Abstract
Natural killer (NK) cells express activating and inhibitory receptors that, in concert, survey cells for proper expression of cell surface major histocompatibility complex (MHC) class I molecules. The mouse cytomegalovirus encodes an MHC-like protein, m157, which is the only known viral antigen to date capable of engaging both activating (Ly49H) and inhibitory (Ly49I) NK cell receptors. We have determined the 3D structure of m157 and studied its biochemical and cellular interactions with the Ly49H and Ly49I receptors. m157 has a characteristic MHC-fold, yet possesses several unique structural features not found in other MHC class I-like molecules. m157 does not bind peptides or other small ligands, nor does it associate with β2-microglobulin. Instead, m157 engages in extensive intra- and intermolecular interactions within and between its domains to generate a compact minimal MHC-like molecule. m157's binding affinity for Ly49I (Kd ≈ 0.2 μM) is significantly higher than that of classical inhibitory Ly49–MHC interactions. Analysis of viral escape mutations on m157 that render it resistant to NK killing reveals that it is likely to be recognized by Ly49H in a binding mode that differs from Ly49/MHC-I. In addition, Ly49H+ NK cells can efficiently lyse RMA cells expressing m157, despite the presence of native MHC class I. Collectively, our results show that m157 represents a structurally divergent form of MHC class I-like proteins that directly engage Ly49 receptors with appreciable affinity in a noncanonical fashion.
Keywords: immune system, molecular recognition
Mouse strains differ in their susceptibility to infection by mouse cytomegalovirus (MCMV), and protection is mediated by natural killer (NK) cells (1). NK cell-mediated resistance to MCMV infection was mapped to a genetic locus on mouse chromosome 6 in a region named the “natural killer gene complex” (1). This region encodes both inhibitory and activating lectin-like Ly49 receptors that, in concert, regulate NK cell activity through recognition of major histocompatibility complex (MHC) class I ligands. In C57BL/6 mice, the receptor encoded by the Ly49H gene (Ly49H) is responsible for controlling MCMV replication in visceral organs (2–4). BALB/c mice do not possess a Ly49H gene, and consequently they restrict MCMV replication; however, transgenic expression of a genomic Ly49H gene in these mice confers resistance to MCMV replication (5). Subsequent studies demonstrated that Ly49H specifically recognizes the m157 glycoprotein encoded by MCMV (6, 7). m157 is expressed on the surface of MCMV-infected cells and anchored through a phosphatidylinositol glycan linker (6, 8). In addition to Ly49H, m157 also binds to at least one inhibitory receptor, Ly49I129/J, expressed in the MCMV-susceptible mouse strain 129/J (6). Structural prediction indicated that m157 is structurally similar to that of an MHC class I molecule (6, 7), despite lacking appreciable sequence similarity.
Class I MHC proteins are membrane-bound heterodimeric proteins consisting of a polymorphic heavy chain noncovalently associated with a conserved, monomorphic β2-microglobulin (β2m) light chain (9). The α1 and α2 domains of the heavy chain are collectively referred to as the “platform” region, composed of two parallel α helices situated on top of a β-sheet foundation. This platform is further supported by the heavy chain's α3 domain and the β2m subunit. It is the groove formed by these two α helices and the β-sheet floor that binds peptide and is recognized by T cell receptors (10). Peptides from foreign sources, such as those produced by viruses during infection, elicit responses from CD8+ T cells and prime the adaptive immune response to prevent further viral spread. Therefore, down-regulating MHC class I expression is one mechanism by which viruses evade T cell-mediated immune detection.
Several MCMV ORFs also encode MHC homologues based on sequence analysis (6, 7), and a crystal structure of one of these ORFs, m144, has confirmed the strong structural similarity to class I MHC (11). The m144 structure revealed many interesting features, such as truncated α1 and α2 helices, a narrowed interhelical groove, and altered β2M interactions. However, m157 is the only MCMV MHC homologue to date whose function has been clarified and cognate host receptors identified. In addition, m157 is not dependent on β2m for expression (6, 7), suggesting an altered 3D architecture from that of m144. Therefore, structural information on m157 will give us deeper insight into how differences we see in viral MHC homolog structures, such as m157 and m144, impact ligand recognition and function. As a first step in elucidating the structural principles underlying Ly49 engagement of m157, we have determined the 3D structure of the ectodomain of m157 to explore its structural relationship with class I MHC molecules, and we have also studied its interaction with the activating and inhibitory NK cell receptors, Ly49H and Ly49I129/J, respectively.
Results
Overall Structure.
