Abstract
Neuronal dynamics unfolding within the cerebral cortex exhibit complex spatial and temporal patterns even in the absence of external input. Here we use a computational approach in an attempt to relate these features of spontaneous cortical dynamics to the underlying anatomical connectivity. Simulating nonlinear neuronal dynamics on a network that captures the large-scale interregional connections of macaque neocortex, and applying information theoretic measures to identify functional networks, we find structure–function relations at multiple temporal scales. Functional networks recovered from long windows of neural activity (minutes) largely overlap with the underlying structural network. As a result, hubs in these long-run functional networks correspond to structural hubs. In contrast, significant fluctuations in functional topology are observed across the sequence of networks recovered from consecutive shorter (seconds) time windows. The functional centrality of individual nodes varies across time as interregional couplings shift. Furthermore, the transient couplings between brain regions are coordinated in a manner that reveals the existence of two anticorrelated clusters. These clusters are linked by prefrontal and parietal regions that are hub nodes in the underlying structural network. At an even faster time scale (hundreds of milliseconds) we detect individual episodes of interregional phase-locking and find that slow variations in the statistics of these transient episodes, contingent on the underlying anatomical structure, produce the transfer entropy functional connectivity and simulated blood oxygenation level-dependent correlation patterns observed on slower time scales.
Keywords: functional MRI, graph theory, neuroanatomy, synchrony
The anatomical connections between regions of the cerebral cortex form a structural network upon which neural activity unfolds. Cortical regions dynamically couple to one another forming functional networks associated with perception, cognition, and action (1–4), as well as during spontaneous activity in the default or resting state (5–12). Functional networks extracted from higher-frequency dynamics (≈10 Hz) undergo rapid reconfiguration, e.g., in perceptual binding (13) or sensorimotor coordination (14). Functional networks extracted from lower-frequency (<0.1 Hz) spontaneous cortical dynamics are organized into anticorrelated clusters (8, 10), and the transient activation of these clusters has been linked to attentional processes (10, 12). Recent analyses suggest that structural and functional brain networks share “small-world” topologies and hub nodes (15–20) and that structural and functional clusters may closely correspond (21, 22). Nevertheless, it remains unclear how functional networks at different time scales relate to one another and to their common structural substrate.
Here we studied mappings between structural and functional networks using a computational approach. A structural network of segregated regions and interregional pathways was obtained from anatomical studies of macaque cortex and collated using the neuroinformatics tool CoCoMac (23). We then simulated the mean activation of each brain region within this network using a nonlinear model of spontaneous neuronal activity (24). From these activity patterns we built maps of interregional interaction by measuring transfer entropy (TE) [a measure designed to capture patterns of directed interaction and information flow (25)], in long (minutes) and intermediate (seconds) data samples. Our most fine-grained temporal measure is the phase locking value (26), which can identify transient synchrony between region pairs on the millisecond scale. For all methods we thresholded the regional interaction maps to produce functional networks whose topologies can be compared with one another and with that of the underlying structural network.
Node centrality is a useful diagnostic for comparing topologies, and so we first identified structural hubs using network measures based on local flow, global centrality, and motif distributions. After constructing functional networks, we found temporally varying network structure at multiple time scales. Using long data samples, TE yields the closest match between structural and functional connections including a close correspondence between structural and functional hubs. On a time scale of seconds, we found that functional networks exhibit significant fluctuations. These fluctuations reflect slow variations in the mean length and frequency of intermittent synchronous episodes. These slow variations, both in functional connectivity and in values of estimated regional blood oxygenation level-dependent (BOLD) signal, were coordinated via anatomical connection patterns, and we defined two major anticorrelated functional clusters. Our model suggests that the cortical resting state is not time-invariant, but instead contains rich and interrelated temporal structure at multiple time scales that is shaped by the underlying structural topology.
Results
Identification of Structural Hubs.
