells.
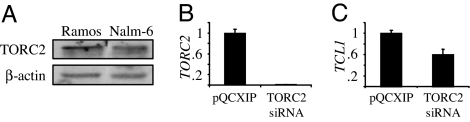
Western blot demonstrated abundant TORC2 in Nalm-6 pre-B and Ramos B cells (Fig. 3A). siRNA knockdown reduced TORC2 expression in Nalm-6 cells by ≈99% (Fig. 3B), whereas Ramos cells did not tolerate multiple TORC2 knockdown strategies (data not shown). TORC2 reduction resulted in a 40% repression in endogenous TCL1 expression in Nalm-6 cells (Fig. 3C). These data support transient TCL1 reporter results in HEK293T cells (Fig. 2) and also recapitulate the extent of TCL1 repression detected in primary GC B cells (11), suggesting a central role for TORC2 in regulating endogenous TCL1 expression in GC B cells.
Fig. 3.
TORC2 inhibition represses TCL1 expression in Nalm-6 pre-B cells. (A) Immunoblot showing relative expression levels of TORC2 in Ramos B cells and Nalm-6 pre-B cells. (B) Real-time quantitative RT-PCR of TORC2 mRNA levels in polyclonal Nalm-6 cells stably expressing TORC2 siRNA. (C) Real-time quantitative RT-PCR of TCL1 mRNA levels in polyclonal Nalm-6 cells stably expressing TORC2 siRNA. Data are representative of two independent experiments and plot the mean ± SD of triplicate measurements.
BCR and CD40L Signaling Mediate TCL1 Repression Independent of CREB Phosphorylation.
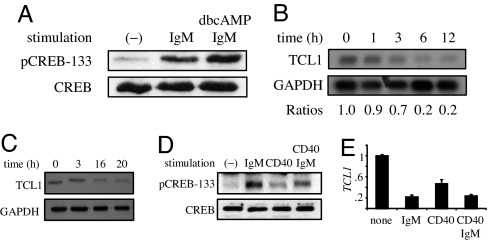
Based on these findings, we postulated that CREB or TORC2 inhibition could repress TCL1 in GC B cells (12, 15). To test this idea, Ramos B cells were stimulated in a manner that resembled a T-dependent GC reaction by signaling through CD40 and the BCR (18). Ramos B cells undergo BCR-mediated apoptosis and provide a model for deletion of self-reactive GC B cells (33). BCR engagement results in CREB phosphorylation (28), indicating potential relevance for CD40 and BCR signaling in regulating TCL1. First, pCREB-133 was determined with anti-IgM to establish that BCR signaling was intact in Ramos cells. As expected, anti-IgM resulted in a significant increase in pCREB-133 that was enhanced by coaddition of cAMP without a change in total CREB protein levels (Fig. 4A). Surprisingly, TCL1 mRNA expression was reduced by ≈80% at 12 h of anti-IgM stimulation (Fig. 4B). TCL1 protein expression was also markedly reduced after 16–20 h of anti-IgM (Fig. 4C). These data support the −424luc reporter results, indicating that enhanced pCREB-133 does not transactivate TCL1. Instead, the data strongly suggest an alternative mechanism for BCR-mediated TCL1 repression.
Fig. 4.
BCR or CD40 signaling activates CREB and represses TCL1 expression. Ramos B cells were treated with 10 μg/ml anti-IgM, 1 μg/ml anti-CD40, or 0.1 mM dbcAMP for the times indicated. (A) Immunoblot for CREB or pCREB-133 at 1 h of stimulation. (B) Northern blot for TCL1 and GAPDH with densitometry quantification. (C) Immunoblot for TCL1 and β-actin. (D) Immunoblot for CREB or pCREB-133 at 1 h of stimulation. (E) Real-time quantitative RT-PCR of TCL1 expression at 6 h of stimulation.
A critical step in the GC reaction occurs when CD40 on B cells binds its ligand, CD154, expressed on T and other cells, which may result in B cell proliferation, survival, or Ig class switch recombination (6). We postulate that this GC signal might, like BCR signaling, also repress TCL1 in Ramos B cells. Consistent with this idea, anti-CD40 stimulation resulted in TCL1 repression (Fig. 4 D and E). When Ramos cells were coincubated with anti-IgM and anti-CD40, TCL1 repression remained at levels seen with either treatment alone, suggesting that these two signaling pathways both use a pCREB-133-independent mechanism to repress TCL1. Together, these data show that activation of key GC-related signaling pathways in Ramos B cells recapitulates the physiologic repression detected for TCL1 during the GC reaction in vivo (12).