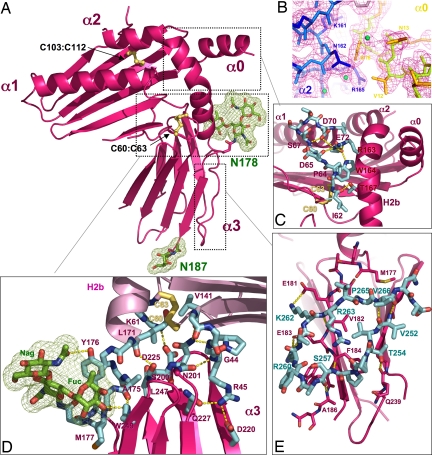
The overall structure of m157 at 2.1 Å [supporting information (SI) Table 1] is reminiscent of the MHC class I-fold comprised of top-mounted α helices supported by a β-sheet platform, which in turn is supported by an Ig α3 domain (12) (Figs. 1A and 2A). m157 is substantially more compact than a classical MHC class I molecule, with dimensions of ≈35 Å × 45 Å × 59 Å compared with 41 Å × 52 Å × 69 Å for a standard MHC class I protein. In the platform region, the α1 and α2 helices are notably shorter and closer together than those found in classical MHC class I molecules that bind peptides (Figs. 1C and 2A Lower). A major deviation from any known MHC-like protein structure is that in m157, because of the addition of a third, top-mounted helix (α0) at its N terminus, there are three, rather than two, “rows” of helices on the platform (Figs. 1 A and C and 2A) (the N-terminal portion of the α2 helix is disordered in our model). Underneath the helices, m157 uses six antiparallel strands to construct the β-sheet floor of the platform, rather than the canonical eight seen in MHC class I (12) and compared with seven found in m144 (11). m157 does not associate with the β2m subunit, which is required for proper stability and expression of nearly all MHC class I-related molecules. There is an extensive network of interactions at the platform–α3 domain interface of m157 that prevents β2m from binding (Fig. 1D).
Fig. 1.
The 3D structure of m157. (A) Ribbon diagram of m157 showing an overall MHC class I-like structural architecture. Disulphide bonds are shown as yellow sticks, and two glycosylation sites are shown in green. (B) The experimental electron density map contoured at 1.5 ó, showing the interactions between the N-terminal α0 helix and the H2b segment of the α2 helix. (C) A network of side- and main-chain (shown in cyan) hydrogen-bonding interactions exists between the α1 and α2 domains. (D) The hinge region between the platform has extensive contacts between the α3 domain loops and the underside of the α1/α2 platform. Additional contacts are contributed by the N-linked glycosylation located at Asn-178 (shown in stick and mesh representation; green). (E) A unique C-terminal extension forms a circular loop that mediates contacts between the two β-sheets of the α3 domain's Ig sandwich (cyan-colored sticks). Hydrogen-bonding interactions are shown as dotted yellow lines, and disulfide bonds are shown in ball and stick representation (yellow).
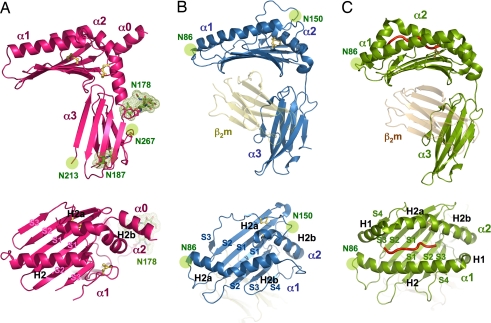
Fig. 2.
The structural relationship of m157 with classical and nonclassical MHC class I molecules. Ribbon diagrams of the 3D structures of m157 (A), H2-T22 (B) (13), and H2-Kb (C) (35) from the side and top views, showing the altered helical and β-strand topology of m157 in relation to nonclassical and classical MHC molecules. H2-T22 and H2-Kb are shown in association with the β2m subunit [yellow (B) and orange (C)]. β-strands are numbered according to their order in the primary amino acid sequence (S1–S4) by domain, and helices are named according to their structural homology to that of the canonical MHC class I structure (12). All structural figures were generated with the program Pymol (36).
The α3 domain is composed of seven antiparallel β-strands arranged into an Ig-fold that maintains the canonical intersheet disulfide bond (Cys-194 to Cys-241) (9). However, the β-strands of m157 in this region are shorter and together form strikingly flatter β-sheets than those seen in canonical Ig-folds (Fig. 1A). The flatness of the sheets reduces their mutual complementarity, which is usually achieved in β-sheet sandwiches through a slight coiling of the sheets around one another (compare m157 α3 domains to other MHC α3 domains in Fig. 2). At the top of the α3 domain, the β-sheets are spread apart to accommodate the insertion of the C-terminal end of the H2b helix, and an extended loop at the C terminus of the α3 domain reinforces the architecture of this Ig-fold (Fig. 1E). The α3 domain is also where all four potential N-linked glycosylation sites are located. Two asparagine residues (Asn-178 and Asn-187) are glycosylated as identified in the model. The electron density is especially clear for Asn-178, where a branched fucose is intimately buried in a hydrophobic pocket located near the hinge between the platform and the stem, providing additional structural support in this region (detailed in Fig. 1D).
Noncanonical Geometry of the α1/α2 Platform Region.
The topology of m157's α helices deviates considerably from that of a classical MHC class I molecule (Fig. 2A) (9). The α1 and α2 helical disposition is more similar to that of the murine nonclassical MHC, T22 (Fig. 2B) (13). The α1 helix of m157 is straight, lacking the characteristic H1 portion of the helix, which, in a classical MHC molecule, extends to the α3 domain (Fig. 2C). The α2 helix of m157, which is traditionally divided into the H1, H2a, and H2b segments in MHC class I, does not contain the H1 segment, whereas the H2b portion of the helix travels much further downward at its C-terminal end, resulting in an extended helix nearly perpendicular to the platform (Fig. 2A Upper). This unique helical section (H2b) mediates crucial contacts that may rigidify the hinge region linking the platform and α3 domain of m157 (Fig. 1C).