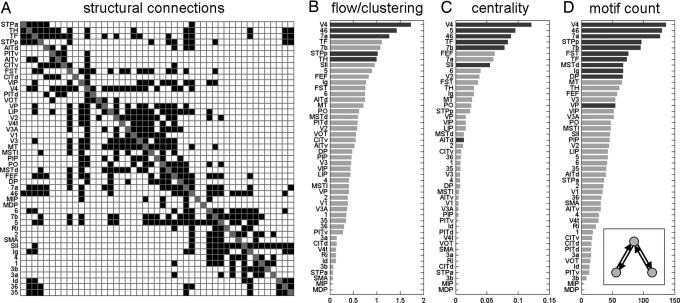
All analyses and simulations were carried out using a connection matrix of macaque neocortex, comprising 47 visual, sensory, and motor areas linked by 505 pathways previously identified by anatomical tracing studies (Fig. 1A). Hubs in brain networks allow for increased levels of information flow between otherwise distant or disconnected nodes, acting as connectors or integrators at central locations within the network. Because all of the hub identification tools we employ depend on the degree distribution of the network, we performed statistical comparisons to degree-matched random and lattice networks using z-scores on a node-by-node basis [see supporting information (SI)]. To estimate the capacity of a node to conduct information flow between its neighboring nodes, we introduced the flow coefficient, a measure of “local centrality” (see SI). It is calculated as the number of actual paths of length 2 divided by the number of all possible paths of length 2 that traverse a central node. Hub regions that act as bridges between different communities of nodes are likely to exhibit small clustering coefficients (16) and large flow coefficients. In macaque neocortex, areas V4, 46, 7a, STPp, and TH (Fig. 1B) are among those with flow-clustering ratios at levels that are significantly increased relative to degree-matched random controls. Betweenness centrality (Fig. 1C) is a more global centrality measure that captures the tendency of a node to lie along the shortest path between pairs of nodes in the network (27). In macaque neocortex, areas V4, 5, 46, TF, 7b, and SII are among those areas with significantly elevated betweenness centrality. Structural motifs (28, 29) provide a complementary means of identifying hubs. In large-scale cortical networks, structural hubs will tend to be reciprocally linked to brain regions that are not directly connected to one another; i.e., they tend to participate in motifs consisting of exactly two reciprocal edges (see Fig. 1D Inset). Fig. 1E shows the motif participation profiles for individual brain regions in macaque neocortex. Areas V4, 46, 7a, STPp, 7b, FST, TE, MSTd, Ig, DP, and VP show significantly increased motif contributions. Reviewing the results of flow centrality, betweenness centrality, and motif approaches to hub identification, we see that only area V4 emerges as highly ranked across all three measures (Fig. 1 B–D) whereas areas 46, 7a, 7b, and STPp show significance in at least two of these three measures.
Fig. 1.
Structural connectivity and network hubs. (A) Large-scale anatomical connection matrix of macaque neocortex. (B) Ranking of areas for flow/clustering ratio. Flow/clustering ratios for each region are compared with those obtained from 1,000 degree-matched, randomized networks. Regions with significantly increased flow/clustering ratio (P < 0.05, uncorrected) are shown in dark gray. (C) Ranking of areas for betweenness centrality. Regions with significantly increased (P < 0.05) centrality are shown in dark gray. (D) Ranking of areas for motif count of the motif class shown in Inset. Dark gray bars indicate that the area shows increased motif counts (z-score > 2) relative to random and lattice controls.
Simulation of Neuronal Dynamics.
To emulate neuronal dynamics within the macaque neocortex we implemented a neural mass model adopted from a conductance-based model of neuronal dynamics (30) for local population activity (31). Units of the model describe local populations of densely interconnected inhibitory and excitatory neurons whose behaviors are determined by voltage- and ligand-gated membrane channels. Sodium and calcium channels display a nonlinear sigmoid-shaped graph of voltage-dependent conductance. Potassium channel conductance is modeled in a more complex manner, exponentially relaxing toward its voltage-dependent state. A medium-scale (mesoscopic) array is then constructed from these local nonlinear populations by introducing long-range pyramidal connections, mimicking glutamate-induced synaptic currents (24, 32). Activity in the system arises purely from nonlinear instabilities. Oscillations are hence spontaneous and self organizing. Spatiotemporal patterns arise through reentrant excitatory–excitatory feedback. In the present case we used the macaque neocortical connectivity to determine the internode coupling. We chose parameter settings that replicate realistic conductances and have previously been reported to show complex, spontaneous activity, including intermittency, phase synchrony, and marginal stability (24, 32). Internode coupling was set to a low value (c = 0.1) at which synchronous dynamics are weakly stable, allowing spontaneous switching between synchronous epochs and desynchronous bursts (32). Under this parameterization the neural interactions in the model reflect a shifting balance between the effects of the structural substrate and of spontaneously arising “self-organizing” patterns that are uninfluenced by task- or input-related factors. The model is described in detail in the SI.
Functional Connectivity at Multiple Time Scales.