BCR and CD40L-Mediated TORC2 Phosphorylation and Cytoplasmic Translocation.
Despite its expression in pre-B and GC-derived B cell lines (Fig. 3A), it was unknown whether primary B cells express TORC2. TORC2 protein expression was robust in all fractionated human tonsil subsets examined, including naïve (CD10+, IgD+), GC (CD10+, IgD−), and memory (CD10−, IgD) B cell subsets (Fig. 5A). TORC2 expression was comparable to the expression level detected in HEK293T cells (Fig. 2C and data not shown).
Fig. 5.
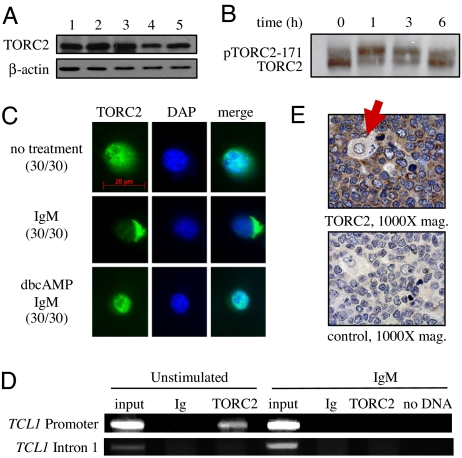
TORC2 responds to BCR signaling by leaving the TCL1 promoter and cytoplasmic translocation. (A) Immunoblot for TORC2 and β-actin in lysates from a mixture of naïve, GC, and memory B cells (lane 1), GC and memory B cells (lane 2), naïve B cells (lane 3), GC B cells (lane 4), and memory B cells (lane 5). (B) Immunoblot for TORC2 showing phosphorylated and nonphosphorylated forms with IgM stimulation of Ramos B cells for the times indicated. (C) Immunofluorescent staining for TORC2 in Ramos cells stimulated with IgM and/or dbcAMP at 3 h. Numbers in parentheses indicate the number of cells showing a similar staining pattern with each condition. (D) ChIP assay with Ramos B cells with or without 10 μg/ml anti-IgM stimulation for 1 h. PCR was performed from immunoprecipitated chromatin fragments by using anti-TORC2 or control anti-Ig Abs and primers from SI Fig. 10. (E) TORC2-specific immunohistochemical stain of GC B cells from human tonsil. Note the presence of an unstained macrophage (denoted by a red arrow) and the exclusive cytoplasmic localization of TORC2.
We postulated that the BCR- or CD40-mediated TCL1 repression in Ramos B cells was from an alteration of TORC2 expression and/or nuclear localization. No significant change in the level of TORC2 protein was detected with anti-IgM incubation, whereas TORC2 was mainly unphosphorylated (active) at rest and underwent robust phosphorylation at Ser-171, as indicated by a previously characterized mobility shift (34), with 1 h of anti-IgM stimulation, followed by a return to baseline at 6 h (Fig. 5B). Immunofluorescence at 3 h of anti-IgM showed that TORC2 was translocated from a mainly nuclear location almost completely into the cytoplasm in 30 of 30 cells examined (Fig. 5C). This translocation was transient, because at 6 h of anti-IgM TORC2 returned to the nucleus in all cells examined (data not shown). Supporting relocalization away from the TCL1 promoter, ChIP demonstrated TORC2 at the TCL1 promoter in unstimulated Ramos cells, whereas after 1 h of anti-IgM stimulation TORC2 was absent from the TCL1 promoter, strongly supporting the immunofluorescence results (Fig. 5D). Enhanced pTORC2–171 and cytoplasmic relocalization are consistent with pCREB-133-independent, TORC2-dependent control of TCL1 and provide a novel mechanism for TCL1 repression in vitro and potentially within GCs in vivo. Supporting this model, robust TORC2 expression was localized in the cytoplasm of activated GC B cells in human tonsil (Fig. 5E).
Stimulation with cAMP results in the nuclear retention of TORC2 even in the presence of additional signals that drive TORC2 out of the nucleus (34). Because cAMP resulted in increased TCL1 expression (Fig. 1), the localization of TORC2 was determined after preincubation with 0.1 mM cAMP followed by 3 h of anti-IgM stimulation. Consistent with results in nonlymphoid cells (34), preincubation with cAMP left TORC2 in the nucleus even with anti-IgM stimulation (Fig. 5C), establishing a signaling hierarchy for regulating TORC2 localization and target gene responses in B cells.