A unique structural feature in m157's platform is the α0 helix at its N terminus (Figs. 1 A–C and 2A Lower). All MHC-like molecules so far contain an N terminus that serves as the first β-strand of the β-sheet platform floor. However, in m157, the N-terminal region has folded into an α-helix that forms a very complementary and tightly packed interface with the side of the α2 helix (Fig. 1B). The resultant three rows of helices give the appearance that the α2 helix is being “presented” in a groove between α0 and α1 (Fig. 2A Lower) in a manner vaguely reminiscent of the manner in which peptides are presented by class I (Fig. 2C Lower). Judging from the interactions between the α0 and α2 helices, it is likely that α0 assists in correct positioning of the α2 helix in lieu of a missing Cys-101-Cys-164 helix-to-floor disulfide bond normally found in both classical and nonclassical MHC (9, 12).
Extensive Interactions Between the Platform and α3 Domain.
The tertiary structure of m157 is composed of an extensive network of contacts between the α1 and α2 helices, between the extended α2 helix and α3 domain, and between the top loops of the α3 domain and the underside of the α1/α2 platform (Fig. 1D). The contact interface between the platform and α3 domain is significantly more extensive than that of a classical MHC class I molecule (excluding β2m) (≈1,265 Å2 of buried surface area compared with that of 240 Å2 in H2-Kb) and also greater than that of ZAG (795 Å2), an MHC class I-related molecule that is also β2m-independent (18). The conformation of the hinge region of m157 is mediated primarily through the lateral support of the H2b segment of the α2 helix (Figs. 1C and 2A), which appears to serve as a side-mounted brace to support the normally flexible connection between the α1/α2 platform and the α3 domain. The platform–α3 interaction is further enhanced by numerous interdomain hydrogen bonds between the connecting loops of the α3 stem region and the surface underneath the α1/α2 platform (Fig. 1D).
As mentioned, m157 is β2m-independent, as are several nonclassical MHC-like proteins such as MIC-A and ZAG (18, 19). In m157, the α3 domain is missing conserved β2m contact residues used in MHC class I, and structural features that likely stabilize it in the absence of β2m also likely prevent β2m from binding to m157. That is, the compactness of m157 and the overall structure of the α1/α2–α3 interaction sterically obstructs accessibility to β2m. This feature stands in contrast to MIC-A, whose α3 domain has no contact with the platform and can adopt multiple conformations relative to the platform (19). In ZAG, there is an enhancement of platform–α3 interactions, but not nearly as extensive as seen in m157, and it was observed that the relative spatial relationship between the platform and α3 domains in ZAG varied within the crystal, suggesting that some degree of flexibility remained (18).
There is an extra 14-residue C-terminal loop (residues 255–268) that extends beyond the conventional Ig domain of the α3 domain. The entire length of this loop is intimately associated with a lateral edge of the α3 domain and curves back to travel perpendicularly across the two β-sheets of the Ig-fold (Fig. 1 A and E). This extended loop appears to provide additional structural enforcement for the Ig domain, whose contact between the β-sheets appears diminished compared with classical MHC α3 domains.
Closed Peptide-Binding Groove.
The unique geometry and configuration of m157's helices in the platform region results in a closed peptide-binding groove (Fig. 2A Lower), reminiscent of those found in certain nonclassical MHC class I molecules, such as the mouse T22 molecule (Fig. 2B), as well as others such as HFE. The rows of helices of m157 are packed closely against each other, with an average interhelical width (as defined by the shortest Cα distance between helices) of ≈7.6 Å between α2 and α1 and 5 Å between α0 and α2. The narrow gap between α1 and α2 helices is nearly half the distance compared to that of a standard MHC class I molecule (≈15.0 Å in the central region of the peptide-binding region) (Fig. 2C), yet compatible with that for T22 (6–10 Å) (Fig. 2B) (13). It is substantially narrower than that of another MCMV-encoded MHC homologue, m144 (≈11.0 Å), which also does not present peptides (11). In addition to the positional proximity, extensive van der Waals' contacts by the residues on the helices have efficiently sealed the interhelical gap, obstructing peptides, or other ligands, from binding between the α helices.
Direct Binding of m157 to Ly49H and Ly49I Receptors.