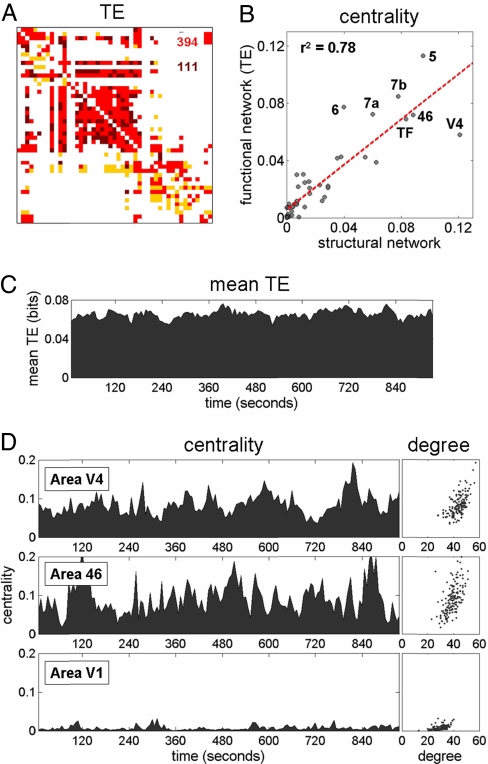
Functional connectivity is defined as the statistical dependence between remote neural processes (33). Here we identified functional connections by using TE (25), an asymmetric information theoretic measure designed to capture directed information flow. We first applied it to long samples of time series data (240,000 time steps; 240 sec). TE values were thresholded to produce binary functional networks (see Fig. 2A). The threshold was chosen such that the total number of connections in a functional network is equal to the number of structural connections. Using long data samples, TE networks and structural networks showed up to 80% overlap, whereas the overlap between structural networks and functional networks extracted with mutual information and wavelet-based tools was lower (see SI).
Fig. 2.
Topology of functional networks and hub dynamics. (A) Example of a functional TE network, calculated using a 240,000 time step data segment (240 sec). The functional matrix has been thresholded to yield a binary network with connection density equal to that of the structural network (505 connections). Comparison between structural and functional networks allows the distinction of true positives (TP functional connections that match existing structural connections; shown in red), as well as false positives (FP functional connections absent in the structural matrix; shown in dark red) and false negatives (FN structural connections absent in the functional network; shown in yellow). Numerals give the count of TP and FP (TP + FP = 505; FP = FN at this threshold). (B) Correlation between centrality of each node within structural and TE functional network (mean of n = 5, obtained from 240-sec data segments): r2 = 0.78. (C) Time course of TE averaged over the entire network: 960,000 time steps (16 min) windowed into 156 overlapping data segments, 30,000 time steps each, consecutively offset by 6,000 time steps (6 sec). (D) Corresponding time course for betweenness centrality for two putative hub regions (area V4, Top, and area 46, Middle) and one non-hub region (area V1, Bottom). Scatter plots at right show the relationship between degree and centrality for these nodes across the 156 functional networks.
We carried out a detailed analysis of structure–function correspondence for several topological measures (see SI). The degrees, flow coefficients, and centrality of nodes within TE networks are strongly correlated with those of nodes within the structural network. For example, betweenness centrality of individual nodes is highly correlated between structural and TE matrices (r2 = 0.78; see Fig. 2B). Weaker correlations for centrality are observed when functional networks are constructed by using mutual information or wavelet methods (r2 = 0.58 and r2 = 0.53, respectively). This suggests that many of the functional hubs identified in neuroimaging studies (18) are likely to be structural hubs as well.
TE matrices computed over large data samples reveal, on average, very close relations between the functional and structural characteristics of brain regions. However, analyses using overlapping time intervals that are shorter while still allowing for unbiased entropy estimation (30,000 time steps; 30 sec with a 6-sec shift between windows) reveal TE patterns as dynamic and time-dependent, indicating gradual reconfiguration of functional connections. These fluctuations occur despite the fact that the underlying structural connections are of constant strength and reflect the marginally stable and itinerant nature of the neuronal dynamics. Fig. 2C shows the variation over time in the mean magnitude of network interactions.
Temporal variations in interaction strength produce changes in the topology of functional networks and thus in the functional centrality of each node. Fig. 2D shows betweenness centrality across time, and also in relation to node degree, for selected areas in the network. For regions that, over longer time periods (see Fig. 2B), exhibit hub characteristics (e.g., areas V4 and 46), analyses over shorter time scales reveal substantial variations in degree and betweenness centrality over wide ranges. The centrality of highly connected non-hub regions (e.g., area V1) remains low. Hence under spontaneous nonlinear dynamics hubs engage and disengage across time (“hub dynamics”).