TCL1 was also repressed after anti-CD40 stimulation (Fig. 4D). To determine whether TORC2 cytoplasmic redistribution was also a mechanism for CD40-mediated TCL1 repression, immunofluorescence was performed. At 3 h of anti-CD40 stimulation, TORC2 was primarily in the cytoplasm in 24 of 30 cells examined (SI Fig. 10). Anti-CD40 stimulation was also unable to translocate TORC2 out of the nucleus after preactivation with cAMP. Combined, the data strongly support the hypothesis that TCL1 expression depends on TORC2 nuclear localization and interaction with the TCL1 promoter and that certain GC signaling pathways repress TORC2 target genes, such as TCL1.
cAMP Protects B Cells from BCR and CD40L-Induced TCL1 Repression and Apoptosis.
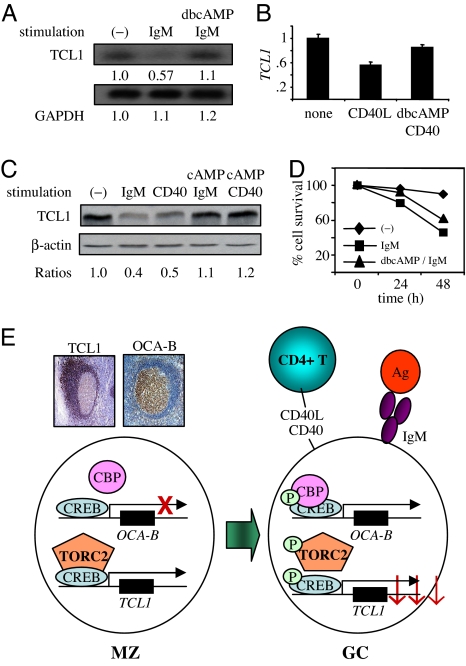
TORC2 control of TCL1 expression predicts that TCL1 will remain at prestimulation levels with cAMP and IgM treatments. Consistent with this prediction, treatment of Ramos B cells with IgM or CD40 resulted in decreased TCL1 mRNA levels, whereas preincubation with cAMP inhibited TCL1 repression (Fig. 6 A and B). At 40 h of stimulation, whereas TCL1 protein was decreased by IgM or CD40 stimulation, preincubation with cAMP blocked TCL1 down-regulation (Fig. 6C). These data reinforce the model for a signaling hierarchy acting on TCL1, modulated at least in part by pTORC2–171 and subcellular localization (Fig. 6).
Fig. 6.
BCR- and CD40-induced TCL1 repression and apoptosis are rescued by cAMP. (A) Northern blot for TCL1 and GAPDH at 6 h of incubation with 10 μg/ml anti-IgM and/or 0.1 mM dbcAMP in Ramos B cells. (B) Real-time quantitative RT-PCR for TCL1 at 6 h of incubation with 1 μg/ml anti-CD40 and/or 0.1 mM dbcAMP in Ramos B cells. (C) Immunoblot for TCL1 and β-actin in lysates from Ramos cells incubated at 40 h with 10 μg/ml anti-IgM, 1 μg/ml anti-CD40, and/or 0.1 mM dbcAMP. Densitometry quantification is shown in Lower. (D) Ramos cell survival measured by annexin V and propidium iodide exclusion with 10 μg/ml anti-IgM and/or 0.1 mM dbcAMP for the times indicated. (E) Model of CREB-TORC2 regulatory node and effects on OCA-B (36) and TCL1 (12, 15, 36) expression levels in mature human B cells during the GC reaction. Immunostain panels show absent OCA-B expression in pre-GC mantle zone (MZ) B cells and high expression in GC B cells, whereas TCL1 is highly expressed in mantle zone B cells and strongly repressed in GC B cells. See Discussion for details.
cAMP inhibition of TCL1 repression suggests that cAMP might protect IgM-treated B cells from death at least partially because TCL1 is a prosurvival oncoprotein with its survival effect linked to BCR signaling (18). Stimulation of Ramos cells with anti-IgM resulted in increased apoptosis, as previously shown (Fig. 6D) (33). Consistent with a predicted prosurvival effect, 0.1 mM dbcAMP preincubation delayed the onset and reduced the extent of apoptosis in Ramos B cells, which could be tumor-promoting within the GC over time (18). These data validate previous results for rescuing Ramos from BCR-induced apoptosis by cAMP with retained TCL1 expression imparting at least a component of this protection (35). These findings also provide a mechanism for enhanced survival fostering lymphomagenesis by enforced TCL1 expression in transgenic mouse GC B cells, or in human GC B cell lymphomas that aberrantly express TCL1, by avoiding BCR-dependent negative selection.
