Abstract
Tau is a highly soluble protein, yet it aggregates abnormally in Alzheimer's disease. Here, we address the question of proteolytic processing of tau and the nucleation of aggregates by tau fragments. We show in neuronal cell models that fragments of the repeat domain of tau containing mutations of FTDP17 (frontotemporal dementia with parkinsonism linked to chromosome 17), produced by endogenous proteases, can induce the aggregation of full-length tau. Fragments are generated by successive cleavages, first N-terminally between K257 and S258, then C-terminally around residues 353–364; conversely, when the N-terminal cleavage is inhibited, no fragmentation and aggregation takes place. The C-terminal truncation and the coaggregation of fragments with full-length tau depends on the propensity for β-structure. The aggregation is modulated by phosphorylation but does not depend on it. Aggregation but not fragmentation as such is toxic to cells; conversely, toxicity can be prevented by inhibiting either aggregation or proteolysis. The results reveal a novel pathway of abnormal tau aggregation in neuronal cells.
Keywords: Alzheimer's disease, paired helical filaments, Tau protein, frontotemporal dementia
The neurofibrillary pathology, based on the aggregation of tau protein into paired helical filaments (PHFs), is a hallmark of Alzheimer's disease (AD) and other tauopathies (1), and the distribution of tau deposits correlates with the loss of neurons (2). The discovery that mutations in the tau gene cause frontotemporal dementia with parkinsonism linked to chromosome 17 (FTDP17) (3) and tau aggregation provided evidence that abnormalities in tau were sufficient to cause neurodegeneration. Despite the development of transgenic mice that recapitulate some of the hallmarks of AD (4, 5), the mechanism of tau aggregation remains enigmatic.
Tau is a natively unfolded protein that shows little tendency to aggregate in physiological buffers (6). However, aggregation can be dramatically accelerated by polyanionic cofactors such as sulfated glycosaminoglycans, RNA, acidic peptides, or fatty acid micelles (7–10). The aggregation is thought to follow a nucleation–elongation pathway (11), whereby an oligomeric nucleus is first formed by self-association of protein subunits, followed by addition of subunits to filament ends. However, several issues remain open in this scheme: (i) Although tau fragments aggregate spontaneously (12), full-length tau is difficult to fibrillize even at high concentration unless aided by polyanions. (ii) Seeding of recombinant tau with Alzheimer PHFs is inefficient without fibrillization inducers. (iii) The nature of the nucleating species of tau fibrils is not known.
A further unsolved question is whether tau aggregation is cytotoxic. Because the occurrence of PHFs correlates with the loss of cognitive functions in AD, it is widely assumed that PHFs are one of the causes of neurodegeneration. On the other hand, neurons bearing PHFs can survive for a long time, and memory deficits in transgenic mice can occur independently of PHFs (13, 14). Thus, further cell models of tau aggregation are needed to clarify the relationship between toxicity and tau aggregation.
A third topic in the tau field is the role of posttranslational modifications in tau aggregation and toxicity. Phosphorylation has been studied extensively because AD tau is hyperphosphorylated (15, 16). By comparison, other modifications have received less attention, notably proteolytic processing (17–23). Although the bulk of AD tau is intact, this issue is important because tau fragments could aggregate more readily and thus trigger aggregation; alternatively, tau fragments could cause toxicity by other mechanisms (24).
Here, we describe the stepwise processing of tau in an inducible neuronal cell model of tauopathy expressing full-length tau with or without the FTDP17 mutation ΔK280, where the fragmentation leads to the aggregation of tau. The fragments are generated in N2a cells by endogenous proteolysis from the tau repeat domain. In vitro, recombinant fragment F3 can aggregate spontaneously and nucleate the aggregation of full-length mutant tau, without requiring fibrillization inducers. In this cell model, the toxicity of tau can be ascribed to its aggregation.
Results
Identification of a Nucleating Fragment for Tau Aggregation in N2a Cells.
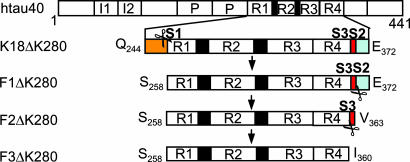
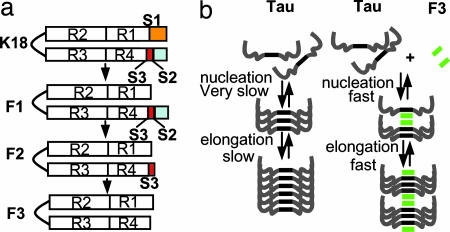
We previously developed a cell model of tauopathy by expressing the mutant tau repeat domain (K18ΔK280; Fig. 1) in N2a cells (21). Besides aggregation in this cell model, we noticed that a fraction of K18ΔK280 was cleaved to generate three small fragments (F1, F2, and F3). They were derived from the same N-terminal cleavage site between K257 and S258 (site S1; Fig. 1). However, the fragments have different C-terminal tails, generated by unknown protease(s). The longest fragment (F1, S258–E372) is soluble and remains in the supernatant after sarkosyl treatment of the cells, whereas the two smaller fragments F2 (ending at V363, site S2) and F3 (ending at I360 or K353, site S3) are prone to aggregation and show up only in the sarkosyl pellet. This observation suggested that the small fragments (F2 or F3) may initiate the aggregation of intact K18ΔK280 in N2a cells. Thus, if the K18 molecule is represented as [N–S1—S3–S2–C], only the fragments F2 = [S1—S3–S2] and F3 = [S1—S3] appear to be efficient at promoting aggregation.
Fig. 1.
Diagram of tau fragments and cleavage sites. The tau isoform htau40 and positions of FTDP17-mutations ΔK280 and P301L are shown at the top. Then repeat domain K18ΔK280 (M-Q244-E372) and fragments F1, F2, and F3 are shown. Scissors indicate cleavage sites behind K257 (site S1) and near residue 360 (sites S2 and S3). To block the cleavage of K18ΔK280, constructs K18ΔK280/257A or 257R were cloned where K257 was replaced either by A257 or R257.