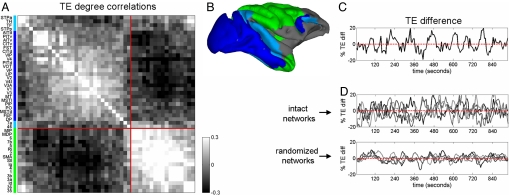
As patterns of functional connections change, network nodes gain and lose functional connections (i.e., change degree). Cross-regional correlations in degree across time reveal the existence of functional clusters that coordinate their intra- and inter-group interactions. Fig. 3A shows the pattern of such correlations obtained for TE networks (average of four simulations), characterized by the presence of two main clusters. One of these clusters contains mostly visual areas, particularly those of the ventral stream, whereas the other contains somatosensory and motor areas as well as portions of the dorsal visual stream. Both clusters consist of brain regions that are largely contiguous on the cortical surface, with one cluster restricted mostly to the occipital and temporal lobes while the other occupies portions of the parietal and frontal lobes (Fig. 3B). Several areas (e.g., parietal area 7a and frontal area 46), although initially grouped with the occipitotemporal cluster, maintain equally strong correlations with members in both clusters, rendering them “intermediate” articulation points that link the two clusters (see also ref. 22). Degree correlations are the result of slow fluctuations in mean TE for the two clusters (Fig. 3C) reflecting alternating periods of elevated information flow within each of the two clusters. In control runs with randomized connection matrices (preserving the in- and out-degree of each node but eliminating the large-scale clustered architecture of the original matrix) we found that, although regional TE fluctuations persisted, between-cluster differences were greatly reduced (Fig. 3D), indicating that clustering in large-scale structural connections is crucial for the generation of large-scale functional anti-correlations within our model.
Fig. 3.
Anticorrelated functional clusters. (A) Degree correlation matrix capturing cross-regional correlations in gains and losses of thresholded functional connections. Matrix represents an average over four 960,000 time step (960-sec) runs, sampled as in Fig. 2 C and D. Cluster analysis yields two main clusters (blue, occipitotemporal; green, parietofrontal). (B) Anatomical location of main clusters in Caret coordinates (51). Areas intermediate in terms of degree correlations are shaded in light blue (areas STPa, STPp, TH, TF, 7a, and 46). (C) Difference in the mean within-cluster TE over time (for the same run shown in Fig. 2 C and D), expressed as percentage of mean signal. (D) TE difference profiles for four runs using intact (Fig. 1A) corticocortical connections (Upper), and four runs carried out in randomized networks preserving in- and out-degree of each node (Lower). Cluster analysis was performed on degree correlation matrices for each separate run to obtain two main clusters. Clusters were then used to calculate differences in mean TE. Standard deviations, expressed as percent baseline TE, are significantly different (two-tailed t test, P < 0.005) for the two groups.
BOLD Correlations and Synchrony.
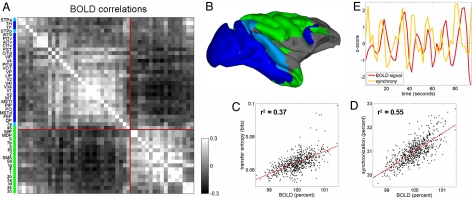
To more closely relate these patterns of functional connectivity to recent neuroimaging work (8, 10, 11) we used the observed changes in membrane potential across time in each brain region, in conjunction with a Balloon–Windkessel hemodynamic model (48–50), to estimate a BOLD signal for each region. After regressing out global BOLD fluctuations (as in ref. 10), pairwise cross-correlations between the residual regional signals again reveal two anticorrelated clusters (Fig. 4 A and B) that correspond closely to those identified on the basis of TE functional connectivity (Fig. 3 A and B).
Fig. 4.
Estimated BOLD signals and relation to synchrony. (A) BOLD correlation matrix, computed from estimated BOLD time series after regressing out global BOLD fluctuations and sampled every 2 sec. Matrix represents an average over four 960,000 time step runs (960 sec, as in Fig. 3). (B) Correlation map obtained by using areas V4 and SII as seed regions, labeling all positively correlated brain regions in either blue (V4) or green (SII). V4 and SII were chosen because they occupy central positions within their respective clusters. Regions not showing positive correlations, or positively correlated with both seed regions, are shown in light blue. (C) Scatter plot of global BOLD signal against variations in aggregate network TE, for corresponding 30-sec data segments obtained from four runs of 960 sec. (D) Scatter plot of global BOLD signal against variations in synchronization. Synchronization is measured as the percentage of phase-locked time steps (in-phase or anti-phase) across all node pairs. (E) BOLD signal and synchrony shown over a 95-sec data segment recorded from area 7a. Synchrony is calculated as the total time that area 7a is phase-locked with any of the other nodes in the network.
