Abstract
Reelin, a large secreted protein implicated in the cortical development of the mammalian brain, is composed of eight tandem concatenations of “reelin repeats” and binds to neuronal receptors belonging to the low-density lipoprotein receptor gene family. We found that both receptor-binding and subsequent Dab1 phosphorylation occur solely in the segment spanning the fifth and sixth reelin repeats (R5–6). Monomeric fragment exhibited a suboptimal level of signaling activity and artificial oligomerization resulted in a 10-fold increase in activity, indicating the critical importance of higher-order multimerization in physiological reelin. A 2.0-Å crystal structure from the R5–6 fragment revealed not only a unique domain arrangement wherein two repeats were aligned side by side with the same orientation, but also the unexpected presence of bound Zn ions. Structure-guided alanine mutagenesis of R5–6 revealed that two Lys residues (Lys-2360 and Lys-2467) constitute a central binding site for the low-density lipoprotein receptor class A module in the receptor, indicating a strong similarity to the ligand recognition mode shared among the endocytic lipoprotein receptors.
Keywords: brain development, x-ray crystallography
Reelin is a large secreted glycoprotein that plays an important role in brain development (1, 2). It is produced by Cajal–Retzius as well as several other neuronal populations, acts on migrating neuronal precursors, and regulates correct cell positioning in the cortex and other brain structures. Reelin is also involved in modulation of synaptic plasticity (3) and is implicated in several brain disorders, including schizophrenia and Alzheimer's disease (4, 5). The response of migrating neurons requires that reelin binds to its receptors, apolipoprotein E receptor 2 (ApoER2) and very-low-density lipoprotein receptor (VLDLR). This binding is followed by tyrosine phosphorylation of cytoplasmic adapter disabled-1 (Dab1) by Src family tyrosine kinases (6, 7). The association of Dab1 with the cytoplasmic receptor tails then initiates a downstream signaling cascade (reviewed in ref. 8).
Reelin protein has a multidomain architecture consisting of a signal sequence, a region similar to F-spondin, another unique region containing an epitope for the CR-50 antibody, and eight tandem repeats consisting of 350–390 amino acid residues [supporting information (SI) Fig. 5A]. Each reelin repeat has an EGF motif at its center flanked by two subrepeats, A and B. We recently solved the crystal structure of the third reelin repeat (R3) at a resolution of 2.05 Å (9). It revealed a horseshoe-like subdomain arrangement in which the two subrepeats made direct contact despite intervention by an EGF motif. The molecular shape of a larger fragment (spanning the third to sixth reelin repeats, R3–6), as analyzed by three-dimensional electron microscopy, consisted of an elongated rod-like structure, suggesting tight packing between consecutive globular repeat domains (9).
The extracellular regions of reelin receptors also possess a multidomain architecture conserved in low-density lipoprotein receptor (LDLR) gene family proteins (SI Fig. 5B). This class of receptors mediates the cellular uptake of lipoproteins and other ligands via a clathrin-dependent endocytotic pathway (10). The N-terminal LDLR class A (LA) module repeat region is also known as a “ligand binding domain,” because it contains major lipoprotein binding activity in the prototypical LDLR (11). The more C-terminally located YWTD β-propeller domain serves as an intramolecular docking site for unliganded LA modules at a low pH (12). It is hypothesized that this pH-dependent intramolecular interaction constitutes the mechanism underlying ligand release in acidic endosomes (13).
Biochemical evidence suggests that the reelin binding activity also resides in the LA repeat regions of ApoER2 and VLDLR (14). As a result, reelin–receptor interaction can be inhibited by Apolipoprotein E (ApoE), a general ligand for LDLR family proteins (15). Furthermore, Andersen et al. (16) have demonstrated that acidic amino acid residues in the first and the third LA modules of ApoER2 play important roles in reelin binding. In contrast to ApoE or other general ligands for LDLR family members, however, reelin is not known to associate with lipids, is recognized by a specific class of receptors only (i.e., ApoER2 and VLDLR), and serves as a signaling molecule rather than an endocytotic cargo. All of this points to a uniqueness of reelin–receptor interaction and subsequent postreceptor events (8).
In contrast to the wealth of genetic and phenotypic information regarding reelin function in brain development, the molecular and mechanistic aspects of reelin recognition by its receptors and its signal transmissions to neurons remains only partially understood. Because of its strong function-blocking abilities, the anti-reelin monoclonal antibody CR-50 developed by Ogawa and coworkers proved very useful in dissecting the biochemical functions of reelin (17, 18). It recognizes a conformation-dependent epitope located in the N-terminal region of reelin (residues 251–407) (15). Despite the inhibition of reelin binding to its receptors by CR-50 (6), simple masking of the receptor-binding site by this antibody is unlikely because a mutant reelin lacking the CR-50 epitope region can associate with cell surface receptors without inducing Dab1 phosphorylation (19). This suggests the possibility that receptor binding and signaling (i.e., Dab1 phosphorylation) activities may occur in different regions of the reelin protein. However, a central fragment devoid of CR-50 epitope (fragment R3–6) was recently shown capable of binding to receptors, of inducing Dab1 phosphorylation in neurons, and (in a slice culture assay) of rescuing the reeler phenotype, suggesting that this region may contain all of the information necessary for reelin's biological activity (20). Resolving this discrepancy requires assays using active reelin proteins with defined physicochemical properties, which have thus far remained unavailable.
In this study, we have characterized the interaction between reelin and its receptor ApoER2 in detail. The crystal structure of the minimum receptor-binding reelin fragment, in combination with a series of mutagenesis, identified amino acid residues involved in the ligand–receptor interaction, which are conserved among vertebrates but not in other animals.