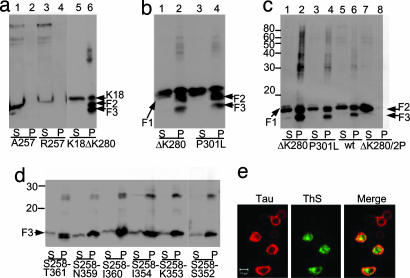
We tested this hypothesis by examining whether the production of the small tau fragments in N2a cells is necessary for the aggregation of the repeat domain K18ΔK280. We mutated the N-terminal cleavage site K257 to A or R to block the thrombin-like protease activity (Fig. 2a). Both mutations abolish the proteolysis of K18ΔK280 at site S1, but surprisingly the cleavages at sites S2 and S3 are suppressed as well, i.e., molecules of the type [N–S1—S3–S2] or [N–S1—S3] are not observed. Thus no small tau fragments (F1, F2, or F3) are detected in the sarkosyl supernatant or pellet (Fig. 2a). Blocking the fragmentation of K18ΔK280 also abolishes aggregation in the N2a cells, as seen by the absence of thioflavin S (ThS) fluorescence and a sarkosyl-insoluble pellet. Thus the cleavage at site S1 is a prerequisite for the cleavage steps at S2 or S3 (generating F2 and F3), and cleavage at site S1 is necessary for efficient aggregation in cells.
Fig. 2.
Tau fragmentation and aggregation. (a–c) Fragmentation precedes aggregation of K18ΔK280. Expression of tau repeat domain or F1 was induced in N2a cells. Lanes labeled P (pellet) denote sarkosyl-insoluble tau species pelletable after sarkosyl extraction; S (supernatant) indicates soluble proteins. (a) K18ΔK280 expressed in N2a cells generates aggregates that appear in the sarkosyl-insoluble pellet (lane 6) and contain a higher molecular weight “smear” and smaller fragments F2 and F3 (F1 is hardly visible because of rapid cleavage to F2). When proteolysis behind K257 is inhibited in A257 and R257 mutants, no fragment F1, no protein pellet, and no fragments F2 or F3 are detected (lanes 2 and 4). (b) Blot analysis of aggregation of K18ΔK280 and K18P301L. In each case the fragmentation accompanies aggregation. Pelleted tau accounts for ≈8–10% of total tau. (c) Fragment F1 and mutants expressed in N2a cells and analyzed by immunoblotting. Because of rapid cleavage, intact F1 is barely visible in soluble fractions (lanes 1, 3, 5, and 7). F1 fragments are cleaved to F2 and F3, which generates aggregates in pellet fractions (lanes 2, 4, and 6; note high molecular weight smear). By contrast, F1ΔK280/2P is only cleaved to F2, which remains in the supernatant (lane 7), but not to F3, and no aggregates are detected (lane 8). The pellets contain ≈17% (F1ΔK280), 10% (F1P301L), 6% (F1 wt), and 0% (F1ΔK280/2P) of total tau. Note that aggregation strongly correlates with the level of F3 (strongest with ΔK280 mutant, absent with ΔK280/2P mutant). (d and e) Identification of a tau aggregation nucleator. (d) F3* fragments expressed in N2a cells with varied C termini, starting at S258 and ending at T361, N359, I360, I354, K353, and S352. Levels of pelleted protein are 15%, 16%, 32%, 20%, 17%, and 8% of total tau, respectively. Note that peptide S258–I360 shows the strongest aggregation and was therefore used in the following experiments (denoted as F3*). (e) Aggregation of F3* (S258–I360) demonstrated by ThS staining. (Left) Tau expression monitored by immunolabeling with antibody K9JA (epitope in repeat domain) and Cy5 secondary antibody. (Center) Aggregation monitored by staining with ThS. (Right) Merged images.
A characteristic feature of the repeat domain K18ΔK280 is its pronounced tendency to aggregate in vitro because of its high propensity for β-structure (25). The same is true for K18 with another FTDP17 mutation, P301L. We asked whether this mutant would show the same fragmentation and aggregation in N2a cells. This is indeed the case and argues that both fragmentation and aggregation are not unique to the K18Δ280 mutation but depend more generally on the β-propensity of the protein (Fig. 2 b and c).
Because fragment F1 shows no aggregation, we suspected that the aggregation of K18ΔK280 is actually promoted by fragments F2 and F3. We tested whether the production of F1 would lead to the generation of F2 and F3, which would then lead to the aggregation of intact K18ΔK280 in N2a cells. To address this question, we cloned three variants of F1, WT F1 (= M–S258–E372), F1ΔK280, and F1P301L, transfected them inducibly into N2a cells, and analyzed their proteolytic products. In all cases, two bands corresponding to F2 and F3 are detected in the sarkosyl pellets, indicating that the production of F1 quickly leads to the efficient C-terminal cleavage to fragments F2 and F3 and subsequent aggregation (Fig. 2c, lanes 2, 4, and 6). The supernatants contain only a weak trace of F1 and part of F2, but no F3, arguing that F3 and not F2 is dominant for rapid aggregation (Fig. 2c, lanes 1, 3, and 5). Next, we tested how the aggregation is regulated by the β-propensity of the fragments. We cloned F1ΔK280/2P, containing prolines within the hexapeptide motifs (I287P and I308P), which act as β-breakers and prevent aggregation (25). When F1ΔK280/2P was transfected into N2a cells, no pellet was generated (Fig. 2c, lane 8), but surprisingly, cleavage of F2 to F3 was halted as well (Fig. 2c, lane 7). We conclude that there is an interdependence between the structure of the hexapeptide motifs in the N-terminal half of K18 and the cleavage at site S3 near the C terminus.