Further investigation reveals that BOLD signal and functional connectivity fluctuations occurring over many seconds are related to one another (Fig. 4C) as well as to the fast synchronization dynamics of the system (Fig. 4D). Synchronous (phase-locked) episodes are first identified by calculating the instantaneous phase difference between the oscillatory dynamics at each node using the phase locking value (26). In our model (see SI), as in other weakly coupled systems (34), phase differences are most commonly found to be near 0 radians (in-phase synchronization) or π radians (anti-phase synchronization). Episodes of synchronization typically last between 50 and 300 msec (see SI), a time scale that is consistent with experimental observations of transient synchronous networks in vivo (3). The total length of all synchronous episodes within a given 30-sec time window is correlated (see SI) with the aggregate TE observed in the network across that period, as well as with the mean BOLD signal across regions (Fig. 4D; r2 = 0.55). Examining BOLD synchrony relations region by region we find that, across consecutive 2-sec time windows, BOLD signal amplitudes are correlated with the time each region spends in synchrony. These correlations are strongest for high-degree nodes (see SI). The relationship is lagged by 2–4 sec because of hemodynamic delays (Fig. 4E).
Functional connectivity, BOLD signal amplitude, and the propensity to synchronize are thus interrelated within our model. The fluctuations in functional connectivity and in BOLD signal that we observe on a slow time scale (≈0.1 Hz) are a result of fluctuations in the aggregate number of transient couplings and decouplings occurring on a far more rapid time scale (≈10 Hz). Region pairs engaged in long synchronous episodes at a consistent phase tend to be strongly functionally connected (with high TE), and their BOLD signals tend to be elevated.
Discussion
Simulating neuronal dynamics on a network of large-scale interregional connections in the macaque cortex allowed us to investigate functional connectivity patterns at multiple time scales and to relate them to one another and to their structural substrate. Fast neuronal dynamics exhibit intermittent synchronization and desynchronization on a time scale of hundreds of milliseconds, enabling the system to continually explore a repertoire of functional microstates (35). Slow variations in the statistics of synchronous coupling give rise to changes in the strength of directed interactions between regions on a time scale of seconds. Our model suggests that spontaneous cortical dynamics exhibit ongoing changes in the pattern and strengths of functional coupling and that these coupling events are in turn related to BOLD signal amplitude. The cortical resting state thus exhibits rich and interrelated spatiotemporal structure at multiple scales.
At the slowest time scale (minutes of data) we find that the aggregate strength of functional couplings between regions is, on average, a good indicator of the presence of an underlying structural link. Thresholded functional networks calculated over long time windows closely matched the original structural connection matrix (Fig. 1A), with 79% of the true anatomical links being recovered from the TE analysis (Fig. 2A and SI) along with several features of global topology such as centrality. The success of the TE approach in matching anatomical connections can be attributed to two factors: (i) its sensitivity to directed (nonreciprocal) interactions and (ii) the ergodicity of the chaotic neuronal dynamics, which allows the high-dimensional probability densities required for the calculation of TE to be well estimated from long time windows.
At an intermediate time scale (6-sec intervals, ≈0.1 Hz) significant fluctuations were observed in the strengths of functional coupling. These fluctuations were correlated in time across regions (Fig. 3), with regions participating in one of two functional clusters. Furthermore, the same anticorrelated functional clusters were found when analyzing BOLD signal time series estimated from regional activity (Fig. 4). One cluster comprises mostly visual regions located in the occipital and temporal lobes whereas the other cluster contains mainly somatomotor and frontal regions. Some regions of high structural centrality [notably areas 46, 7a, 7b, and STPp, previously labeled “associational areas” (36)] participated in both clusters, whereas others (e.g., V4 and SII) occupied central positions within their respective clusters.
At the fastest time scale analyzed (≈10 Hz) we observed intermittent synchronization and desynchronization between brain regions. Signals were typically locked at 0 or π radians phase difference for between 50 and 300 msec. At weak interregional coupling, global synchronization never occurs. Instead the system's dynamics consists of a very large set of metastable states similar to what is observed in other classes of high-dimensional chaos [e.g., chaotic itinerancy (37)] that exhibit multiscale temporal structure (38).
The link between fluctuations in transfer entropy and, at a similar time scale, in resting-state fMRI time-series appears to be mediated by the relationship between the synchronization of neuronal dynamics and the mean activity of neuronal populations. This observation is consistent with recent simulations (39), which showed that increased synchronization cannot be divorced from increases in mean firing-rate. Importantly, it suggests that empirical fMRI signals may reflect the time-varying fast synchronization of population dynamics.
Our model currently does not incorporate any time variation in sensory inputs or in the efficacy of anatomical links, and yet we find spontaneous neuronal dynamics that are structured at multiple time scales. The model therefore suggests that the combination of fast and intermittently synchronizing neuronal dynamics and a clustered anatomical substrate may account for two recently observed phenomena: (i) large-scale organization and the existence of hubs in functional connectivity networks (17–20) and (ii) functional clusters that are anticorrelated on a time scale of seconds (8, 10) and possibly bridged (10) via areas in parietal cortex (e.g., area 7a) and frontal cortex (e.g., area 46). On the basis of the model, we predict that individual differences in the strength and location of resting state functional clusters will correlate strongly with individual differences in the topology and efficacy of large-scale corticocortical pathways now detectable using neuroimaging technologies (40).