Discussion
The GC B cell gene regulatory program supports the generation of B cells with a robust Ab repertoire and foils the development of potentially oncogenic B cells. This regulatory program must suppress cell survival genes, such as TCL1, to foster negative selection and promote apoptosis. Our work exposed a regulatory circuit in which GC B cell signaling through CD40 and the BCR activates pCREB-133-responsive genes, such as OCA-B and BCL2 (36, 37), and also represses certain CREB-dependent genes, of which TCL1 is an important example, through Ser-171 phosphorylation and nuclear exclusion of TORC2. We have shown that TCL1 is a direct target of the CREB-TORC2 complex and that repression depends on pTORC2–171 but not pCREB-133. Our data provide a model for constitutive expression of TCL1 by CREB-TORC2 and Sp1 (22) in developing B cells until they enter the GC follicle center, where down-modulation provided by GC signaling phosphorylates TORC2 to drive it from the nucleus (Fig. 6E).
The TCL1 reporter shows high basal activity in many different cell types including those that do not express endogenous TCL1 (22). Our results support a mechanism for high basal promoter activity dependent on CREB levels and TORC2 levels and location. Interestingly, expression of a CREB Ser-133-Ala mutant did not reduce TCL1 promoter function, as anticipated, but instead supported promoter activity alone or in combination with TORC2. This result indicates that genes within the GC B cell regulatory program, such as TCL1, may be CREB-dependent, but CBP- or p300-independent, a prediction supported by reported data showing that CBP or p300 individually is not required for peripheral B cell function or development (38).
The reduction of TCL1 expression in Nalm-6 cells with >95% TORC2 repression was ≈40% (Fig. 3C), which appears modest when compared with the decrease in TCL1 levels after anti-IgM and CD40 stimulation (Fig. 4E). There are several possible explanations for this result, including that generation of stable TORC2 RNAi-expressing cells with puromycin selection could have eliminated TCL1-low-targeted cells, because TCL1-reduced Nalm-6 cells have a significant survival disadvantage (unpublished results). Also, TCL1 regulators in addition to TORC2, such as Sp1, may be impacted by GC-related IgM or CD40 signaling. It is important to stress that the 40% reduction in TCL1 actually parallels the extent of TCL1 reduction in primary GC B cells, suggesting that TORC2 controls this component of TCL1 gene expression.
The pCREB-133-independent circuit, mediated by the activity of the essential CREB coactivator TORC2 as shown here for TCL1, likely constitutes a new program of GC B cell regulation. Until this report, the only established physiologic role for TORC2 control of CREB-dependent gene expression was in cell metabolism (32, 34, 39–41). Our results provide a mechanism for constitutive CREB activation of certain genes, including TCL1, in the absence of cAMP stimulation in B cells (31). TCL1 is an example of survival gene that is constitutively activated by a CREB-TORC2 complex independent of inducible CREB phosphorylation.
TORC2 dependency resolves the paradoxical effect of CREB-dependent TCL1 repression by BCR stimulation in Ramos B cells. BCR engagement markedly increased pCREB-133, yet TCL1 was repressed. We discovered that pTORC2–171 was translocated out of the nucleus with CD40 or BCR activation, repressing TCL1 expression. TORC2 was previously shown to traffic in and out of the nucleus in response to metabolic signaling (32, 41). Now we report that GC-related CD40 and BCR signaling also results in pTORC2–171 translocation out of the nucleus, providing an example, along with OCA-B, of coactivator control of the GC B cell regulatory package (18, 42). These results suggest, like BCL6 repression of p53 to permit somatic hypermutation and class switch recombination of Ig genes (43), a critical role for CD40- and/or BCR-mediated pTORC2–171 emigration from the nucleus to facilitate FAS elimination of detrimental GC B cells (18) by TCL1 repression (Fig. 6E). Either CD40 or BCR signaling alone initiates apoptosis and negative selection in the absence of cosignaling (6, 33, 44). However, FAS-susceptible TCL1-repressed GC B cells can be rescued from death by TCL1-independent increases in the expression of BCL2L1 (Bcl-XL) and CFLAR (Flip) (45–47). Aberrant TCL1 expression in TCL1-transgenic mice or by defective TORC2-mediated TCL1 repression favors GC B cell survival independent of antigenic rescue (18) to help drive malignant transformation.