Because fragments F2 and F3 can nucleate the aggregation of K18ΔK280, we next sought to confirm this observation by cloning the smallest fragment (F3) and characterize its aggregation in cells. F3 occurs in two main species, S258–I360 or S258–L353 (21). We therefore cloned these two peptides and variants differing by one residue at the C terminus (S258–I354, S258–S352, S258–T361, S258–N359). The constructs were expressed in N2a cells for 5 days, and all of them led to sarkosyl-insoluble aggregates (Fig. 2d). Among them, peptide S258–I360 was the most potent one in terms of aggregate formation, generating ≈32% aggregates in the sarkosyl pellet. Therefore, peptide S258–I360 (F3ΔK280) was used in the following experiments and will be denoted for short as F3* (by analogy, we abbreviate htau40ΔK280 as Tau*). The PHF-like aggregation of F3* in N2a cells was confirmed by ThS staining (Fig. 2e).
Fragment F3* Nucleates the Aggregation of Full-Length Tau in N2a Cells.
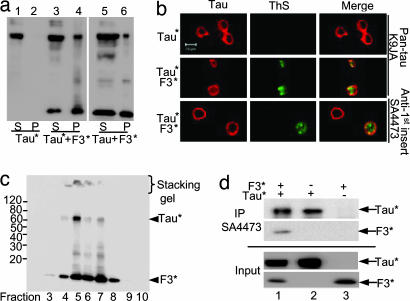
Because F3* is prone to aggregation and able to nucleate the aggregation of the repeat domain K18ΔK280 in cells, we tested whether it is also able to initiate the aggregation of full-length Tau with or without the ΔK280 mutation. When WT Tau is expressed alone in N2a cells, no protein is detected in the sarkosyl pellet (data not shown). This observation is in agreement with the general experience that overexpression of WT full-length tau alone does not lead to aggregation (26), suggesting that there must be nucleating factors in cells. The same inability to aggregate is observed with Tau* alone, even though it carries the ΔK280 mutation that increases the aggregation potential (Fig. 3a, lane 2). In striking contrast, when Tau* is coexpressed with F3*, both components begin to coaggregate (Fig. 3a, lane 4). The same holds for the P301L mutant of full-length tau [supporting information (SI) Fig. 6 Upper]. Thus, the aggregation of the full-length tau initiated by F3* is not unique for the specific FTDP17 mutation ΔK280. To verify that the higher band detected in the sarkosyl pellets represents the insoluble intact Tau* (rather than F3* oligomers), we applied antibody SA4473. It is directed against the first insert of tau near the N terminus (exon 2), and thus recognizes full-length Tau* but not F3* fragments, whereas the pan-tau antibody K9JA (epitope in repeat domain) recognizes both full-length Tau* and F3* (SI Fig. 6a Lower). Both antibodies detect Tau* in the pellet, proving the aggregation of full-length tau. Similar experiments were done to demonstrate the aggregation of WT full-length Tau (htau40wt) under the influence of the nucleating fragment F3*. Because the intrinsic propensity for β-structure and aggregation is much lower in WT tau (25), it is critical to optimize the concentration available for driving the aggregation process nucleated by F3* (e.g., by increasing doxycyclin; Fig. 3a, lane 6). The results illustrate that, in principle, aggregation of Tau in cells obeys similar rules as aggregation in vitro and can be driven by a combination of high concentration, β-conformation, heterologous nucleation (e.g., by F3*), and aggregation time (a parameter that is limited in cell culture studies).
Fig. 3.
Interaction between Tau and fragment F3*. (a and b) Aggregation of full-length Tau nucleated by tau fragment F3*. (a) Blot analysis (antibody K9JA) of aggregation of full-length Tau* (htau40ΔK280) induced by F3*. Without the fragment, Tau* remains in supernatant (lanes 1 and 2), but the fragment induces pelletable aggregates comprising both components (lanes 3 and 4). The same result, but with lesser efficiency, is obtained with WT full-length Tau, i.e., no aggregation for Tau alone (not shown) but aggregate formation in the presence of F3* (lanes 5 and 6). (b) Aggregation of full-length mutant tau (Tau*) in inducible N2a cells demonstrated by ThS staining. (Left) tau expression monitored by immunolabeling with antibody K9JA (Top and Middle) or SA4473 (epitope in the first N-terminal insert of tau) and Cy5 secondary antibody (Bottom). (Center) Formation of aggregates monitored by ThS staining; note that it occurs only in the presence of F3* fragments. (Right) Merged images. The staining with antibody SA4473 confirms that full-length tau is present in aggregates. For analogous results on P301L mutants see SI Fig. 6. (c and d) Interaction of F3* with Tau* in N2a cells. (c) Separation of sarkosyl pellet from N2a cells by iodixanol gradient centrifugation, demonstrating coaggregation of full-length Tau* with F3*. Note that Tau* appears only in fractions enriched in F3* and that higher aggregates are present in the stacking gel in fractions 4–7. (d) Interaction of F3* with Tau* by coimmunoprecipitation. F3* expressed alone or coexpressed with Tau* in N2a cells was pulled down with antibody SA4473 and analyzed by SDS/PAGE followed by immunoblotting with K9JA antibody. Note that part of F3* was pulled down by SA4473 when it was coexpressed with Tau*.
The aggregation of full-length tau (ΔK280 or P301L mutants) initiated by F3 fragments (ΔK280 or P301L mutants) was further assessed by ThS staining in N2a cells. No ThS signal was detected when full-length mutant tau was expressed alone (Fig. 3b and SI Fig. 6b Top), but when tau mutants were expressed together with mutant F3*, ThS staining was detected in ≈12% of the cells (Fig. 3b Middle and Bottom and SI Fig. 6b Middle and Bottom). To exclude the possibility that the aggregates are only composed of F3*, we applied again antibody SA4473 that only recognizes full-length tau. The label colocalized substantially with the ThS staining, demonstrating that full-length tau indeed contributes to aggregates containing β-structure in N2a cells (Fig. 3b and SI Fig. 6b Bottom).