Methods
Connection Data Set.
We examine a large-scale anatomical data set, referred to in this article as “macaque neocortex,” consisting of a binary connection matrix of brain regions (listed in SI) connected by interregional pathways. Macaque neocortex (Fig. 1A) is an updated network matrix generated following the parcellation scheme of Felleman and Van Essen (36), including visual, somatosensory and motor cortical regions as well as their interconnections (which had not been included previously). The data were manually collated in the CoCoMac database from published tracing studies according to standard procedures (23, 41). Subsequently, all relevant data were translated algorithmically to the Felleman and Van Essen map using coordinate-independent mapping (42, 43). After resolution of redundant and inconsistent results a binary connection matrix with n = 47 nodes (vertices) and K = 505 edges (connections) was generated.
Graph Theory Methods.
All graph-theoretic analyses were conducted on binary matrices. Where necessary, weighted matrices were binarized by thresholding in such a manner as to maintain a constant connection density across networks. The clustering coefficient of a node (15) is calculated as the number of all existing connections between the node's neighbors divided by the number of all possible such connections. In analogy to the clustering coefficient, we define the flow coefficient as the number of all paths of length 2 linking neighbors of a central node that pass through the node, divided by the total number of all possible such paths (see SI).
Central nodes in a network are those that have structural or functional importance, for example, by serving as way stations for network traffic (analogous to bridges or connectors) or by influencing many other nodes via short paths (Fig. 1C). The betweenness centrality of a node is defined as the fraction of shortest paths between all pairs of nodes that travel through that node (27) and was calculated here by using algorithms developed by D. Gleich (www.stanford.edu/∼dgleich/programs/matlab_bgl; ref. 44).
Structural motifs (subgraphs) of size M consist of M nodes and a set of edges (maximally M2 − M, for directed graphs, minimally M − 1 with connectedness ensured). In this study we analyzed macaque neocortex for motifs of size M = 3 (Fig. 1D).
Statistical evaluation of our results requires the design of appropriate null models (45). Such models are not uniquely defined, as statistical comparisons may be carried out relative to many different random models that preserve particular subsets of structural parameters. In this study we generated populations of control (“random”) networks (n = 1,000) using a Markov switching algorithm that preserves degree sequences (46). Lattice networks used as additional controls for data shown in Fig. 1D were created as previously described (16).
TE.
TE was estimated (47) from Gaussian-resampled time series (see SI) discretized into one of 32 bins of uniform width. Sampling bias was corrected by subtraction of a baseline value for each node pair. The baseline for each pair was equal to the TE value obtained from time-shifted data, at a single shift of 4,000 time steps or averaging over shifts of 3,000, 3,500, 4,000, 4,500 and 5,000 time steps (see SI).
TE (25) quantifies the deviation from the generalized Markov property: p(xt+1|xt) = p(xt+1|xt,yt) where xt is the bin assignment of time series x at time t. If conditioning on Yt alters the transition probabilities of Xt, then the assumption of a Markov process is invalid. The incorrectness of the assumption is expressed by the TE, formulated as the Kullback–Leibler entropy:
where the index Y → X indicates the influence of Y on X. TE is nonsymmetric and can thus detect the directed exchange of information (e.g., information flow or causal influence) between x and y.
BOLD Signal Estimation.
Closely following ref. 48 in employing the Balloon–Windkessel hemodynamic model (49, 50), we estimated a BOLD signal for each region based on the local neuronal activity output by our neural mass model. The estimated BOLD signal was calculated independently for each region, disregarding potential effects due to blood flow between spatially adjacent brain regions. Equations and parameters relating neuronal activity and vasodilatory signal with blood inflow, volume, and deoxyhemoglobin content are taken directly from ref. 48. The main model input, “neuronal activity,” is taken to be the absolute value of the time derivative of the mean excitatory membrane potential within each brain region (glutamate turnover).
Supplementary Material
Acknowledgments
We thank S. Newman, R. McIntosh, L. Pessoa, G. Tononi, and A. Vespignani for helpful comments and J. Roberts for technical assistance with BOLD modeling. R.K. acknowledges support by the Deutsche Forschungsgemeinschaft (KO 1560/6-2). C.J.H., R.K., M.B., and O.S. were supported by the J. S. McDonnell Foundation.
Abbreviations
- TE
transfer entropy
- BOLD
blood oxygenation level-dependent.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0701519104/DC1.