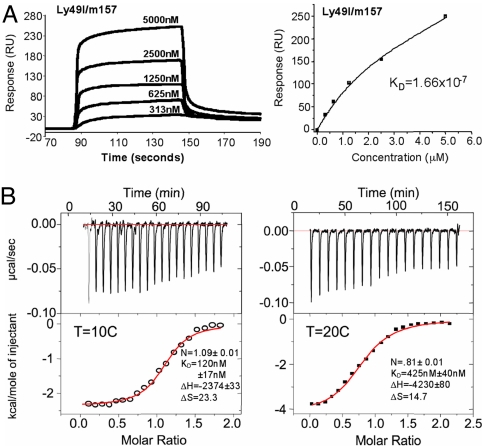
To investigate the interaction between Ly49 receptors and m157, surface plasmon resonance (SPR) and isothermal titration calorimetry (ITC) measurements were performed by using insect cell-expressed m157, as well as the full-length extracellular domains of Ly49H, Ly49I129/J, and Ly49IB6 receptors. Consistent with the earlier biochemical studies, m157 does not bind to Ly49IB6 (negative control and data not shown), but demonstrates robust binding to the Ly49I129/J inhibitory receptor (6, 7) (Fig. 3) both through SPR and ITC measurements. The Kd of Ly49I129/J with m157 was determined to be 166 nM through SPR and 120 nM from the ITC measurements, with a stoichiometry of one m157 to one Ly49I dimer. By comparison, the reported dissociation constants (Kd) of Ly49 inhibitory receptors with their standard MHC class I ligands range from 1 to 80 μM (20, 21). Therefore, Ly49I129/J binds m157 with appreciably higher affinity (approximately five times more) than that of Ly49s with their cognate self-MHC receptors. Although we have also demonstrated m157 binding to the Ly49H receptor (SI Fig. 6), technical difficulties associated with a baseline drift prevented calculation of a precise equilibrium Kd, but the approximate Kd is roughly 1 μM, similar to Ly49 affinities for cognate self-MHC. Interestingly, from these experiments, it appears that m157 binds to Ly49I with a 2- to 5-fold higher affinity compared to Ly49H. Although it is tempting to speculate on the functional relevance of this affinity difference between the activating (Ly49H) and inhibitory (Ly49I) receptors, we reserve further discussion until we can obtain more precise Ly49H binding data given the technical caveats we mentioned.
Fig. 3.
m157 binds to the inhibitory Ly49I with higher affinity than classical MHC/Ly49 interactions. (A) SPR sensogram traces (Left) and equilibrium-binding curves (Right) are shown of m157 binding to inhibitory Ly49I129 receptor. Ly49I was immobilized to a Biacore streptavidin chip via an engineered biotinylation site at the N terminus, thus ensuring appropriate orientation of the receptor on the chip surface. Concentrations of analyte (m157) are as indicated. Kd, equilibrium dissociation constant. (B) ITC traces of Ly49I and m157 at 10°C (Left) and 20°C (Right). (Upper) Rate of heat released as a function of time from injections of Ly49I into a cell containing m157. (Lower) Fitted parameters using nonlinear least-squares fitting (solid line) of the integrated areas under the respective peaks in Upper, plotted against the molar ratio of Ly49I and m157.
Expression of m157 Enables Killing of MHC Class I+ Cells.
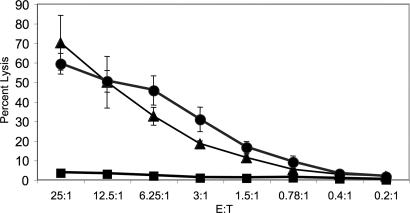
In some circumstances, the expression of MHC class I can protect cells from lysis by NK cells. To address whether m157 can render cells susceptible to NK cell-mediated cytotoxicity in the presence of class I MHC on the potential target, the MHC class I-positive RMA lymphoma cell line was stably transfected with m157 (containing a FLAG epitope on the N terminus to confirm expression). C57BL/6 NK cells efficiently killed m157+ RMA cells in vitro, whereas the parental RMA cells were not lysed (Fig. 4). Killing of the m157-bearing RMA cells was as efficient as killing of RMA cells expressing ligands for the activating NKG2D receptors, which was used as a positive control (22). Thus, expression of m157 renders MHC class I-positive cells, normally resistant to NK cell attack, susceptible to detection and elimination by NK cells containing Ly49H-activating receptors.
Fig. 4.
Expression of m157 on RMA cells overcomes class I-mediated inhibition of NK cell cytolysis. RMA tumor cells, which are protected from lysis by NK cells because of their expression of MHC class I, were transfected with m157. These m157-bearing RMA cells (filled triangles) were efficiently lysed by C57BL/6 NK cells expressing the activating Ly49H receptor. Similarly, RMA cells transfected with a ligand for the activating NKG2D receptor (filled circles) (22) were also efficiently killed by NK cells. By contrast, parental untransfected RMA cells (filled squares) were resistant to NK cell-mediated lysis because of their expression of MHC class I, consistent with prior studies. NK cell-mediated cytolysis assays were performed at various effector to target (E:T) ratios (x axis), and cytolysis was measured as percentage-specific lysis (y axis).
Discussion
The recognition of MHC class I on the cell surface by NK cell inhibitory receptors is one of the primary immune safety mechanisms against viruses and tumors that have down-regulated MHC class I to avoid detection by CD8+ T cells, a theory put forth in the “missing self” hypothesis (23). A rational consequence of this system, at least for viruses, would be the expression of a decoy ligand for these NK inhibitory receptors, provided that these decoy ligands are not recognized by host αβ–TCR+ T cells. MCMV encodes m157, a GPI-linked cell surface protein with a putative MHC class I-like structure. m157 was first identified for its reactivity with the Ly49H-activating receptor, expression of which on NK cells confers resistance to MCMV infection. m157 also is a ligand for the Ly49I inhibitory receptor in the 129/J strain mice (Ly49129/J), suggesting that this viral protein was initially intended to function as an inhibitory ligand expressed in place of the down-regulated MHC class I molecules on MCMV-infected cells.