To confirm the coaggregation of nucleating fragments with full-length tau by an independent approach we used iodixanol density gradient centrifugation to separate the sarkosyl pellets from cells cotransfected with F3* and Tau*. Full-length Tau* only appeared in fractions enriched in F3* (Fig. 3c, fractions 4–7), whereas F3* (which can aggregate by itself) was detected in several further fractions (Fig. 3c, fractions 3 and 8). This observation confirms that the aggregation of Tau* is nucleated by F3*. Furthermore, we tried to verify the interaction of F3* with full-length Tau* in cells by preparing lysates from N2a cells expressing either Tau* alone, F3* alone, or coexpressing both proteins. The lysates were probed by the tau antibody SA4473 (recognizing only Tau*) for coimmunoprecipitation, followed by Western blot analysis with pan-tau antibody K9JA. When F3* was coexpressed with Tau*, F3* was coimmunoprecipitated with Tau* by antibody SA4473, confirming the interaction of the two components in cells (Fig. 3d Upper, lane 1). By comparison, when Tau* was expressed alone it was immunoprecipitated, but not F3* alone (Fig. 3d Upper, lanes 2 and 3). Corresponding inputs of the cell extracts are shown in Fig. 3d Lower, lanes 1–3.
The nature of the products of coaggregation was tested in vitro by electron microscopy and ThS fluorescence, which is sensitive to the cross-β structure of the filaments. In the case of Tau* alone, the ThS signal in solution without heparin rises very slowly (SI Fig. 7a, dashed line), and the resulting aggregates are largely amorphous (SI Fig. 7b). However, addition of F3* causes a rapid increase in the ThS signal (SI Fig. 7a, solid line), corresponding to a largely filamentous population of aggregates (SI Fig. 7c). This observation is consistent with the assumption that F3* enables both partners to adopt the structure required for PHF assembly in the absence of polyanionic cofactors. To prove the seeding capacity of F3* directly, we first induced the aggregation of F3*, followed by sonication to break the aggregates into smaller fragments. These seeds were used to initiate the aggregation of full-length Tau or Tau*, resulting in the rapid appearance of PHFs (SI Fig. 7 d–f).
Phosphorylation of Soluble and Aggregated Tau.
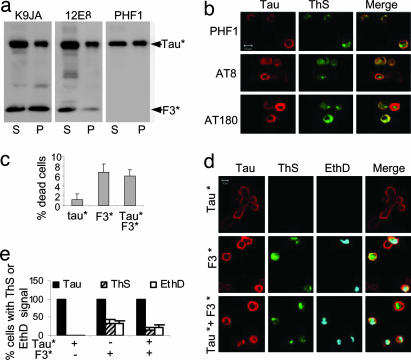
Because phosphorylation has been suggested to play a role in tau aggregation (15, 16), we examined whether phosphorylation is linked to the aggregation of Tau* induced by F3*. We coexpressed Tau* with F3* in N2a cells for 1–5 days and detected the phosphorylation with a panel of phosphorylation-dependent antibodies (PHF1, AT180, AT8, and 12E8). The total soluble and aggregated tau was determined by the phosphorylation-independent pan-tau antibody K9JA (Fig. 4a Left). Phosphorylation in the repeats (12E8 epitope, p-S262 and p-S356) was detected mainly in the supernatant, both for F3* and Tau* (Fig. 4a Center), consistent with the fact that phosphorylation at these sites inhibits tau aggregation (27). Phosphorylation in the C-terminal tail (PHF-1 epitope, p-Ser-396 plus p-Ser-404) occurs in Tau* in the supernatant and the pellet, suggesting that this type of phosphorylation may have a somewhat stimulating influence on tau aggregation (note that the PHF-1 epitope is not present in F3*; Fig. 4a Right). The epitopes in the proline-rich region upstream of the repeats, i.e., AT180 (p-T231 plus p-S235) and AT8 (p-S202 plus p-T205) showed phosphorylation only by immunofluorescence of the cells (Fig. 4b) but not in blots after extraction, presumably because of a high activity of phosphatases. As a control of protein concentration, the pan-tau antibody K9JA detected both F3* and Tau* in the supernatant and the pellet. The data suggest that the phosphorylation in the repeat domain has a noticeable negative effect on the aggregation of tau in the N2a cells, whereas phosphorylation at the PHF1 epitope appears to support aggregation to some extent (for quantification see SI Fig. 8).
Fig. 4.
Aggregation and cell toxicity. (a and b) Aggregation and phosphorylation of Tau* in the presence of F3* in N2a cells. (a) Blot analysis of aggregation of full-length Tau* induced by F3*, using pan-tau antibody K9JA (Left) or phosphorylation-dependent antibodies 12E8 (Center) and PHF1 (Right). Note that phosporylation occurs both in supernatant and pellet and at different sites because of the activity of different kinases (e.g., MARK, GSK-3β). (b) Phosphorylation and aggregation of full-length Tau* induced by F3* in N2a cells, demonstrated by ThS staining. (Left) Tau expression monitored by immunolabeling with phosphorylation-dependent antibodies PHF1, AT8, or AT180 and Cy5 secondary antibody. (Center) Aggregates monitored by staining with ThS. (Right) Merged images. (c–e) Cell toxicity of tau aggregation. N2a cells transfected with tau construct F3*, Tau*, or cotransfected with F3* plus Tau* were induced for 2 days. (c) LDH release measured as an indicator of cell death, calculated as the percent of total LDH (media plus lysates). (Left) N2a cells expressing Tau*. (Center) F3*. (Right) Tau*+F3*. Note that expression of F3* alone or coexpression of F3* and Tau* strongly increases toxicity. (d) Cell death monitored by nuclear staining with EthD. Tau expression determined by immunolabeling with antibody K9JA (red), tau aggregation by ThS staining (green), and cell death by EthD staining (cyan). Note that cell death (blue) occurs preferentially in cells with aggregates (green). (e) Quantitation of ThS- or EthD-positive cells, showing that aggregation (induced by F3*) correlates with toxicity.