Results
Determination of the Receptor-Binding Unit Within Reelin.
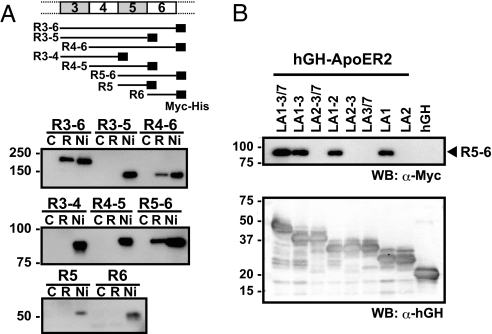
To further narrow down the receptor-binding site within the region corresponding to the third through sixth reelin repeats (R3–6) (20), we first prepared a series of constructs containing various numbers of repeats (Fig. 1A). All fragments containing both the fifth and sixth reelin repeats (R3–6, R4–6, and R5–6) bound to hGH-ApoER2, whereas no binding was observed in those fragments lacking the sixth reelin repeats (R3–5, R3–4, and R4–5) (Fig. 1A). Furthermore, fragments comprised of a single repeat (i.e., R5 and R6) did not bind to the receptor. It is possible, however, that the low expression levels of these single-domain fragments precluded detection of the binding. Mutated two-domain fragments in which R5 or R6 was replaced by R3 (R3R6 and R5R3), or even mutually swapped (R6R5), also did not exhibit any binding activity (SI Fig. 6B). This strongly suggests that the consecutive presence of fifth and sixth reelin repeats is required for receptor-binding activity. The same R5–6 region was shown to be essential for recognition by the reelin receptor VLDLR (SI Fig. 7B). By employing a similar truncation strategy, we next narrowed down the reelin-binding site within the receptor ectodomain. We first confirmed that the LA module region of ApoER2, but not the YWTD β-propeller region, was capable of binding reelin (SI Fig. 8). We then determined that the minimum segment required for reelin recognition by the ApoER2 ectodomain corresponded to the first LA module segment (Fig. 1B).
Fig. 1.
Determination of minimum regions essential for reelin–receptor interaction. (A) A series of deletion constructs (shown schematically at the top) of reelin containing a C-terminal tag (black rectangle) were expressed in 293T cells and subjected to a solid-phase binding assay, using hGH-ApoER2 fusion protein. Culture supernatants were incubated with receptor-immobilized (lane R) or unmodified (lane C) beads and bound proteins were analyzed by Western blot analysis, using anti-Myc antibody. Ni-NTA agarose pull-down (lane Ni) was used to estimate the amounts of total recombinant fragments present in the medium. (B) LA module fragments of ApoER2 extracellular domain were expressed as hGH fusion protein, captured on anti-hGH antibody beads, and incubated with culture supernatants containing the R5–6 fragment. Bound R5–6 fragments were detected by anti-Myc antibody (Upper). The same membranes were reprobed with anti-hGH antisera to compare the expression levels of the different constructs (Lower).
Biological Activity of R5–6.
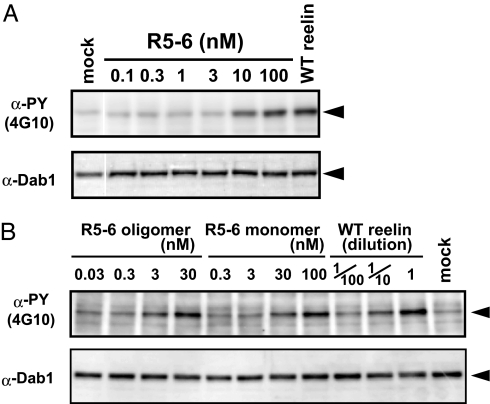
Previous studies have demonstrated that recombinant full-length reelin protein expressed in mammalian cells binds to cell surface receptors and induces tyrosine phosphorylation of Dab1 (21). However, quantitative assessments of such activity have proven difficult, because highly purified and stable reelin samples were not available. We therefore purified an R5–6 fragment from the culture supernatant and tested its ability to activate signaling pathways in cultured neurons. As shown in Fig. 2A, the addition of a purified reelin R5–6 fragment to neurons induced the tyrosine phosphorylation of Dab1 in a dose-dependent manner, confirming that this fragment is capable of triggering the biological signal. R5–6 also induced Dab1 degradation after prolonged incubation, another hallmark of reelin signaling (22, 23) (SI Fig. 9A). In addition, internalization of R5–6 was observed in neurons (SI Fig. 9B), further confirming that the R5–6 is biologically active. Despite successfully emulating the biological activity of full-length reelin, the R5–6 fragment proved much less effective than its full-length counterpart in transducing signals; indeed, it required ≈500 times higher concentration to achieve a Dab1 phosphorylation level comparable with that induced by WT reelin, based on the estimated concentration of WT reelin in the culture supernatant. By using size-exclusion chromatography and analytical ultracentrifugation, we confirmed that R5–6 is monomeric in solution (SI Fig. 10). To address the possibility that the low activity of the R5–6 fragment stemmed from its monomeric state in solution, we next tested the activity of the artificially oligomerized R5–6 fragment. To this end, the R5–6 fragment was first site-specifically biotinylated by using a free cysteine (i.e., Cys-2101) and then incubated with streptavidin, which is a tetrameric protein (24). The resulting oligomers (up to tetramer-sized) were separated from the monomer by gel-filtration chromatography and concentrated (SI Fig. 11, hatched area) and subjected to the Dab1 phosphorylation assay. As shown in Fig. 2B, the R5–6 oligomer showed a marked increase of activity over that of the biotinylated monomer. Densitometric analysis of the blots revealed that the oligomer possessed at least 10 times higher activity than the monomer, based on the molar concentration with respect to monomers. Quantitative comparison with the WT reelin is difficult, however, because WT reelin samples are constituted from unpurified culture supernatants. Indirect quantification of the WT reelin by Western blot analysis, using a WT reelin-alkaline phosphatase fusion protein as a standard, revealed that the supernatant of the transiently transfected cells contained roughly 300–1,000 pM reelin (M.H., unpublished data). Therefore, our data indicate that there still remains an activity gap of 30- to 100-fold between the R5–6 oligomer and the WT reelin multimer. Nevertheless, our results indicate that the low activity of the R5–6 fragment is partly due to its monovalency, and that homo-oligomerization greatly enhances signaling activity.