To understand the structural principles underlying the recognition of m157 by Ly49 receptors, we determined the 3D structure of m157 to 2.1-Å resolution and investigated its interaction with the recombinant Ly49H, Ly49I129/J, and Ly49IB6 receptors. m157 bears gross architectural resemblance to the MHC family (9), but with major deviations. Overall, the m157 structure has more extensive intra- and intermolecular contacts within and between domains, respectively, than MHC class I, resulting in a more compact structure. m157 possesses a three-helix “sandwich” on the platform because of the addition of a short α0 helix at the N terminus. m157 does not bind peptide because of a virtually sealed peptide-binding groove, nor does it associate with β2m for stable cell surface expression. There are several secondary structural motifs that are present in m157, potentially to compensate for the absence of β2m, including an extended α2 helix (H2b) that serves as the center scaffold of the molecule, and an extensive network of interactions at the platform–α3 domain interface. Lacking the peptide and β2m subunit normally required by other classical MHC class I molecules, m157 thus represents a “minimal” MHC that can be properly expressed without additional cellular resources, an ideal molecule to promote immune evasion.
m157's 3D structure is distinctly MHC-like; however, several key features suggest it may engage Ly49 receptors in a novel fashion. The available structural knowledge indicates that Ly49 receptors interact with MHC class I molecules through a “site 2,” a composite of the β2m subunit, the α3 domain, and residues located on the underside of the α1/α2 platform (20, 24, 25) (Fig. 5). Given that m157 does not associate with β2m and the α3 domain has been repositioned with respect to its MHC counterpart because of structural constraints, it is unlikely that Ly49 receptors will be able to associate with m157 after the conventional paradigm of Ly49–MHC interactions.
Fig. 5.
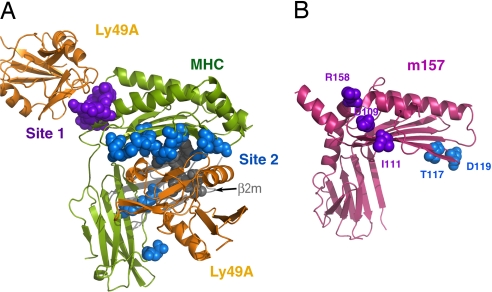
m157 mutants that abrogate Ly49H-mediated viral resistance do not superimpose with the canonical Ly49 binding sites on MHC class I. (A) Crystal structure of a representative Ly49–MHC complex (Ly49A/H-2Dd) (20, 24), with the residues of the MHC α-chain that involve Ly49 interaction presented in sphere style (purple, site 1; blue, site 2). (B) m157 mutations identified in previous reports, which are likely to express properly on the cell surface but fail to engage Ly49H (26, 27), were mapped onto the 3D structure (blue from ref. 26 and purple from ref. 27). Both models are in the same orientation with respect to the platform region. Collectively, they reveal the two surface areas on m157 that are likely to involve Ly49H recognition, which are distinct from the conventional MHC–Ly49 interfaces.
Recent studies have indicated that certain mutations in the m157 gene resulted in immune evasion of MCMV from the NK cells of infected B6 mice, which are normally resistant to such a virus (26, 27). This escape ability is largely attributed to the failure of mutant m157 to engage Ly49H-activating receptors. Many of the mutations abrogate cell surface expression of m157 in these studies; however, there are a number of strains that contain mutations that maintain the global architecture and cell-surface expression, but might disrupt the specific recognition site of Ly49H.
French et al. (26) identified five mutant strains of MCMV resistant to Ly49H-mediated detection in which full-length m157 is likely to be expressed. Two of these mutants, delThr117 and Asp119Tyr, do not appear to cause any major structural disruptions, and thus might provide insight into the location of Ly49H binding. These mutations are located on the underside of the platform domain, on the connecting loop of the second and third β-strands of the α2 domain, in an area that would contact β2m in a canonical MHC class I molecule (Fig. 5). In a complementary study, Voigt et al. (27) also characterized strains of MCMV expressing m157 mutants not capable of stimulating Ly49H+ reporter cells. Three of these strains encode m157 molecules with high homology to the m157 structure presented here and contain three mutations located on the surface of m157 that could affect Ly49H binding: Asp109Asn, Ile111Leu, and Arg158Gln. They are located near the N termini of the second β-strand and the H2b helix segment, near the site 1 region of the Ly49–MHC-I interaction (Fig. 5).
When mapped collectively onto the m157 structure (Fig. 5), these mutations cluster into two areas: one in close proximity to the N-terminal α0 helix and the other located near the putative β2m–MHC interface (Fig. 5). Because m57 does not associate with β2m and there is significant positional variation in the α3 domain (in relation to the platform) between m157 and a MHC class I molecule, the original physiologically relevant site 2 Ly49–MHC interface, as revealed in previous crystal structures (20, 24), no longer exists in m157. From our mutational model, it is therefore possible that a homodimeric form of Ly49H contacts both of these sites, and thus suggests a form of Ly49 engagement that may bridge sites 1 and 2, but that deviates substantially from the conventional paradigm of Ly49–MHC interactions.