The phosphorylation and distribution of the aggregates in cells is illustrated in Fig. 4b. In all cases, the distribution of Tau* and F3* (recognized by antibodies PHF1, AT8, or AT180) overlaps partly with ThS staining for aggregated tau, indicating that soluble and aggregated tau coexist in the cells, independently of phosphorylation. Note that the somewhat variable nature of the localization of tau staining and ThS staining could be explained if some of the aggregates consist predominantly of F3*; in addition, the epitope of antibody K9JA lies in the repeat domain, which becomes partially occluded in the aggregates and therefore is less accessible to the antibody.
Toxicity of Tau Aggregation.
In AD brains, the aggregation of tau correlates with the degeneration of neurons (2), but it is a matter of debate whether the toxicity is caused by tau aggregation as such or other events (4, 5). Because aggregation was detected in N2a cells overexpressing F3* alone or together with Tau*, the cell models are suitable to address the issue of toxicity. We investigated whether tau expression or aggregation was toxic by using the lactate dehydrogenase (LDH) release assay. The results show that expression of Tau*, which cannot aggregate in N2a cells, has little effect on cell viability (Fig. 4c). In contrast, the expression of F3* alone or together with Tau*, which forms aggregates in N2a cells, causes a dramatic increase of LDH release. This argues that tau aggregation is toxic, rather than tau expression. This observation is consistent with our earlier results (21) showing that only aggregatable variants of the Tau repeat domain show toxicity, whereas disruption of β-structure renders them nontoxic (see Fig. 2c, lanes 7 and 8). However, in an attempt to observe the effect of tau aggregation on cytotoxicity directly, we used a modified LIVE-DEAD assay in combination with ThS staining, where cytotoxicity is demonstrated by nuclear staining with ethidium homodimer (EthD) and tau aggregation by ThS staining. To show the full-length tau aggregates, we used antibody SA4473 in N2a cells coexpressing Tau* and F3*. ThS staining was detected in cells solely expressing F3* or coexpressing F3* and Tau*, whereas no ThS signal was detected in cells expressing only Tau* (Fig. 4d). Notably, the majority of ThS-positive cells (70–80%) were also labeled with EthD (Fig. 4d Middle and Bottom). These results underscore that tau aggregation is cytotoxic. The quantitation (Fig. 4e) shows that expression of Tau* by itself generates no ThS or EthD signal (for tau aggregation and toxicity), F3* by itself generates both signals at similar levels (≈30%), indicating that aggregation is highly correlated with toxicity, and Tau* plus F3* is similar, except that the level of dying cells is lower (≈15%), presumably because the level of aggregation is lower as well.
Discussion
Tau is a natively unfolded soluble protein that does not aggregate in physiological conditions. On the other hand, tau aggregates are hallmarks of several brain diseases termed tauopathies, including AD (1). It is unclear why tau forms insoluble fibers in neurons in these diseases. Posttranslational modifications of tau such as phosphorylation, proteolytic processing, or interactions with cofactors have been considered as promotors for aggregation (28). Despite extensive efforts to uncover the effects of tau phosphorylation on aggregation, the issue is still controversial. Likewise, the role of the interaction with nucleating cofactors, e.g., polyanions such as sulfated glycosaminoclycans, RNA, or acidic proteins such as tubulin or α-synuclein remains poorly understood (29). Regarding proteolytic processing, it is well established that the repeat domain of tau, after removal of the N-terminal and C-terminal domains, aggregates into PHFs more readily than the full-length protein. This is due to the exposure of aggregation-prone hexapeptide motifs (6, 12), suggesting that the N- or C-terminal tails are inhibitory to aggregation. However, it has been unclear how this cleavage and aggregation may occur in neurons. The degradation of tau is thought to take place through the ubiquitin–proteasome pathway (30, 31), but other mechanisms contribute as well, such as N-terminal shortening by aminopeptidases (20) or lysosomal proteases (32, 33). Several types of tau truncation have been reported in cells or brain tissue, for example, behind E391 by an unknown protease (17), behind D421 by caspase 3 (18, 19), cleavage sites in the N-terminal half by calpain generating a 17-kDa fragment (23), or fragments generated by thrombin-like proteases (21, 22). Truncated tau fragments containing the repeat domain tended to show enhanced aggregation (12, 34); however, it remained unclear how intracellular fragmentation might cause the aggregation of full-length tau observed in AD.
Here, we describe an endogenous cascade of tau processing and conformational changes that reveals some of the principles of tau aggregation in cells. The major element is the sequential cleavage of the tau repeat domain by endogenous proteases that exposes fragments containing the β-forming hexapeptide motifs (sequential steps 1–3 in Scheme 1, and diagrammed in Fig. 5a). The fragments F2 and F3 readily aggregate by themselves, but more importantly, they nucleate and coaggregate with full-length tau (Scheme 1, step 4). The rate of aggregation depends on the type of fragment (F3 fastest), rates of proteolysis (generation of F1 is slow), FTDP17 mutations that enhance β-structure (e.g., ΔK280, P301L), and the concentration of fragments. The buildup of aggregates gradually leads to toxicity (Scheme 1, step 5).
Scheme 1.
Cascade of tau processing and aggregation. K18 is cleaved in N2a cells by a thrombin-like protease to generate F1 (a slow step), which is then successively cleaved by unknown proteases to give rise to F2 (a fast step) and F3 (a medium step). F3 can nucleate the aggregation of the full-length tau. Tau aggregation slowly causes cytotoxicity.
Fig. 5.