Fig. 2.
Dab1 phosphorylation activity. (A) Mouse cortical neurons were incubated for 20 min with the purified R5–6 fragment at the indicated concentration or with culture supernatant from the cells transfected with (WT) or without (mock) full-length reelin. Phosphorylated and total Dab1 were detected by Western blot analysis, using 4G10 and anti-Dab1, respectively. (B) The biotinylated R5–6 fragment was oligomerized by streptavidin (see SI Fig. 11) and tested for Dab1 phosphorylation activity, using cultured neurons. The concentrations of R5–6 fragments are shown in nM with respect to monomers, using a molecular mass value of 85,000; the concentrations of WT reelin are expressed as the dilution fold of the culture supernatant.
Crystal Structure of R5–6.
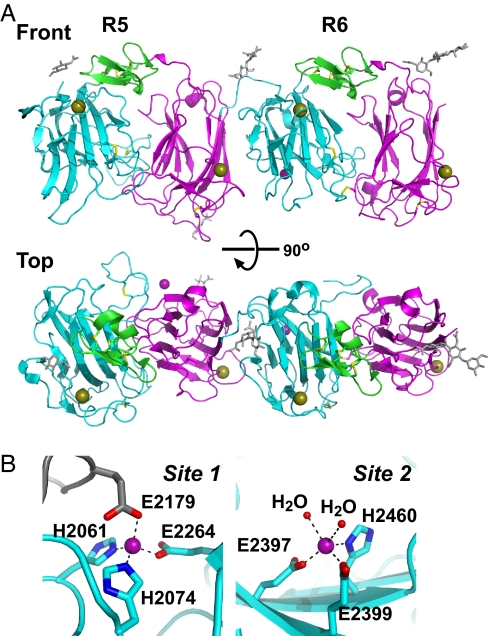
To gain insight into the molecular architecture of the reelin segment that mediates reelin–receptor interaction at the atomic level, we determined the crystal structure of R5–6 at a resolution of 2.0 Å (SI Fig. 12 and SI Table 1). The crystal contained one R5–6 molecule per asymmetric unit, and the final model contained residues 1956–2423 and 2427–2663 of the R5–6 protein and four N-glycan chains. The overall structure of each repeat was practically identical to that of the previously determined R3, in that the three subdomains (subrepeat A, EGF, and subrepeat B) were arranged in a horseshoe-like manner to form a compact globular structure (Fig. 3A). Both R5 and R6 contained one Ca ion bound per subrepeat in the position equivalent to the Ca2+ site found in the subrepeat B of R3 (9), suggesting that Ca binding was the common denominator in reelin subrepeats.
Fig. 3.
Crystal structure of R5–6. (A) Overall structure of R5–6 shown in a ribbon model, and viewed from two different angles. Subdomains in each reelin repeat are differently colored: subrepeat A (cyan), EGF (green), and subrepeat B (magenta). Ca and Zn ions are shown as gold and purple spheres, respectively. Disulfide bonds (yellow) and four N-glycan chains (gray) are shown in stick models. All structural images were prepared with PyMOL (http://pymol.sourceforge.net). (B) Zn-binding sites. Close-up view of site 1 (Left) and site 2 (Right) are shown. Both Zn2+ (purple spheres) were coordinated by His and Glu side chains of an R5–6 molecule (cyan). In site 1, a Glu side chain from the symmetry mate molecule (gray) completed the coordination, whereas in site 2, two water molecules (red spheres) were bound.
The most striking feature of the R5–6 structure is the arrangement of the two reelin repeats. As shown in Fig. 3A and in SI Fig. 13, R5 and R6 are arranged side by side, connected by a short linker segment, with no space between them. The relative positioning of the two repeats is very unusual in that the two domains are related by an almost perfect translational movement, with no bends or twists occurring at the junction. The interface between the two repeat domains contains hydrophobic contacts and buries a total 1,480-Å2 solvent-accessible surface area, suggesting that it is relatively stable. This unique repeat arrangement is consistent with the flattened, rod-like structure of the R3–6 fragment determined by electron microscopy (9) and lends strong support to the contention that reelin repeats are packed tightly in a tandem fashion within the R3–6 segment.