A delicate balance of signaling through the activating and inhibitory receptors modulates the final response of NK cells. That recognition of MHC class I on the target cells can suppress NK cell attack was first suggested based on the observation that NK cells preferentially killed RMA/S, a class I MHC-deficient variant of the RMA T cell lymphoma, the basis for the “missing self” hypothesis of NK cell recognition (23). MCMV has devised mechanisms to down-regulate expression of class I MHC in the infected cells presumably to escape detection by CMV-specific CD8+ cytotoxic T lymphocytes. Three proteins encoded by MCMV (m04, m06, and m152) function to quench MHC class I expression of the host cell (28). Because cells infected with MCMV have diminished MHC class I expression, they are more susceptible to attack by NK cells. Yet it is unclear whether Ly49H+ NK cells only initiate a cytolytic response when there is a reduced level of MHC class I expression, and, indeed, how NK cells would respond in the presence of both activating and inhibitory ligands. We tested these issues by expressing m157 on the cell surface of RMA, a prototypic tumor that is protected from NK cell-mediated cytotoxicity by the expression of MHC class I (23). NK cells from B6 mice efficiently lysed m157-bearing RMA, but not parental RMA, demonstrating that m157 enables NK cells to destroy these targets despite the presence of inhibitory signals. Prior studies demonstrated that the expression of ligands for the activating NKG2D receptor on MHC class I-bearing RMA cells renders these cells susceptible to NK cell-mediated cytotoxicity (22). The ability of m157 to render MHC class I-positive cells susceptible to NK cells indicates that NKG2D is not unique in this aspect. Rather, it suggests that activating NK cell receptors that engage ligands of sufficient affinity, avidity, or abundance on a target cell are capable of overcoming the inhibitory signal conferred by MHC class I. This is advantageous, particularly against viruses that do not down-regulate the expression of MHC class I.
Materials and Methods
Protein Expression and Purification.
The extracellular portion of the mature form of m157 (6) (residues 1–285) was fused with a C-terminal hexahistidine tag and cloned into the pAcGP67A transfer vector (PharMingen, San Diego, CA). This construct was used to produce recombinant baculovirus via recombination with the baculovirus genome (Sapphire Baculovirus; Orbigen, San Diego, CA) after transfection into Sf9 insect cells. To produce recombinant m157, Hi5 cells (Invitrogen, Carlsbad, CA) cultured in Insect-Xpress media (Cambrex, Charles City, IA) were incubated for 72 h with the recombinant virus, and the secreted protein was purified from the supernatant with Ni-NTA beads (Qiagen, Valencia, CA). m157 was eluted from the Ni-NTA beads by using elution buffer (HBS-Imidazole: 10 mM Hepes, pH 7.2, 150 mM NaCl, 0.02% NaAzide + 200 mM Imidazole). m157 was purified by size-exclusion chromatography by using a Superdex 200 gel filtration column equilibrated with HBS as an elution buffer.
Crystallization and Data Collection.
Purified m157 was concentrated to 2.5 mg/ml in its HBS elution buffer. Crystallization screens were carried out in the sitting drop format at 20°C. Initial leads identified in various low molecular weight polyethylene glycols were refined, and the final crystallization conditions for growing large single crystals were 35% PEG400, 200 mM NH4Ac, Mes (pH 6). The crystals belong to space group C2 with the following unit cell dimensions: a, 120.0 Å; b, 43.8 Å; c, 62.3 Å; α, 90°; β, 101.5°; γ, 90°. X-ray data sets were measured on a 3 × 3 ADSC CCD at Beamline 8.2.1 at the Advanced Light Source, Lawrence Berkeley National Laboratory. Crystals were frozen directly in their mother liquor. Complete data sets to 1.9 Å (native) and 2.0 Å (Hg derivative) were collected by using 1° oscillations over 120 frames. The data were indexed, integrated, and scaled with the programs MOSFLM (29) and SCALA (30). Data statistics are shown in SI Table 1.
Structure Determination and Refinement.
The crystal structure of m157 was determined by a single isomorphous replacement with anomalous scattering method (SI Table 1) using mercury (anomalous signal collected at high-energy remote) as a derivative. Experimental phasing was calculated by using program SHARP (31), followed by solvent flattening/flipping with SOLOMON (30). Automated model tracing was done with ARP/wARP (32), and structural refinement was performed by using CNS (33). The following sets of residues do not have clear electron density and, therefore, are excluded from our final model: 1–7, 53–54, 132–146, 188–190, 211–214, and 269–285.
SPR and Microcalorimetry.