References
- 1.Bressler SL. Brain Res Rev. 1995;20:288–304. doi: 10.1016/0165-0173(94)00016-i. [DOI] [PubMed] [Google Scholar]
- 2.Gray CM, Singer W. Annu Rev Neurosci. 1995;18:555–586. doi: 10.1146/annurev.ne.18.030195.003011. [DOI] [PubMed] [Google Scholar]
- 3.Varela F, Lachaux J-P, Rodriguez E, Martinerie J. Nat Rev Neurosci. 2001;2:229–239. doi: 10.1038/35067550. [DOI] [PubMed] [Google Scholar]
- 4.Buszáki G, Draguhn A. Science. 2004;304:1926–1929. doi: 10.1126/science.1099745. [DOI] [PubMed] [Google Scholar]
- 5.Buzsáki G. Rhythms of the Brai. Oxford: Oxford Univ Press; 2006. [Google Scholar]
- 6.Raichle ME, MacLeod AM, Snyder AZ, Powers WJ, Gusnard DA, Shulman GL. Proc Natl Acad Sci USA. 2001;98:676–682. doi: 10.1073/pnas.98.2.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gusnard DA, Raichle ME. Nat Rev Neurosci. 2001;2:685–694. doi: 10.1038/35094500. [DOI] [PubMed] [Google Scholar]
- 8.Greicius MD, Krasnow B, Reiss AL, Menon V. Proc Natl Acad Sci USA. 2003;100:253–258. doi: 10.1073/pnas.0135058100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fox MD, Snyder AZ, Zacks JM, Raichle ME. Nat Neurosci. 2006;9:23–25. doi: 10.1038/nn1616. [DOI] [PubMed] [Google Scholar]
- 10.Fox MD, Snyder AZ, Vincent JL, Corbetta M, Van Essen DC, Raichle ME. Proc Natl Acad Sci USA. 2005;102:9673–9678. doi: 10.1073/pnas.0504136102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fox MD, Corbetta M, Snyder AZ, Vincent JL, Raichle ME. Proc Natl Acad Sci USA. 2006;103:10046–10051. doi: 10.1073/pnas.0604187103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mason MF, Norton MI, Van Horn JD, Wegner DM, Grafton ST, Macrae CN. Science. 2007;315:393–395. doi: 10.1126/science.1131295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodriguez E, George N, Lachaux J-P, Martinerie J, Renault B, Varela FJ. Nature. 1999;397:430–433. doi: 10.1038/17120. [DOI] [PubMed] [Google Scholar]
- 14.Brovelli A, Ding M, Ledberg A, Chen Y, Nakamura R, Bressler SL. Proc Natl Acad Sci USA. 2004;101:9849–9854. doi: 10.1073/pnas.0308538101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Watts DJ, Strogatz SH. Nature. 1998;393:440–442. doi: 10.1038/30918. [DOI] [PubMed] [Google Scholar]
- 16.Sporns O, Zwi J. Neuroinformatics. 2004;2:145–162. doi: 10.1385/NI:2:2:145. [DOI] [PubMed] [Google Scholar]
- 17.Salvador R, Suckling J, Coleman MR, Pickard JD, Menon D, Bullmore E. Cereb Cortex. 2005;15:1332–1342. doi: 10.1093/cercor/bhi016. [DOI] [PubMed] [Google Scholar]
- 18.Achard S, Salvador R, Whitcher B, Suckling J, Bullmore E. J Neurosci. 2006;26:63–72. doi: 10.1523/JNEUROSCI.3874-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stam CJ, Jones BF, Nolte G, Breakspear M, Scheltens P. Cereb Cortex. 2006;17:92–99. doi: 10.1093/cercor/bhj127. [DOI] [PubMed] [Google Scholar]
- 20.Bassett DS, Meyer-Lindenberg A, Achard S, Duke T, Bullmore E. Proc Natl Acad Sci USA. 2006;103:19518–19523. doi: 10.1073/pnas.0606005103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sporns O, Tononi G, Edelman GM. Cereb Cortex. 2000;10:127–141. doi: 10.1093/cercor/10.2.127. [DOI] [PubMed] [Google Scholar]
- 22.Zemanová L, Zhou C, Kurths J. Physica D. 2006;224:202–212. [Google Scholar]
- 23.Kötter R. Neuroinformatics. 2004;2:127–144. doi: 10.1385/NI:2:2:127. [DOI] [PubMed] [Google Scholar]
- 24.Breakspear M, Terry J, Friston K. Network Comp Neural Syst. 2003;14:703–732. [PubMed] [Google Scholar]
- 25.Schreiber T. Phys Rev Lett. 2000;85:464–464. doi: 10.1103/PhysRevLett.85.461. [DOI] [PubMed] [Google Scholar]