Another unexpected feature of the R5–6 structure was the presence of bound Zn ions. Two Zn ions were clearly identified on the molecular surface, coordinated in a tetrahedral or a pentahedral geometry by His and Glu residues. The first Zn ion (referred to as site 1), which was located at a crystal packing interface involving R5, was liganded by His-2061, His-2074, and Glu-2264 from one molecule and Glu-2179 from the packing mate (Fig. 3B Left). Ligands for the second Zn-binding site (site 2) in R6 included Glu-2397, Glu-2399, His-2460, and two water molecules (Fig. 3B Right). The bond distances for the metal-ligand interactions were in reasonable agreement with the average values reported for Zn2+-binding proteins (SI Table 2). Because the buffer used during the crystallization and purification did not contain any Zn compounds, these Zn2+ clearly originated with the protein sample and probably bound during biosynthesis. The bridging of the two R5–6 molecules by the site-1 Zn2+ was likely caused by the high concentration of the fragment under the crystallization conditions, because the analytical ultracentrifugation data suggests that R5–6 was largely monomeric in solution (SI Fig. 10). We therefore concluded that reelin R5 and R6 both bind one Zn2+ by three side chains from specific His and Glu residues. Under the physiological condition, the fourth (and the fifth for site 2) ligand is most likely provided by water molecules. At this point, we do not know whether these newly found Zn ions play any role in reelin signaling. We mutated the Zn-coordinating His residues in the context of the full-length reelin and tested their activity. These mutants (H2060Y and H2460A) exhibited normal binding to ApoER2 and induced Dab1 phosphorylation in cultured neurons (SI Fig. 14), suggesting that the Zn ions are not essential for receptor binding.
Identification of Lys Residues Involved in Receptor Binding.
Using surface plasmon resonance analysis, Andersen et al. (16) demonstrated that partially purified reelin can bind to an ectodomain fragment of ApoER2 immobilized on a sensor chip. Curiously, using amino-coupling chemistry, and with the receptor in the solution phase, they failed to detect the same interaction when reelin protein was immobilized on the surface (16). We reasoned that this failure was due to the chemical modification of Lys residues, because biotinylation of amino groups on R5–6 also resulted in the reduction of ApoER2-binding ability (data not shown). In fact, when we immobilized the R5–6 fragment on the sensor chip, using a different type of chemistry (i.e., via a Cys residue), it efficiently mediated the concentration-dependent binding of ApoER2 ECΔ4–6 (SI Fig. 15). This observation strongly suggests that Lys residue(s) in the R5–6 segment plays a critical role in receptor binding.
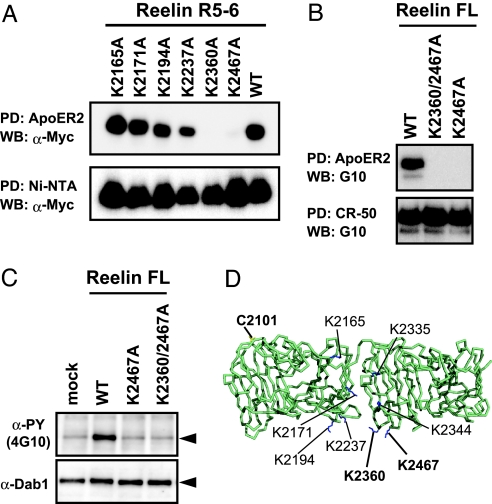
To determine which Lys residue is essential for recognition by receptors, we designed a series of Lys → Ala mutants of R5–6 fragment and tested their receptor-binding ability. There were 19 exposed Lys residues distributed rather evenly on the surface of the R5–6 fragment (SI Fig. 13). Based on the critical requirement of the consecutive R5-R6 segment for this activity, we assumed that the receptor recognized R5 and R6 simultaneously. We therefore mutated only the Lys residues located near the repeat boundary, namely, K2165, K2171, K2194, K2237, K2335, K2344, K2360, and K2467 (Fig. 4D).
Fig. 4.
Mapping the amino acid residues critical for receptor binding. (A) The binding of R5–6 mutants to hGH-ApoER2 EC was analyzed by a solid-phase binding assay. Receptor-bound (Upper) and Ni-captured (Lower) fragments were detected by Western blot analysis, using anti-Myc antibody. (B) Full-length reelin samples, with the indicated mutations, were tested for receptor-binding activity (Upper). CR-50 pull down confirmed that roughly equal amounts of reelin were present in the culture supernatants (Lower). (C) Dab1 phosphorylation activities of the full-length reelin mutants. (D) Lys residues mutated in this study and the single free cysteine residue are shown in blue and yellow stick models, respectively, on the Cα backbone of R5–6.
Of the eight Lys→Ala mutants tested for their expression in 293T cells, six were well secreted in the medium, suggesting that their overall fold is not severely affected by the mutation (Fig. 4A Lower). Two mutants (K2335A and K2344A) showed very low level of expression, and hence were omitted from further analysis. The receptor-binding activities of the expressed mutant R5–6 fragments were assessed by using a receptor pull-down assay. As shown in Fig. 4A, mutation of either K2360 or K2467 abolished ApoER2-binding activity, whereas each of the other mutants exhibited a binding activity comparable with that of WT R5–6. Furthermore, these residues were mutated in the context of full-length reelin and the resultant single (K2467A) or double (K2360A/K2467A) mutants were tested for their respective activity. As is clearly evident in Fig. 4 B and C, these mutants could not bind to the receptors nor could they induce Dab1 phosphorylation. In addition to ApoER2, we also confirmed that the full-length reelin with the K2467A mutation could not bind to VLDLR (SI Fig. 7C). We concluded that these two clustered Lys residues, particularly K2467, were critical for the biological activity of reelin.