Our results suggest that pTORC2–171 is an important determinant of GC B cell fate. It will be interesting to identify additional TORC2 target genes that may also be repressed to facilitate the GC reaction. Introduction of a dominant negative TORC2 expression construct harboring a Ser-171→Ala-171 mutation into B cells with anti-IgM or anti-CD40 stimulation could further define the role of TORC2 in negative selection and B cell oncogenesis. Unfortunately, multiple attempts and strategies for overexpressing TORC2 in B cells so far have been unsuccessful because transduced cells typically underwent apoptosis (data not shown).
There already is a recognized role for hyperactive CREB-dependent gene regulation in cancer (48). CREB overexpression associates with a poor outcome in acute myeloid leukemia, and CREB transgenic mice develop a myeloproliferative disease (49, 50). Hyperactive CREB also associates with overexpression of BCL2 in follicular lymphoma (51). Our data suggest that defects in a CREB-TORC2 regulatory node, which likely initiates TCL1 repression in GC B cells, could be responsible for cases of GC-experienced lymphomas that maintain oncogenic TCL1 expression.
Materials and Methods
Human Cell Lines, Tissues, and Chemicals.
HEK293T, Jurkat, Nalm-6, and Ramos cell lines were maintained in RPMI medium 1640 with 10% FBS and antibiotics. Primary B-CLL were maintained as described (29). Fresh tonsil was sorted into mature B cell subsets and verified as described (12). Tonsil sections were retrieved from the pathology archives at University of California, Los Angeles. Human materials were used under institutional review board-approved protocols. dbcAMP and AICAR were from Sigma (St. Louis, MO).
Expression Vectors and Retroviruses.
The −424luc TCL1 reporter gene was previously described (22). An altered −424luc reporter was generated (QuikChange kit; Stratagene, La Jolla, CA) to disrupt the CRE-like half-site by a GACGT→GATCT mutation. CREB, mCREB (Ser-133→Ala-133), and TORC2 expression plasmids were from M. Montminy (The Salk Institute, La Jolla, CA). A TORC2 siRNA hairpin oligonucleotide (sequence available upon request) was cloned 3′ of an H1 promoter-modified pQCXIP (Clontech, Mountain View, CA) expression vector (S. Smale, University of California, Los Angeles). Retrovirus was generated in HEK293T cells grown in DMEM with 10% FBS and antibiotics by cotransfection with pCL-AMPHO using FuGENE 6 (Roche, Basel, Switzerland). Viral supernatant was collected 48 and 72 h after transfection, filtered, and stored at 4°C. Nalm-6 pre-B cells (1 × 105 per well) were incubated with 1 ml of virus supplemented with 2 μl of polybrene and centrifuged at 1,800 × g for 1 h at 30°C, which was repeated once the following day. One day after repeat infection, puromycin (1 μg/ml) was started and GFP expression was monitored. TORC2 RNA depletion was determined by Western blot.
RNA Analysis.
Total RNA was extracted by using TRIzol (Life Technologies, Carlsbad, CA). Northern blot for TCL1 and GAPDH was as described (12). cDNA was made by using the SuperScript First-Strand Synthesis System (Invitrogen, Carlsbad, CA). Real-time quantitative RT-PCR (SYBRgreen) was performed by using an Applied Biosystems 7700 sequence detector as described (52). Expression was normalized to a 36B4 control. Primer and probe sequences are available upon request.
Protein Analysis and Abs.
Western blots were as described (12), with the following modifications. Between 20 and 40 μg of whole-cell lysate for each sample was separated by 8% SDS/PAGE and transferred to a nitrocellulose membrane. Blocked membranes were incubated with CREB (1:1,000; Santa Cruz Biotechnology, Santa Cruz, CA), pCREB-133 (1:1,000; Upstate, Billerica, MA), TORC2 (1:3,000; M. Montminy), TCL1 (1:7,500), and β-actin (1:5,000; Sigma, St. Louis, MO) antiserum in TBST and 5% milk overnight. Cell stimulations were performed with anti-CD40 (K. Zhang, University of California, Los Angeles) or anti-IgM (Jackson ImmunoResearch, West Grove, PA) Abs.
Apoptosis Assay.
Apoptosis was measured by annexin V binding and propidium iodide permeability (BD Pharmingen, Franklin Lakes, NJ). Flow cytometry was performed on a Coulter Elite (Beckman Coulter), and data were analyzed by using FCS Express v2.0 (DeNovo Software).
Immunofluorescence Microscopy.