Model of relationship between tau fragmentation and aggregation. (a) Sequential cleavage of the tau repeat domain, first at site S1, later at sites S2 and S3. The latter sites become accessible for cleavage only after the cleavage at S1. Fragments F2 and especially F3 have a strong tendency for aggregation. (b) Aggregation of full-length tau alone (Left) would be too slow to be observable, but fragment F3 causes nucleation and coassembly of F3 with full-length tau (Right).
Two features are noteworthy: The dependence of aggregation on fragment type is similar in cells and in vitro, so that the properties of tau alone suffice to explain the reaction sequence, without the need to postulate other nucleating elements such as cellular polyanions. In particular, full-length mutant Tau* (which normally requires polyanions for aggregation) can be coaggregated with mutant fragment F3* in vitro even in the absence of polyanions. This is explained by the strong cooperativity between the β-forming motifs in full-length tau and the fragments.
Second, there is a curious dependence of C-terminal cleavages (sites S2 and S3; Scheme 1, steps 2 and 3) on the prior N-terminal cleavage at site S1 (Scheme 1, step 1), indicating that the N-terminal header (up to K257) may protect the region around residue 360. It suggests that the repeat domain is folded up to bring the ends close to one another (Fig. 5a). Even more remarkable is the fact that the cleavage at site S3 (Scheme 1, step 3) near the middle of repeat 4 depends on the β-propensity of the hexapeptide motifs at the beginning of repeats 2 and 3, which can be disrupted by proline mutations (as in the ΔK280/2P mutant; see Fig. 1). One interpretation is that the β-sheet interaction between the two hexapeptide motifs causes a conformation that makes the site S3 accessible to the protease. The example shows how the β-propensity of the repeats determines not only the formation of β-sheets in the aggregates, but also the generation of amyloidogenic fragments.
Some of the above considerations are incorporated into the reaction diagram of Fig. 5b, illustrating the roles of sequential cleavage and folding. It illustrates that tau aggregation follows a nucleation–elongation mechanism whose rate-limiting step is the formation of an oligomeric nucleus (11). As with other polymerization reactions of this kind it can be accelerated by preformed seeds (nuclei or filaments). Tau fragments containing the repeat domain are particularly effective in nucleation because they encode the structural template (hexapeptide motifs with β-structure) (12). A small fraction of such seeds could thus generate PHFs from full-length tau, consistent with the observation that Alzheimer PHFs contain mainly full-length tau of all isoforms (35, 36). The question is, however: Is this assembly principle realized in cells? If yes, what is the nucleating fragment? What proteases are responsible for its generation, and why and when do they become activated? In the case of the N2a cell system, we show that endogenous proteases can generate the fragments capable of nucleating full-length tau.
In AD, tau hyperphosphorylation occurs together with aggregation, and there is an ongoing debate on how these two properties are related (15, 16). Thus we asked whether the aggregation of the full-length FTDP tau initiated by the small fragments is linked to phosphorylation. On the level of tau fragments, the only notable sites are the KXGS motifs (S262, S293, S324, S356) whose phosphorylation is inhibitory to aggregation, and correspondingly the phospho-fragments are found mostly in the supernatant (21, 27). More interestingly, with the aggregation of full-length tau we can now ask which of the many potential sites become phosphorylated in the cells. It turns out that all of the sites tested so far become phosphorylated to some extent, including several proline-directed sites. They are distributed both between soluble and aggregated tau, suggesting that none of the sites make an all-or-nothing difference to aggregation. Whereas phosphorylation in the repeat domain tends to inhibit, other sites show a tendency for supporting aggregation, e.g., the PHF-1 site, consistent with the observations of others (34).
Finally, we comment on the issue of tau toxicity. The assumption that tau aggregates in AD are toxic to neurons was challenged by recent findings showing that cognitive deficits were unrelated to tau aggregates (13, 14). The assessment of tau toxicity is complicated by the fact that the extent and reversibility of tau aggregates depends on their stages, influence of Aβ, and types of aggregate (oligomers, fibers) (5). At any rate, in our cell model, toxicity is clearly related to the appearance of tau aggregates. The toxicity can be suppressed by switching off the tau expression, using nonaggregating tau variants, or inhibiting N-terminal proteolysis. However, it is of particular interest that aggregation can be avoided by inhibiting the proteases that generate the amyloidogenic fragments. This may open a novel window to suppress tau pathology in cells.
Materials and Methods
Cell Culture and Transfection.
The selection of a Tet-On, G418-resistant N2a cell line has been described (21). Tet-on inducible cells were cultured in Eagle's MEM with 10% defined FBS, 2 mM glutamine, 0.1% nonessential amino acids, and 600 μg/ml G418. The DNA encoding different fragments of the tau repeat domain (F1wt, F1ΔK280, F1P301L, F3ΔK280, F3P301L) was inserted into the bidirectional vector pBI-5 between ClaI and SalI restriction sites (pBI-5 is an unpublished derivative of pBI-2) (37). Effectene (Qiagen, Hilden, Germany) was used for transfection. Expression of tau was induced by 1 μg/ml doxycycline in medium for 1–5 days, and medium was changed every 2 days.
Biochemical Assays.
For solubility assays, cells were collected by centrifugation at 1,000 × g for 5 min. The levels and solubility of different tau constructs were determined by sarkosyl extraction following ref. 38 (for details see SI Text). Supernatant and sarkosyl-insoluble pellet samples were analyzed by Western blotting. The amount of material loaded for supernatant and sarkosyl-insoluble pellet represented ≈0.5 and 15% of the total material present in the supernatant and pellet, respectively (the ratio between supernatant and sarkosyl insoluble pellet was always 1:30). For quantification of tau levels, the Western blots were probed with pan-tau antibody K9JA (DAKO, Glostrup, Denmark) and analyzed by densitometry.
Immunofluorescence.