Discussion
Reelin receptors belong to a class of membrane proteins called LDLR-related proteins. This protein family now includes nine members and bind many ligands including the protease-protease inhibitor complex, viruses, signaling molecules, and lipoproteins (25). Most of the known ligands of LDLR-related proteins bind to a structural module in the receptor ectodomain called the LA module. In many cases, basic residues are known to be involved in receptor binding. Conserved basic amino acid residues within ApoE amino acids 140–150 are crucial for interaction with LDLR, and a site-directed mutagenesis study has demonstrated that Lys-143 and Lys-146 are absolutely necessary (26). Two Lys residues in a receptor-binding domain of α2-macroglobulin have also been shown to be involved in binding to the LA module portion of LRP (27, 28). Recent structural studies have revealed the ligand-recognition mechanism of LA modules at an atomic level (12, 29, 30). The most striking feature of the ligand-recognition mode was that in all cases Lys residues on structurally unrelated and dissimilar ligands were recognized by the conserved Ca-coordinating acidic residues and an aromatic residue from the LA modules (29). The present study reveals that reelin also falls into this class of ligands in which limited numbers of Lys residues (K2360 and K2467) are specifically recognized by an LA module. These residues are present in repeat 6, and located at the “bottom tip” near the noncovalent interface with the preceding repeat 5. There are two possibilities regarding the role of repeat 5 in receptor binding: it may stabilize the folded conformation of repeat 6, thereby exerting an indirect role in receptor recognition; or it may provide direct contacts to LA1 (note that the two Lys residues are located adjacent to the repeat 5). This point awaits future clarification via structural determination of the reelin–receptor complex.
The critical involvement of Lys residues in reelin binding to the receptor strongly suggests that the recognition mode is shared by other LDLR family ligands. Some differences should also exist, however, because unlike RAP or ApoE, which generally recognizes LA-containing receptors, reelin can only interact with LA modules in ApoER2 and VLDLR. This specificity must be achieved by interactions involving non-Lys residues on the reelin side and specific residues in LA1 on the receptor side. It is also possible that residues outside LA1 contribute to binding. In fact, Andersen et al. (16) have reported that both LA1 and LA3 of ApoER2 are important for reelin recognition. When an LA1–4 structure model was aligned with the R5–6 structure such that the Lys-2467 pointed toward the “necklace” site in the extreme most N-terminal LA module, as described by Fisher et al. (29), the second and the third LA modules were in a position permitting direct interaction with repeat 5 (SI Fig. 16). We speculate that the LA3 (and possibly LA2) module may provide an additional binding affinity and specificity by interacting weakly with repeat 5, albeit not to an extent sufficient to support reelin binding by itself. Our results, which show (i) that the R5–6 segment is sufficient for the induction of Dab1 phosphorylation and (ii) that the Lys-mutant of the full-length reelin is devoid of receptor-binding and Dab1-phosphorylating activities, suggest that the R5–6 segment represents the major and most physiologically important receptor-binding region. However, the presence of additional functional sites outside the R5–6 region cannot be ruled out.
Determination of the crystal structure of R5–6 led to the identification of previously undescribed Zn2+ binding sites on the reelin surface. Sequence analysis alone cannot detect the presence of such sites, underscoring the power of direct structure determination. In the deposited structure database, there are many protein structures that have Zn ions bound on their surface, referred to as “interface Zn sites” (31). Unlike the protein-bound Zn ions that participate in the enzymatic activities of proteins or in cluster formations, many of these are introduced during crystallization, and hence their physiological relevance remains uncertain. In contrast, R5–6 is loaded with Zn ions upon biosynthesis, which may indicate the authenticity of such Zn2+ sites. Although we do not know whether natural full-length reelin is also loaded with Zn2+, there is no reason to speculate otherwise, considering that the brain has the highest Zn2+ content of all organs (32). Increasing evidence shows that Zn ions in neuronal tissues play important roles in both physiological and pathological settings (33). Because we could not observe any effects on either the receptor-binding or signaling activities of reelin by mutating Zn-coordinating His residues, the physiological relevance of Zn2+ sites in reelin remains unclear. However, it is interesting to note that reelin protein has been implicated in neurological disorders such as schizophrenia and Alzheimer's disease, in which Zn ions are also suspected of playing a role (4, 5).
In the present study, we have clearly shown that oligomer formation greatly enhanced the signaling capability of the receptor-binding fragment. Because CR-50 inhibits reelin oligomerization by binding to its N-terminal region (34), we believe that the inhibition by CR-50 mainly stems from the blockade of reelin multimerization required for efficient signaling. Nonetheless, we cannot exclude the possibility of long-range steric hindrance. Ligand oligomerization can affect the signaling activity by two different mechanisms: by receptor clustering or by an avidity effect. Strasser et al. (35) have postulated that receptor clustering is the major mechanism underlying reelin signaling. However, our results that monomeric R5–6 can trigger Dab1 phosphorylation, albeit weakly, argues against the notion that ligand multimerization is prerequisite to signaling activity. In general, ligand multimerization can result in an increase (of several orders) in the apparent affinity toward receptors (36, 37). In fact, such a disparity in affinity may partly be responsible for the apparent lack of reelin signaling in the presence of CR-50. Therefore, it is likely that both of these mechanisms contribute to the efficient signaling capacity of WT reelin. In addition, there seems to be more than receptor clustering at work in providing the necessary cellular pathway for neuronal activation by reelin.
Recently sequenced genomes for ascidians (Ciona intestinalis) and lancelet (Branchiostoma floridae) revealed that these chordate species have reelin in their genome. In both organisms, the putative reelin protein displays a typical domain arrangement, including eight reelin repeats. In both cases, however, the C-terminal basic peptide region is missing. Reelin is also found in the genome of sea urchins (38), indicating that the emergence of reelin predates that of chordates. Interestingly, both of the critical Lys residues are completely conserved among all vertebrate species but are replaced by unrelated amino acids in those lower organisms that lack layered architecture in the primitive brain or the brain itself (SI Fig. 17). This fact strongly suggests that the evolutional acquisition of receptor-binding ability by reelin has played a pivotal role in developing a complex brain structure.