Ramos cells were plated on polylysine coverslips, washed, fixed with paraformaldehyde, washed, blocked with 0.2% BSA, incubated with anti-TORC2 Ab, washed, incubated with anti-rabbit-FITC, and washed again. Cells were counterstained with DAPI. Microscopy was performed by using a Zeiss Axioskop 2 plus microscope with a Plan-APOCHROMAT ×63/1.0 oil objective. Images were acquired by using a Zeiss Axiocam camera and Axiovision version 3.01 software.
Immunohistochemistry.
Formalin-fixed paraffin-embedded sections of human tonsil were stained with TORC2 antiserum (1:1,000) by using immunoperoxidase techniques as described (12, 15).
SI.
For more information on additional assays, ChIP, and transient transfection, see SI Methods.
Supplementary Material
Acknowledgments
We thank Marc Montminy, Reuben J. Shaw (The Salk Institute), Susan Hedrick (The Salk Institute), and Steve Smale for extensive discussions, manuscript review, and numerous reagents. This work was supported by the National Institutes of Health Grants PNEY018228, GM073981, CA90571, and CA107300 (to M.A.T.) and GM85841 and GM040185 (to R.W.), National Research Service Award GM07185 (to A.I.K. and M.S.), and a University Research Engineering and Technology Institutes grant from the National Aeronautics and Space Administration to the University of California, Los Angeles, Institute for Cell Mimetic Studies (NCC 2-1364 to M.A.T.). M.A.T. is a Scholar of the Leukemia and Lymphoma Society.
Abbreviations
- GC
germinal center
- dbcAMP
dibutyryl cAMP
- mCREB
mutant CREB
- AICAR
5-aminoimidazole-4-carboxamide riboside.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0704170104/DC1.
References
- 1.Kuppers R, Klein U, Hansmann ML, Rajewsky K. N Engl J Med. 1999;341:1520–1529. doi: 10.1056/NEJM199911113412007. [DOI] [PubMed] [Google Scholar]
- 2.Manser T. J Immunol. 2004;172:3369–3375. doi: 10.4049/jimmunol.172.6.3369. [DOI] [PubMed] [Google Scholar]
- 3.McHeyzer-Williams LJ, Driver DJ, McHeyzer-Williams MG. Curr Opin Hematol. 2001;8:52–59. doi: 10.1097/00062752-200101000-00010. [DOI] [PubMed] [Google Scholar]
- 4.Kuppers R. Nat Rev Cancer. 2005;5:251–262. doi: 10.1038/nrc1589. [DOI] [PubMed] [Google Scholar]
- 5.Shapiro-Shelef M, Calame K. Nat Rev Immunol. 2005;5:230–242. doi: 10.1038/nri1572. [DOI] [PubMed] [Google Scholar]
- 6.Rothstein TL. Cell Res. 2000;10:245–266. doi: 10.1038/sj.cr.7290053. [DOI] [PubMed] [Google Scholar]
- 7.Carey GB, Donjerkovic D, Mueller CM, Liu S, Hinshaw JA, Tonnetti L, Davidson W, Scott DW. Immunol Rev. 2000;176:105–115. doi: 10.1034/j.1600-065x.2000.00502.x. [DOI] [PubMed] [Google Scholar]
- 8.Adachi M, Suematsu S, Kondo T, Ogasawara J, Tanaka T, Yoshida N, Nagata S. Nat Genet. 1995;11:294–300. doi: 10.1038/ng1195-294. [DOI] [PubMed] [Google Scholar]
- 9.Laine J, Kunstle G, Obata T, Sha M, Noguchi M. Mol Cell. 2000;6:395–407. doi: 10.1016/s1097-2765(00)00039-3. [DOI] [PubMed] [Google Scholar]
- 10.Pekarsky Y, Koval A, Hallas C, Bichi R, Tresini M, Malstrom S, Russo G, Tsichlis P, Croce CM. Proc Natl Acad Sci USA. 2000;97:3028–3033. doi: 10.1073/pnas.040557697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Teitell MA. Nat Rev Cancer. 2005;5:640–648. doi: 10.1038/nrc1672. [DOI] [PubMed] [Google Scholar]
- 12.Said JW, Hoyer KK, French SW, Rosenfelt L, Garcia-Lloret M, Koh PJ, Cheng TC, Sulur GG, Pinkus GS, Kuehl WM, et al. Lab Invest. 2001;81:555–564. doi: 10.1038/labinvest.3780264. [DOI] [PubMed] [Google Scholar]