Affinity measurements were performed in HBS-P buffer with a Biacore 3000 (Biacore AB, Uppsala, Sweden). Recombinant Ly49 receptors (Ly49IB6, Ly49I129/J, and Ly49H) with genetically engineered N-terminal biotinylation sites (Avidity, Denver, CO) were produced in insect cells by using the baculovirus expression system and isolated and purified as described above for m157. Biotinylation of the Ly49 recombinant protein was performed according to the manufacturer's protocol (Avidity) and subsequently purified over gel filtration. Six hundred response units of each receptor were immobilized onto the surface of a streptavidin-coated chip (Biacore). To conduct equilibrium-binding measurements, baculovirus-expressed m157 (described above) was injected over four flow cells at 10°C, the first an empty reference and the remaining three each containing one of the immobilized Ly49 receptors. The measurements were conducted over a range of m157 concentrations (Fig. 3), and the amount of binding was calculated as the difference in the response at equilibrium in the Ly49 and control flow cells. Equilibrium Kd values were determined assuming a 1:1 interaction (A + B = AB). Response levels at equilibrium were plotted against protein concentration and calculated via a nonlinear least-squares fitting of the Langmuir binding equation: AB = BxABmax/(Kd + B). They were further confirmed by linear Scatchard plot analysis by using Origin 6.0 software (MicroCal; OriginLab Corp., Northhampton, MA). ITC was performed on a VP-ITC (MicroCal; OriginLab Corp.) at 10°C and 20°C. The proteins used were identical to those used for SPR measurements and purified in HBS buffer (pH 7.2) immediately before use. Ly49I was injected at a concentration of 23 μM (dimeric) into a cell containing 5 μM of m157, giving a ratio of ≈4.6:1 (receptor to ligand), ensuring ligand saturation. Binding isotherms were fit by nonlinear least-squares fitting by using the single-site model provided by OriginLab Corp. (MicroCal). Equilibrium Kd were calculated as the reciprocal of KA.
Cell Lines and Transfectants.
The RMA (H-2b-positive) and RMA/S (H-2b-negative) T cell lymphoma cell lines (kindly provided by J. Ryan, University of California, San Francisco, CA) were cultured in RPMI 1640 medium containing 10% FCS, 2 mM glutamine, 50 units/ml penicillin, and 50 μg/ml streptomycin. Stable transfectants of RMA expressing m157 on cell surface were made as previously described (22).
51Cr-Release Assay.
NK cell preparation and standard 4-h cytotoxicity assays were carried out as previously described (34).
Supplementary Material
Acknowledgments
We thank the staff members at both Stanford Linear Accelerator Center and Advanced Light Source (Berkeley, CA) for their assistance with data collection. This work was supported by a Cancer Research Institute Postdoctoral Fellowship (E.J.A.), a National Institutes of Health-Immunology postdoctoral training grant (E.J.A.), and National Institutes of Health Grants A1065504 (to K.C.G.) and AI068129 (to L.L.L.). L.L.L. is an American Cancer Society Research Professor.
Abbreviations
- ITC
isothermal titration calorimetry
- NK
natural killer
- MHC
major histocompatibility complex
- MCMV
mouse cytomegalovirus
- β2m
monomorphic β2-microglobulin
- SPR
surface plasmon resonance.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2NYK).
This article contains supporting information online at www.pnas.org/cgi/content/full/0703735104/DC1.
References
- 1.Scalzo AA, Fitzgerald NA, Wallace CR, Gibbons AE, Smart YC, Burton RC, Shellam GR. J Immunol. 1992;149:581–589. [PubMed] [Google Scholar]
- 2.Brown MG, Dokun AO, Heusel JW, Smith HR, Beckman DL, Blattenberger EA, Dubbelde CE, Stone LR, Scalzo AA, Yokoyama WM. Science. 2001;292:934–937. doi: 10.1126/science.1060042. [DOI] [PubMed] [Google Scholar]
- 3.Daniels KA, Devora G, Lai WC, O'Donnell CL, Bennett M, Welsh RM. J Exp Med. 2001;194:29–44. doi: 10.1084/jem.194.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee SH, Girard S, Macina D, Busa M, Zafer A, Belouchi A, Gros P, Vidal SM. Nat Genet. 2001;28:42–45. doi: 10.1038/ng0501-42. [DOI] [PubMed] [Google Scholar]
- 5.Lee SH, Zafer A, de Repentigny Y, Kothary R, Tremblay ML, Gros P, Duplay P, Webb JR, Vidal SM. J Exp Med. 2003;197:515–526. doi: 10.1084/jem.20021713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arase H, Mocarski ES, Campbell AE, Hill AB, Lanier LL. Science. 2002;296:1323–1326. doi: 10.1126/science.1070884. [DOI] [PubMed] [Google Scholar]