The present study contributes to a better understanding of the molecular mechanism implicated in the reelin signaling pathway at an atomic level, particularly in its initial phase involving ligand–receptor interaction. Because the current methodology for studying reelin signaling is limited, however, our understanding about the signaling mechanism beyond Dab1 phosphorylation remains poor at best. In fact, Dab1 is not a reelin pathway-specific signaling molecule but is rather thought to be a general facilitator of the receptor trafficking targeted at cytoplasmic NPXY motifs (39). It is possible that Dab1 exerts its effect by controlling receptor trafficking (and hence, reelin binding/internalization kinetics), in addition to acting as a classical “adapter molecule” that passes on the signal to the next cytosolic molecule such as Nckβ and phosphatidylinositol-3 kinase (8). Therefore, identification of a reelin-specific postreceptor event(s) seems crucial for a mechanistic understanding of the reelin pathway. In light of the analogy to the LDL receptor pathway, it may be worthwhile to explore the fate of internalized reelin. It is tempting to speculate on the possible involvement of Zn2+ homeostasis in brain development, particularly in relation to the newly found Zn-binding property of reelin reported in this study.
Materials and Methods
Protein Expression and Purification.
For transient transfection, 293T cells were transfected with 1 μg of expression plasmid DNAs (SI Methods), using Fugene 6 (Roche, Indianapolis, IN). For stable expression, CHO lec 3.2.8.1 cells (provided by P. Stanley, Albert Einstein College of Medicine, Bronx, NY) (40) were transfected with plasmids encoding reelin R5–6 with a Myc tag at the C terminus (R5–6-Myc). This was accomplished by electroporation, plated on 96-well plates and selected for resistance against 1.5 mg/ml G418 (Gibco, Carlsbad, CA). The clone with the highest secreted levels of the reelin R5–6 fragment was cultured in roller bottles (Corning Glassworks, Corning, NY). The R5–6 fragment was purified from culture supernatants by ammonium sulfate precipitation and Ni-NTA agarose chromatography and then further purified by gel-filtration chromatography with a Superdex 200 GL column. For stable expression of ApoER2 ECΔ4-6, 293S-GnT1− cells (provided by H. G. Khorana, Massachusetts Institute of Technology, Cambridge, MA) (41) were transfected with plasmid encoding hGH-ApoER2 ECΔ4–6, plated on 96-well plates, and selected for resistance against 0.5 μg/ml puromycin. The hGH-ApoER2 ECΔ4–6 fragment was purified from culture supernatants by ammonium sulfate precipitation and Ni-NTA agarose chromatography and treated with TEV protease to remove the hGH portion. The cleaved ApoER2 ECΔ4–6 fragment was purified by gel-filtration chromatography on a Superdex 200 GL column.
Crystallization, Data Collection, and Structural Determination.
For crystallization of R5–6, a tag at the C terminus was cleaved by overnight incubation with TEV protease at room temperature. Untagged protein was purified by using gel filtration and concentrated to 15 mg/ml. Initial crystallization conditions were determined by using a Topaz 1.96 chip (Fluidigm, South San Francisco, CA). Protein solution (3 μl) was screened against the Index screening kit (Hampton Research, Aliso Viejo, CA). Diffraction-quality crystals were obtained through vapor-diffusion in hanging drops containing equal volumes of protein and the well solution containing 4–7% PEG 3350, 75–140 mM ammonium acetate, and 100 mM Hepes (pH 7.0).
Before data collection, crystals were cryoprotected with Paratone-N (Hampton Research) and flash-frozen in liquid nitrogen. Diffraction data for structural determination were collected at SPring-8 BL41-XU. Data sets for assignment of the Zn ions were collected at PF BL-5 (see SI Methods). All data sets were processed with HKL2000 (42). Initial phases were determined by the molecular replacement method. The atomic coordinates of subrepeat B (R3B) were extracted from the crystal structure of R3 [Protein Data Bank (PDB) code 2DDU] and used as the search model. The program MOLREP (43) located four R3B molecules in the asymmetric unit, which corresponded to the positions of R5A, R5B, R6A, and R6B. The solution was subjected to phase improvement and automated model-building with ARP/wARP software (44). The resulting model was corrected and fit into the electron density, using O software (45), followed by translation/libration/screw (TLS) and restraint refinement with REFMAC5 (46). Several rounds of refinement resulted in an R-factor of 17.9% and a free R factor of 21.9%. Refinement statistics are summarized in SI Table 1.
Solid-Phase Binding Assay.
A solid-phase binding assay was carried out as described in ref. 9. Briefly, a monoclonal antibody against hGH (HGH-B; American Type Culture Collection, Manassas, VA) was immobilized onto CNBr-activated-Sepharose 4B (Amersham Pharmacia Biotech, Little Chalfont, U.K.) according to the manufacturer's protocol. Various extracellular regions of human ApoER2 fused to the C terminus of hGH were transiently expressed in 293T cells. Cell culture supernatant containing hGH fusion proteins was mixed with HGH-B immobilized Sepharose and incubated at 4°C for 1 h. Receptor-captured beads were briefly washed and incubated with cell culture supernatants containing reelin fragments at 4°C for 2 h. After washing with 20 mM Tris buffer (pH 7.5), containing 150 mM NaCl and 2 mM CaCl2, proteins retained on beads were eluted and separated on SDS/PAGE, and transferred to a PVDF membrane. The PVDF membranes were blocked with 5% BSA in 20 mM Tris (pH 8.0), 150 mM NaCl, and 0.05% Tween20 and then probed with anti-Myc antibody.
Dab1 Phosphorylation Assay.