Inducible N2a cells were either singly transfected with pBI5 plasmids encoding tau fragments or cotransfected with pBI5 plasmids encoding full-length tau. After 1 day, cells were induced to express tau with 1 μg/ml doxycycline for 2–5 days (or more, if higher concentrations were needed). The cells on the coverslips were fixed with 4% paraformaldehyde in PBS for 15 min, then permeabilized with 80% MeOH for 6 min at −20°C, incubated with 0.1% ThS for 5 min, and washed three times in ethanol (50%). Samples were incubated with antibody K9JA or SA4473 in 5% goat serum (PBS). The secondary anti-rabbit antibody labeled with Cy5 was also diluted with 5% goat serum in PBS and incubated for 45 min. The cells were washed twice with PBS, once with water, and mounted. Confocal microscopy was done with a LSM510 microscope (Zeiss, Oberkochen, Germany) using a ×63 objective.
Density Gradient Centrifugation.
A discontinuous density gradient of the nonionic medium iodixanol was created by layering different concentrations of iodixanol in a centrifugation tube (from bottom to top: 450 μl of 50%, 45%, 40%, 35%, 30%, 20%, 10%, and 5%). Samples were applied in a volume of 80 μl, and centrifugation was performed in a Sorvall TV865 rotor (Kendro Laboratory Products, Langenselbold, Germany) in an ultracentrifuge (Optima 80K; Beckman Coulter, Fullerton, CA) for 3 h at 350,000 × g at 4°C. Fractions were collected from the bottom of the tubes and numbered from high to low density.
Cytotoxicity Assays.
Cytotoxicity was assessed by an LDH assay kit (Roche Applied Science, Indianapolis, IN) or a LIVE-DEAD assay kit (Molecular Probes, Eugene, OR). LDH activity was measured spectrophotometrically at 492 nm. Cell death was calculated as percent of LDH released into medium, compared with total LDH obtained after total cell lysis. After 1 day of doxycycline-induced protein expression, the medium with 10% serum was exchanged for medium with 1% serum (after washing with PBS), and after 1 additional day the medium was collected for LDH determination. For the LIVE–DEAD assay, N2a cells seeded on the coverslips were induced to express tau constructs for 2 days. EthD (5 mM; Molecular Probes) was added to the medium to a final concentration of 2 μM and incubated at 37°C for 30 min. Cells were fixed with 4% paraformaldehyde in PBS for 15 min and processed for immunofluorescence.
Immunoprecipitation.
Transfected N2a cells were rinsed twice with ice-cold PBS, lysed in RIPA buffer (50 nm Tris·HCL, pH 8.0/150 mM NaCl/10% glycerol/1% Nonidet P-40/1% sodium deoxycholate/0.1% SDS, 1 mM PMSF/1 mM DTT/10 μg/ml leupeptin) supplemented with protease inhibitors (Roche, Basel, Switzerland) and incubated on ice for 30 min. After centrifugation, lysates were precleared with protein G-Sepharose beads (Roche) for 1 h at 4°C. The lysates were incubated with antibody SA4473 overnight with constant rotation at 4°C. Proteins were precipitated for 2 h with G-Sepharose beads. After centrifugation, the beads were washed twice with cold PBS, resuspended in Laemmli sample buffer, and analyzed by SDS/PAGE followed by Western blotting with tau antibody.
Supplementary Material
Acknowledgments
We thank Dr. P. Davies (Albert Einstein College, Bronx, NY) and P. Seubert (Elan Pharma, South San Francisco, CA) for antibodies PHF-1 and 12E8. This work was supported by Deutsche Forschungsgemeinschaft.
Abbreviations
- AD
Alzheimer's disease
- FTDP17
frontotemporal dementia with parkinsonism linked to chromosome 17
- LDH
lactate dehydrogenase
- EthD
ethidium homodimer
- PHF
paired helical filament
- ThS
thioflavin S.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0703676104/DC1.
References
- 1.Lee VM, Goedert M, Trojanowski JQ. Annu Rev Neurosci. 2001;24:1121–1159. doi: 10.1146/annurev.neuro.24.1.1121. [DOI] [PubMed] [Google Scholar]
- 2.Braak H, Braak E. Acta Neuropathol (Berl) 1991;82:239–259. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- 3.Spillantini MG, Goedert M. Trends Neurosci. 1998;21:428–433. doi: 10.1016/s0166-2236(98)01337-x. [DOI] [PubMed] [Google Scholar]
- 4.Lee VM, Trojanowski JQ. J Alzheimer's Dis. 2006;9:257–262. doi: 10.3233/jad-2006-9s328. [DOI] [PubMed] [Google Scholar]
- 5.LaFerla FM, Oddo S. Trends Mol Med. 2005;11:170–176. doi: 10.1016/j.molmed.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 6.Wille H, Drewes G, Biernat J, Mandelkow E-M, Mandelkow E. J Cell Biol. 1992;118:573–584. doi: 10.1083/jcb.118.3.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perez M, Valpuesta JM, Medina M, Montejo de Garcini E, Avila J. J Neurochem. 1996;67:1183–1190. doi: 10.1046/j.1471-4159.1996.67031183.x. [DOI] [PubMed] [Google Scholar]
- 8.Goedert M, Jakes R, Spillantini MG, Hasegawa M, Smith MJ, Crowther RA. Nature. 1996;383:550–553. doi: 10.1038/383550a0. [DOI] [PubMed] [Google Scholar]
- 9.Kampers T, Friedhoff P, Biernat J, Mandelkow E-M. FEBS Lett. 1996;399:344–349. doi: 10.1016/s0014-5793(96)01386-5. [DOI] [PubMed] [Google Scholar]