- 13.Virgilio L, Narducci MG, Isobe M, Billips LG, Cooper MD, Croce CM, Russo G. Proc Natl Acad Sci USA. 1994;91:12530–12534. doi: 10.1073/pnas.91.26.12530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Narducci MG, Pescarmona E, Lazzeri C, Signoretti S, Lavinia AM, Remotti D, Scala E, Baroni CD, Stoppacciaro A, Croce CM, Russo G. Cancer Res. 2000;60:2095–2100. [PubMed] [Google Scholar]
- 15.Teitell M, Damore MA, Sulur GG, Turner DE, Stern MH, Said JW, Denny CT, Wall R. Proc Natl Acad Sci USA. 1999;96:9809–9814. doi: 10.1073/pnas.96.17.9809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoyer KK, French SW, Turner DE, Nguyen MT, Renard M, Malone CS, Knoetig S, Qi CF, Su TT, Cheroutre H, et al. Proc Natl Acad Sci USA. 2002;99:14392–14397. doi: 10.1073/pnas.212410199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rothstein TL, Wang JK, Panka DJ, Foote LC, Wang Z, Stanger B, Cui H, Ju ST, Marshak-Rothstein A. Nature. 1995;374:163–165. doi: 10.1038/374163a0. [DOI] [PubMed] [Google Scholar]
- 18.Shen RR, Ferguson DO, Renard M, Hoyer KK, Kim U, Hao X, Alt FW, Roeder RG, Morse HC, III, Teitell MA. Blood. 2006;108:1991–1998. doi: 10.1182/blood-2006-02-001354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chan FK, Chen A, Winoto A. J Immunol. 1998;161:4252–4256. [PubMed] [Google Scholar]
- 20.Hiromura M, Suizu F, Narita M, Kinowaki K, Noguchi M. J Biol Chem. 2006;281:27753–27764. doi: 10.1074/jbc.M602420200. [DOI] [PubMed] [Google Scholar]
- 21.Pekarsky Y, Santanam U, Cimmino A, Palamarchuk A, Efanov A, Maximov V, Volinia S, Alder H, Liu CG, Rassenti L, et al. Cancer Res. 2006;66:11590–11593. doi: 10.1158/0008-5472.CAN-06-3613. [DOI] [PubMed] [Google Scholar]
- 22.French SW, Malone CS, Shen RR, Renard M, Henson SE, Miner MD, Wall R, Teitell MA. J Biol Chem. 2003;278:948–955. doi: 10.1074/jbc.M207166200. [DOI] [PubMed] [Google Scholar]
- 23.Kiss C, Nishikawa J, Takada K, Trivedi P, Klein G, Szekely L. Proc Natl Acad Sci USA. 2003;100:4813–4818. doi: 10.1073/pnas.0730710100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klein U, Tu Y, Stolovitzky GA, Keller JL, Haddad J, Jr, Miljkovic V, Cattoretti G, Califano A, Dalla-Favera R. Proc Natl Acad Sci USA. 2003;100:2639–2644. doi: 10.1073/pnas.0437996100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quandt K, Frech K, Karas H, Wingender E, Werner T. Nucleic Acids Res. 1995;23:4878–4884. doi: 10.1093/nar/23.23.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blois JT, Mataraza JM, Mecklenbrauker I, Tarakhovsky A, Chiles TC. J Biol Chem. 2004;279:30123–30132. doi: 10.1074/jbc.M402793200. [DOI] [PubMed] [Google Scholar]
- 27.Mayr B, Montminy M. Nat Rev Mol Cell Biol. 2001;2:599–609. doi: 10.1038/35085068. [DOI] [PubMed] [Google Scholar]
- 28.Xie H, Rothstein TL. J Immunol. 1995;154:1717–1723. [PubMed] [Google Scholar]
- 29.Herling M, Patel KA, Khalili J, Schlette E, Kobayashi R, Medeiros LJ, Jones D. Leukemia. 2006;20:280–285. doi: 10.1038/sj.leu.2404017. [DOI] [PubMed] [Google Scholar]
- 30.Zanesi N, Aqeilan R, Drusco A, Kaou M, Sevignani C, Costinean S, Bortesi L, La Rocca G, Koldovsky P, Volinia S, et al. Cancer Res. 2006;66:915–920. doi: 10.1158/0008-5472.CAN-05-3426. [DOI] [PubMed] [Google Scholar]
- 31.Conkright MD, Canettieri G, Screaton R, Guzman E, Miraglia L, Hogenesch JB, Montminy M. Mol Cell. 2003;12:413–423. doi: 10.1016/j.molcel.2003.08.013. [DOI] [PubMed] [Google Scholar]