A Dab1 phosphorylation assay was performed as described in refs. 9 and 39. Briefly, cortical neurons were obtained from an embryonic day 15 ICR mouse and cultured for 3–4 days. Recombinant proteins in serum-free Opti-MEM (Invitrogen, Carlsbad, CA) or culture supernatant from the cells transfected with full-length reelin were added to the neurons and incubated for 20 min at 37°C. Total cell lysate was prepared in SDS/PAGE sample buffer and separated by 7.5% SDS/PAGE. Proteins were transferred to a PVDF membrane and probed with anti-phosphotyrosine antibody 4G10 (Upstate Biotechnology, Lake Placid, NY) or with anti-Dab1 (Chemicon, Temecula, CA). Immunoprecipitation of Dab1 before the blotting with 4G10 produced essentially the same results.
Supplementary Material
Acknowledgments
We thank Drs. N. Igarashi, N. Matsugaki, and Y. Yamada of Photon Factory and Drs. M. Kawamoto and N. Shimizu of SPring-8 BL-41XU for their help with x-ray data collection; Keiko Sudou, Keiko Tamura-Kawakami, and Emiko Mihara for their excellent technical support; Mayumi Morimoto for preparation of the manuscript, Miyo Sakai for performing analytical ultracentrifugation; and Dr. Fumio Arisaka for help in interpreting the sedimentation velocity data. This work was partly supported by the Grant-in-Aid for Scientific Research (A) from the Ministry of Education, Culture, Sports, Science and Technology of Japan (MEXT), by the Grant-in-Aid for Scientific Research on Priority Areas from MEXT, and by the Protein 3000 Project grant from MEXT.
Abbreviations
- LDLR
low-density lipoprotein receptor
- ApoE
apolipoprotein E
- ApoER2
ApoE receptor 2
- Dab1
disabled-1
- LA
LDLR class A
- VLDLR
very LDLR.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structural factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2E26).
This article contains supporting information online at www.pnas.org/cgi/content/full/0700438104/DC1.
References
- 1.D'Arcangelo G, Miao GG, Chen SC, Soares HD, Morgan JI, Curran T. Nature. 1995;374:719–723. doi: 10.1038/374719a0. [DOI] [PubMed] [Google Scholar]
- 2.Tissir F, Goffinet AM. Nat Rev Neurosci. 2003;4:496–505. doi: 10.1038/nrn1113. [DOI] [PubMed] [Google Scholar]
- 3.Herz J, Chen Y. Nat Rev Neurosci. 2006;7:850–859. doi: 10.1038/nrn2009. [DOI] [PubMed] [Google Scholar]
- 4.Fatemi SH. Int Rev Neurobiol. 2005;71:179–187. doi: 10.1016/s0074-7742(05)71008-4. [DOI] [PubMed] [Google Scholar]
- 5.Botella-Lopez A, Burgaya F, Gavin R, Garcia-Ayllon MS, Gomez-Tortosa E, Pena-Casanova J, Urena JM, Del Rio JA, Blesa R, Soriano E, et al. Proc Natl Acad Sci USA. 2006;103:5573–5578. doi: 10.1073/pnas.0601279103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.D'Arcangelo G, Homayouni R, Keshvara L, Rice DS, Sheldon M, Curran T. Neuron. 1999;24:471–479. doi: 10.1016/s0896-6273(00)80860-0. [DOI] [PubMed] [Google Scholar]
- 7.Hiesberger T, Trommsdorff M, Howell BW, Goffinet A, Mumby MC, Cooper JA, Herz J. Neuron. 1999;24:481–489. doi: 10.1016/s0896-6273(00)80861-2. [DOI] [PubMed] [Google Scholar]
- 8.Stolt PC, Bock HH. Cell Signal. 2006;18:1560–1571. doi: 10.1016/j.cellsig.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 9.Nogi T, Yasui N, Hattori M, Iwasaki K, Takagi J. Embo J. 2006;25:3675–3683. doi: 10.1038/sj.emboj.7601240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeon H, Blacklow SC. Annu Rev Biochem. 2005;74:535–562. doi: 10.1146/annurev.biochem.74.082803.133354. [DOI] [PubMed] [Google Scholar]
- 11.Strickland DK, Gonias SL, Argraves WS. Trends Endocrinol Metab. 2002;13:66–74. doi: 10.1016/s1043-2760(01)00526-4. [DOI] [PubMed] [Google Scholar]
- 12.Rudenko G, Henry L, Henderson K, Ichtchenko K, Brown MS, Goldstein JL, Deisenhofer J. Science. 2002;298:2353–2358. doi: 10.1126/science.1078124. [DOI] [PubMed] [Google Scholar]
- 13.Beglova N, Jeon H, Fisher C, Blacklow SC. Mol Cell. 2004;16:281–292. doi: 10.1016/j.molcel.2004.09.038. [DOI] [PubMed] [Google Scholar]
- 14.Koch S, Strasser V, Hauser C, Fasching D, Brandes C, Bajari TM, Schneider WJ, Nimpf J. Embo J. 2002;21:5996–6004. doi: 10.1093/emboj/cdf599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.D'Arcangelo G, Nakajima K, Miyata T, Ogawa M, Mikoshiba K, Curran T. J Neurosci. 1997;17:23–31. doi: 10.1523/JNEUROSCI.17-01-00023.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andersen OM, Benhayon D, Curran T, Willnow TE. Biochemistry. 2003;42:9355–9364. doi: 10.1021/bi034475p. [DOI] [PubMed] [Google Scholar]