- 7.Smith HR, Heusel JW, Mehta IK, Kim S, Dorner BG, Naidenko OV, Iizuka K, Furukawa H, Beckman DL, Pingel JT, et al. Proc Natl Acad Sci USA. 2002;99:8826–8831. doi: 10.1073/pnas.092258599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tripathy SK, Smith HR, Holroyd EA, Pingel JT, Yokoyama WM. J Virol. 2006;80:545–550. doi: 10.1128/JVI.80.1.545-550.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Madden DR. Annu Rev Immunol. 1995;13:587–622. doi: 10.1146/annurev.iy.13.040195.003103. [DOI] [PubMed] [Google Scholar]
- 10.Bjorkman PJ, Parham P. Annu Rev Biochem. 1990;59:253–288. doi: 10.1146/annurev.bi.59.070190.001345. [DOI] [PubMed] [Google Scholar]
- 11.Natarajan K, Hicks A, Mans J, Robinson H, Guan R, Mariuzza RA, Margulies DH. J Mol Biol. 2006;358:157–171. doi: 10.1016/j.jmb.2006.01.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bjorkman PJ, Saper MA, Samraoui B, Bennett WS, Strominger JL, Wiley DC. Nature. 1987;329:506–512. doi: 10.1038/329506a0. [DOI] [PubMed] [Google Scholar]
- 13.Wingren C, Crowley MP, Degano M, Chien Y, Wilson IA. Science. 2000;287:310–314. doi: 10.1126/science.287.5451.310. [DOI] [PubMed] [Google Scholar]
- 14.Burmeister WP, Huber AH, Bjorkman PJ. Nature. 1994;372:379–383. doi: 10.1038/372379a0. [DOI] [PubMed] [Google Scholar]
- 15.Zeng Z, Castano AR, Segelke BW, Stura EA, Peterson PA, Wilson IA. Science. 1997;277:339–345. doi: 10.1126/science.277.5324.339. [DOI] [PubMed] [Google Scholar]
- 16.Wang CR, Castano AR, Peterson PA, Slaughter C, Lindahl KF, Deisenhofer J. Cell. 1995;82:655–664. doi: 10.1016/0092-8674(95)90037-3. [DOI] [PubMed] [Google Scholar]
- 17.Lebron JA, Bennett MJ, Vaughn DE, Chirino AJ, Snow PM, Mintier GA, Feder JN, Bjorkman PJ. Cell. 1998;93:111–123. doi: 10.1016/s0092-8674(00)81151-4. [DOI] [PubMed] [Google Scholar]
- 18.Sanchez LM, Chirino AJ, Bjorkman P. Science. 1999;283:1914–1919. doi: 10.1126/science.283.5409.1914. [DOI] [PubMed] [Google Scholar]
- 19.Li P, Willie ST, Bauer S, Morris DL, Spies T, Strong RK. Immunity. 1999;10:577–584. doi: 10.1016/s1074-7613(00)80057-6. [DOI] [PubMed] [Google Scholar]
- 20.Dam J, Guan R, Natarajan K, Dimasi N, Chlewicki LK, Kranz DM, Schuck P, Margulies DH, Mariuzza RA. Nat Immunol. 2003;4:1213–1222. doi: 10.1038/ni1006. [DOI] [PubMed] [Google Scholar]
- 21.Wang J, Whitman MC, Natarajan K, Tormo J, Mariuzza RA, Margulies DH. J Biol Chem. 2002;277:1433–1442. doi: 10.1074/jbc.M110316200. [DOI] [PubMed] [Google Scholar]
- 22.Cerwenka A, Baron JL, Lanier LL. Proc Natl Acad Sci USA. 2001;98:11521–11526. doi: 10.1073/pnas.201238598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karre K, Ljunggren HG, Piontek G, Kiessling R. Nature. 1986;319:675–678. doi: 10.1038/319675a0. [DOI] [PubMed] [Google Scholar]
- 24.Tormo J, Natarajan K, Margulies DH, Mariuzza RA. Nature. 1999;402:623–631. doi: 10.1038/45170. [DOI] [PubMed] [Google Scholar]
- 25.Michaelsson J, Achour A, Rolle A, Karre K. J Immunol. 2001;166:7327–7334. doi: 10.4049/jimmunol.166.12.7327. [DOI] [PubMed] [Google Scholar]
- 26.French AR, Pingel JT, Wagner M, Bubic I, Yang L, Kim S, Koszinowski U, Jonjic S, Yokoyama WM. Immunity. 2004;20:747–756. doi: 10.1016/j.immuni.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 27.Voigt V, Forbes CA, Tonkin JN, Degli-Esposti MA, Smith HR, Yokoyama WM, Scalzo AA. Proc Natl Acad Sci USA. 2003;100:13483–13488. doi: 10.1073/pnas.2233572100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wagner M, Gutermann A, Podlech J, Reddehase MJ, Koszinowski UH. J Exp Med. 2002;196:805–816. doi: 10.1084/jem.20020811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leslie AGW. Joint CCP4 + ESF-EAMCB Newsletter on Protein Crystallography. 1992;(no 26) [Google Scholar]
- 30.Collaborative Computational Project. Acta Crystallogr. 1994;D50:760–763. [Google Scholar]
- 31.de la Fortelle E, Bricogne G. Methods Enzymol. 1997;276:472–494. doi: 10.1016/S0076-6879(97)76073-7. [DOI] [PubMed] [Google Scholar]
- 32.Perrakis A, Morris R, Lamzin VS. Nat Struct Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 33.Brunger AT, Adams P, Clore M, Gros P, Nilges M, Read R. Acta Crystallogr. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 34.Ogasawara K, Yoshinaga SK, Lanier LL. J Immunol. 2002;169:3676–3685. doi: 10.4049/jimmunol.169.7.3676. [DOI] [PubMed] [Google Scholar]
- 35.Rudolph MG, Shen LQ, Lamontagne SA, Luz JG, Delaney JR, Ge Q, Cho BK, Palliser D, McKinley CA, Chen J, et al. J Immunol. 2004;172:2994–3002. doi: 10.4049/jimmunol.172.5.2994. [DOI] [PubMed] [Google Scholar]
- 36.Delano WL. The Pymol Molecular Graphics System. Palo Alto, CA: DeLano Scientific LLC; 2003. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.