- 32.Koo SH, Flechner L, Qi L, Zhang X, Screaton RA, Jeffries S, Hedrick S, Xu W, Boussouar F, Brindle P, et al. Nature. 2005;437:1109–1111. doi: 10.1038/nature03967. [DOI] [PubMed] [Google Scholar]
- 33.Tsubata T, Wu J, Honjo T. Nature. 1993;364:645–648. doi: 10.1038/364645a0. [DOI] [PubMed] [Google Scholar]
- 34.Screaton RA, Conkright MD, Katoh Y, Best JL, Canettieri G, Jeffries S, Guzman E, Niessen S, Yates JR, III, Takemori H, et al. Cell. 2004;119:61–74. doi: 10.1016/j.cell.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 35.Sakata N, Kawasome H, Terada N, Johnson GL, Gelfand EW. J Allergy Clin Immunol. 2000;105:522–531. doi: 10.1067/mai.2000.104251. [DOI] [PubMed] [Google Scholar]
- 36.Stevens S, Ong J, Kim U, Eckhardt LA, Roeder RG. J Immunol. 2000;164:5306–5312. doi: 10.4049/jimmunol.164.10.5306. [DOI] [PubMed] [Google Scholar]
- 37.Wilson BE, Mochon E, Boxer LM. Mol Cell Biol. 1996;16:5546–5556. doi: 10.1128/mcb.16.10.5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu W, Fukuyama T, Ney PA, Wang D, Rehg J, Boyd K, van Deursen JM, Brindle PK. Blood. 2006;107:4407–4416. doi: 10.1182/blood-2005-08-3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Al-Hakim AK, Goransson O, Deak M, Toth R, Campbell DG, Morrice NA, Prescott AR, Alessi DR. J Cell Sci. 2005;118:5661–5673. doi: 10.1242/jcs.02670. [DOI] [PubMed] [Google Scholar]
- 40.Canettieri G, Koo SH, Berdeaux R, Heredia J, Hedrick S, Zhang X, Montminy M. Cell Metab. 2005;2:331–338. doi: 10.1016/j.cmet.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 41.Shaw RJ, Lamia KA, Vasquez D, Koo SH, Bardeesy N, Depinho RA, Montminy M, Cantley LC. Science. 2005;310:1642–1646. doi: 10.1126/science.1120781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Teitell MA. Trends Immunol. 2003;24:546–553. doi: 10.1016/j.it.2003.08.002. [DOI] [PubMed] [Google Scholar]
- 43.Phan RT, Dalla-Favera R. Nature. 2004;432:635–639. doi: 10.1038/nature03147. [DOI] [PubMed] [Google Scholar]
- 44.Eeva J, Pelkonen J. Apoptosis. 2004;9:525–531. doi: 10.1023/B:APPT.0000038032.22343.de. [DOI] [PubMed] [Google Scholar]
- 45.Schneider TJ, Grillot D, Foote LC, Nunez GE, Rothstein TL. J Immunol. 1997;159:4834–4839. [PubMed] [Google Scholar]
- 46.Wang J, Lobito AA, Shen F, Hornung F, Winoto A, Lenardo MJ. Eur J Immunol. 2000;30:155–163. doi: 10.1002/1521-4141(200001)30:1<155::AID-IMMU155>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 47.Zhang X, Li L, Choe J, Krajewski S, Reed JC, Thompson C, Choi YS. Cell Immunol. 1996;173:149–154. doi: 10.1006/cimm.1996.0260. [DOI] [PubMed] [Google Scholar]
- 48.Conkright MD, Montminy M. Trends Cell Biol. 2005;15:457–459. doi: 10.1016/j.tcb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 49.Shankar DB, Cheng JC, Kinjo K, Federman N, Moore TB, Gill A, Rao NP, Landaw EM, Sakamoto KM. Cancer Cell. 2005;7:351–362. doi: 10.1016/j.ccr.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 50.Shankar DB, Cheng JC, Sakamoto KM. Cancer. 2005;104:1819–1824. doi: 10.1002/cncr.21401. [DOI] [PubMed] [Google Scholar]
- 51.Arcinas M, Heckman CA, Mehew JW, Boxer LM. Cancer Res. 2001;61:5202–5206. [PubMed] [Google Scholar]
- 52.Laffitte BA, Repa JJ, Joseph SB, Wilpitz DC, Kast HR, Mangelsdorf DJ, Tontonoz P. Proc Natl Acad Sci USA. 2001;98:507–512. doi: 10.1073/pnas.021488798. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.