- 17.Nakajima K, Mikoshiba K, Miyata T, Kudo C, Ogawa M. Proc Natl Acad Sci USA. 1997;94:8196–8201. doi: 10.1073/pnas.94.15.8196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ogawa M, Miyata T, Nakajima K, Yagyu K, Seike M, Ikenaka K, Yamamoto H, Mikoshiba K. Neuron. 1995;14:899–912. doi: 10.1016/0896-6273(95)90329-1. [DOI] [PubMed] [Google Scholar]
- 19.Kubo K, Mikoshiba K, Nakajima K. Neurosci Res. 2002;43:381–388. doi: 10.1016/s0168-0102(02)00068-8. [DOI] [PubMed] [Google Scholar]
- 20.Jossin Y, Ignatova N, Hiesberger T, Herz J, Lambert de Rouvroit C, Goffinet AM. J Neurosci. 2004;24:514–521. doi: 10.1523/JNEUROSCI.3408-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Benhayon D, Magdaleno S, Curran T. Brain Res Mol Brain Res. 2003;112:33–45. doi: 10.1016/s0169-328x(03)00032-9. [DOI] [PubMed] [Google Scholar]
- 22.Bock HH, Jossin Y, May P, Bergner O, Herz J. J Biol Chem. 2004;279:33471–33479. doi: 10.1074/jbc.M401770200. [DOI] [PubMed] [Google Scholar]
- 23.Arnaud L, Ballif BA, Cooper JA. Mol Cell Biol. 2003;23:9293–9302. doi: 10.1128/MCB.23.24.9293-9302.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chaiet L, Wolf FJ. Arch Biochem Biophys. 1964;106:1–5. doi: 10.1016/0003-9861(64)90150-x. [DOI] [PubMed] [Google Scholar]
- 25.Herz J, Bock HH. Annu Rev Biochem. 2002;71:405–434. doi: 10.1146/annurev.biochem.71.110601.135342. [DOI] [PubMed] [Google Scholar]
- 26.Zaiou M, Arnold KS, Newhouse YM, Innerarity TL, Weisgraber KH, Segall ML, Phillips MC, Lund-Katz S. J Lipid Res. 2000;41:1087–1095. [PubMed] [Google Scholar]
- 27.Dolmer K, Gettins PG. J Biol Chem. 2006;281:34189–34196. doi: 10.1074/jbc.M604389200. [DOI] [PubMed] [Google Scholar]
- 28.Nielsen KL, Holtet TL, Etzerodt M, Moestrup SK, Gliemann J, Sottrup-Jensen L, Thogersen HC. J Biol Chem. 1996;271:12909–12912. doi: 10.1074/jbc.271.22.12909. [DOI] [PubMed] [Google Scholar]
- 29.Fisher C, Beglova N, Blacklow SC. Mol Cell. 2006;22:277–283. doi: 10.1016/j.molcel.2006.02.021. [DOI] [PubMed] [Google Scholar]
- 30.Verdaguer N, Fita I, Reithmayer M, Moser R, Blaas D. Nat Struct Mol Biol. 2004;11:429–434. doi: 10.1038/nsmb753. [DOI] [PubMed] [Google Scholar]
- 31.Maret W. In: Handbook of metalloproteins. Messerschmidt A, Bode W, Cygler M, editors. Chichester, UK: Wiley; 2004. pp. 432–441. [Google Scholar]
- 32.Weiss JH, Sensi SL, Koh JY. Trends Pharmacol Sci. 2000;21:395–401. doi: 10.1016/s0165-6147(00)01541-8. [DOI] [PubMed] [Google Scholar]
- 33.Frederickson CJ, Koh JY, Bush AI. Nat Rev Neurosci. 2005;6:449–462. doi: 10.1038/nrn1671. [DOI] [PubMed] [Google Scholar]
- 34.Utsunomiya-Tate N, Kubo K, Tate S, Kainosho M, Katayama E, Nakajima K, Mikoshiba K. Proc Natl Acad Sci USA. 2000;97:9729–9734. doi: 10.1073/pnas.160272497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Strasser V, Fasching D, Hauser C, Mayer H, Bock HH, Hiesberger T, Herz J, Weeber EJ, Sweatt JD, Pramatarova A, et al. Mol Cell Biol. 2004;24:1378–1386. doi: 10.1128/MCB.24.3.1378-1386.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carman CV, Springer TA. Curr Opin Cell Biol. 2003;15:547–556. doi: 10.1016/j.ceb.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 37.Mammen M, Choi S-K, Whitesides GM. Angew Chem Int Ed. 1998;37:2754–2794. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2754::AID-ANIE2754>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 38.Sodergren E, Weinstock GM, Davidson EH, Cameron RA, Gibbs RA, Angerer RC, Angerer LM, Arnone MI, Burgess DR, Burke RD, et al. Science. 2006;314:941–952. doi: 10.1126/science.1133609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morimura T, Hattori M, Ogawa M, Mikoshiba K. J Biol Chem. 2005;280:16901–16908. doi: 10.1074/jbc.M409048200. [DOI] [PubMed] [Google Scholar]
- 40.Stanley P. Mol Cell Biol. 1989;9:377–383. doi: 10.1128/mcb.9.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reeves PJ, Callewaert N, Contreras R, Khorana HG. Proc Natl Acad Sci USA. 2002;99:13419–13424. doi: 10.1073/pnas.212519299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 43.Vagin A, Teplyakov A. J Appl Cryst. 1997;30:1022–1025. [Google Scholar]
- 44.Perrakis A, Harkiolaki M, Wilson KS, Lamzin VS. Acta Crystallogr D Biol Crystallogr. 2001;57:1445–1450. doi: 10.1107/s0907444901014007. [DOI] [PubMed] [Google Scholar]
- 45.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 46.Murshudov GN, Vagin AA, Dodson EJ. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.