- 10.King ME, Ahuja V, Binder LI, Kuret J. Biochemistry. 1999;38:14851–14859. doi: 10.1021/bi9911839. [DOI] [PubMed] [Google Scholar]
- 11.Friedhoff P, von Bergen M, Mandelkow E-M, Davies P, Mandelkow E. Proc Natl Acad Sci USA. 1998;95:15712–15717. doi: 10.1073/pnas.95.26.15712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.von Bergen M, Friedhoff P, Biernat J, Heberle J, Mandelkow E-M, Mandelkow E. Proc Natl Acad Sci USA. 2000;97:5129–5134. doi: 10.1073/pnas.97.10.5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Santacruz K, Lewis J, Spires T, Paulson J, Kotilinek L, Ingelsson M, Guimaraes A, DeTure M, Ramsden M, McGowan E, et al. Science. 2005;309:476–481. doi: 10.1126/science.1113694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andorfer C, Acker C, Kress Y, Hof P, Duff K, Davies P. J Neurosci. 2005;25:5446–5454. doi: 10.1523/JNEUROSCI.4637-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iqbal K, Alonso Adel C, Chen S, Chohan MO, El-Akkad E, Gong CX, Khatoon S, Li B, Liu F, Rahman A, et al. Biochim Biophys Acta. 2005;1739:198–210. doi: 10.1016/j.bbadis.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 16.Kosik KS, Shimura H. Biochim Biophys Acta. 2005;1739:298–310. doi: 10.1016/j.bbadis.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 17.Novak M, Kabat J, Wischik CM. EMBO J. 1993;12:365–370. doi: 10.1002/j.1460-2075.1993.tb05665.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gamblin TC, Chen F, Zambrano A, Abraha A, Lagalwar S, Guillozet AL, Lu M, Fu Y, Garcia-Sierra F, LaPointe N, et al. Proc Natl Acad Sci USA. 2003;100:10032–10037. doi: 10.1073/pnas.1630428100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rissman RA, Poon WW, Blurton-Jones M, Oddo S, Torp R, Vitek MP, LaFerla FM, Rohn TT, Cotman CW. J Clin Invest. 2004;114:121–130. doi: 10.1172/JCI20640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karsten SL, Sang TK, Gehman LT, Chatterjee S, Liu JK, Lawless GM, Sengupta S, Berry RW, Pomakian J, Oh HS, et al. Neuron. 2006;51:549–560. doi: 10.1016/j.neuron.2006.07.019. [DOI] [PubMed] [Google Scholar]
- 21.Khlistunova I, Biernat J, Wang YP, Pickhardt M, von Bergen M, Gazova Z, Mandelkow E, Mandelkow M. J Biol Chem. 2006;281:1205–1214. doi: 10.1074/jbc.M507753200. [DOI] [PubMed] [Google Scholar]
- 22.Arai T, Guo JP, McGeer PL. J Biol Chem. 2005;280:5145–5153. doi: 10.1074/jbc.M409234200. [DOI] [PubMed] [Google Scholar]
- 23.Park SY, Ferreira A. J Neurosci. 2005;25:5365–5375. doi: 10.1523/JNEUROSCI.1125-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.King ME, Kan HM, Baas PW, Erisir A, Glabe CG, Bloom GS. J Cell Biol. 2006;175:541–546. doi: 10.1083/jcb.200605187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.von Bergen M, Barghorn S, Li L, Marx A, Biernat J, Mandelkow E-M, Mandelkow E. J Biol Chem. 2001;276:48165–48174. doi: 10.1074/jbc.M105196200. [DOI] [PubMed] [Google Scholar]
- 26.Baum L, Seger R, Woodgett JR, Kawabata S, Maruyama K, Koyama M, Silver J, Saitoh T. Brain Res Mol Brain Res. 1995;34:1–17. doi: 10.1016/0169-328x(95)00111-5. [DOI] [PubMed] [Google Scholar]
- 27.Schneider A, Biernat J, von Bergen M, Mandelkow E, Mandelkow E-M. Biochemistry. 1999;38:3549–3558. doi: 10.1021/bi981874p. [DOI] [PubMed] [Google Scholar]
- 28.Stoothoff WH, Johnson GV. Biochim Biophys Acta. 2005;1739:280–297. doi: 10.1016/j.bbadis.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 29.Lee VM, Giasson BI, Trojanowski JQ. Trends Neurosci. 2004;27:129–134. doi: 10.1016/j.tins.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 30.Shimura H, Schwartz D, Gygi SP, Kosik KS. J Biol Chem. 2004;279:4869–4876. doi: 10.1074/jbc.M305838200. [DOI] [PubMed] [Google Scholar]
- 31.Petrucelli L, Dickson D, Kehoe K, Taylor J, Snyder H, Grover A, De Lucia M, McGowan E, Lewis J, Prihar G, et al. Hum Mol Genet. 2004;13:703–714. doi: 10.1093/hmg/ddh083. [DOI] [PubMed] [Google Scholar]
- 32.Bednarski E, Lynch G. J Neurochem. 1996;67:1846–1855. doi: 10.1046/j.1471-4159.1996.67051846.x. [DOI] [PubMed] [Google Scholar]
- 33.Nixon RA, Mathews PM, Cataldo AM. J Alzheimer's Dis. 2001;3:97–107. doi: 10.3233/jad-2001-3114. [DOI] [PubMed] [Google Scholar]
- 34.Abraha A, Ghoshal N, Gamblin TC, Cryns V, Berry RW, Kuret J, Binder LI. J Cell Sci. 2000;113:3737–3745. doi: 10.1242/jcs.113.21.3737. [DOI] [PubMed] [Google Scholar]
- 35.Goedert M, Spillantini MG, Cairns NJ, Crowther RA. Neuron. 1992;8:159–168. doi: 10.1016/0896-6273(92)90117-v. [DOI] [PubMed] [Google Scholar]
- 36.Lee VM, Balin BJ, Otvos L, Trojanowski JQ. Science. 1991;251:675–678. doi: 10.1126/science.1899488. [DOI] [PubMed] [Google Scholar]
- 37.Baron U, Freundlieb S, Gossen M, Bujard H. Nucleic Acids Res. 1995;23:3605–3606. doi: 10.1093/nar/23.17.3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Greenberg SG, Davies P. Proc Natl Acad Sci USA. 1990;87:5827–5831. doi: 10.1073/pnas.87.15.5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.