- 26.Lachaux J-P, Rodriguez E, Martinerie J, Varela FJ. Hum Brain Mapp. 1999;8:194–208. doi: 10.1002/(SICI)1097-0193(1999)8:4<194::AID-HBM4>3.0.CO;2-C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Freeman LC. Sociometry. 1977;40:35–41. [Google Scholar]
- 28.Milo R, Shen-Orr S, Itzkovitz S, Kashtan N, Chklovskii D, Alon U. Science. 2002;298:824–827. doi: 10.1126/science.298.5594.824. [DOI] [PubMed] [Google Scholar]
- 29.Sporns O, Kötter R. PLoS Biol. 2004;2:1910–1918. doi: 10.1371/journal.pbio.0020369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morris C, Lecar H. Biophys J. 1981;35:193–213. doi: 10.1016/S0006-3495(81)84782-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Larter R, Speelman B, Worth RM. Chaos. 1999;9:795–804. doi: 10.1063/1.166453. [DOI] [PubMed] [Google Scholar]
- 32.Breakspear M, Williams L, Stam K. J Comput Neurosci. 2004;16:49–68. doi: 10.1023/b:jcns.0000004841.66897.7d. [DOI] [PubMed] [Google Scholar]
- 33.Friston KJ. Hum Brain Mapp. 1994;2:56–78. [Google Scholar]
- 34.Ermentrout GB, Kleinfeld D. Neuron. 2001;29:33–44. doi: 10.1016/s0896-6273(01)00178-7. [DOI] [PubMed] [Google Scholar]
- 35.Bressler SL, Tognoli E. Int J Psychophysiol. 2006;60:139–148. doi: 10.1016/j.ijpsycho.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 36.Felleman DJ, Van Essen DC. Cereb Cortex. 1991;1:1–47. doi: 10.1093/cercor/1.1.1-a. [DOI] [PubMed] [Google Scholar]
- 37.Kaneko K, Tsuda I. Chaos. 2003;13:926–936. doi: 10.1063/1.1607783. [DOI] [PubMed] [Google Scholar]
- 38.Atay FM, Jost J. Complexity. 2004;10:17–22. [Google Scholar]
- 39.Chawla D, Lumer ED, Friston KJ. Neural Comput. 2000;12:2805–2821. doi: 10.1162/089976600300014737. [DOI] [PubMed] [Google Scholar]
- 40.Hagmann P, Thiran J-P, Jonasson L, Vandergheynst P, Clarke S, Maeder P, Meuli R. NeuroImage. 2003;19:545–554. doi: 10.1016/s1053-8119(03)00142-3. [DOI] [PubMed] [Google Scholar]
- 41.Stephan KE, Kamper L, Bozkurt A, Burns GA, Young MP, Kötter R. Philos Trans R Soc London B. 2001;356:1159–1186. doi: 10.1098/rstb.2001.0908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stephan KE, Zilles K, Kötter R. Philos Trans R Soc London B. 2000;355:37–54. doi: 10.1098/rstb.2000.0548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kötter R, Wanke E. Philos Trans R Soc London B. 2005;360:751–766. doi: 10.1098/rstb.2005.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gleich D. MatlabBGL. Stanford, CA: Stanford University Institute for Computational and Mathematical Engineering; 2006. Version 1.01. [Google Scholar]
- 45.Artzy-Randrup Y, Stone L. Phys Rev E. 2005;72 doi: 10.1103/PhysRevE.72.056708. 056708. [DOI] [PubMed] [Google Scholar]
- 46.Maslov S, Sneppen K. Science. 2002;296:910–913. doi: 10.1126/science.1065103. [DOI] [PubMed] [Google Scholar]
- 47.Lungarella M, Ishiguro K, Kuniyoshi Y, Otsu N. Int J Bifurcat Chaos. 2007;17:1–19. [Google Scholar]
- 48.Friston KJ, Harrison L, Penny W. NeuroImage. 2003;19:1273–1302. doi: 10.1016/s1053-8119(03)00202-7. [DOI] [PubMed] [Google Scholar]
- 49.Mandeville JB, Marota JJ, Ayata C, Zararchuk G, Moskowitz MA, Rosen B, Weisskoff RM. J Cereb Blood Flow Metab. 1999;19:679–689. doi: 10.1097/00004647-199906000-00012. [DOI] [PubMed] [Google Scholar]
- 50.Buxton RB, Wong EC, Frank LR. Magn Reson Med. 1998;39:855–864. doi: 10.1002/mrm.1910390602. [DOI] [PubMed] [Google Scholar]
- 51.Van Essen DC, Dickson J, Harwell J, Hanlon D, Anderson CH, Drury HA. J Am Med Inform Assoc. 2001;41:1359–1378. doi: 10.1136/jamia.2001.0080443. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.