Abstract
Eukaryotic parasites are a leading cause of morbidity and mortality worldwide, yet little is known about the genetic basis of their virulence. Here, we present a forward genetic screen to study pathogenesis in the protozoan parasite Toxoplasma gondii. By using modified signature-tagged mutagenesis, the growth of 6,300 T. gondii insertional mutants was compared in cell culture and murine infection to identify genes required specifically in vivo. One of the 39 avirulent mutants is disrupted in a divergent ortholog of the regulator of chromosome condensation 1 (RCC1), which is critical for nuclear trafficking in model systems. Although this RCC1 mutant grows similar to wild type in standard tissue culture conditions, it is growth-impaired under nutrient limitation. Genetic complementation of mutant parasites with the T. gondii RCC1 gene fully restores both virulence in mice and growth under low-nutrient conditions. Further analysis shows that there is a significant defect in nuclear trafficking in the RCC1 mutant. These findings suggest that the rate of nuclear transport is a critical factor affecting growth in low-nutrient conditions in vivo and in vitro. Additionally, we observed that although RCC1 proteins are highly conserved in organisms from humans to yeast, no protozoan parasite encodes a characteristic RCC1. This protein divergence may represent a unique mechanism of nucleocytoplasmic transport. This study illustrates the power of this forward genetics approach to identify atypical virulence mechanisms.
Keywords: nuclear transport, Toxoplasma
Protozoan parasites such as Plasmodium spp., Entamoeba histolytica, Trypanosoma spp., and Leishmania spp. cause some of the most devastating human diseases (1). Recent completion of genome sequences from these parasites makes large-scale screens to uncover parasite virulence factors attractive, but such analysis is currently not possible in these genetically recalcitrant organisms. In contrast, the apicomplexan parasite Toxoplasma gondii is haploid, is amenable to genetic manipulation, and can be studied in a natural host mouse model (2, 3). T. gondii is an obligate intracellular parasite that infects nearly one-third of the human population, and is well known for causing congenital infections and encephalitis in immunocompromised persons (4, 5). During infection, the parasite initially replicates rapidly in any nucleated cell, disseminating throughout the host. T. gondii then converts to a slow-growing encysted form that allows the parasite to establish a lifelong chronic infection, primarily in brain and muscle tissue (4, 6). Given its medical importance and relatively amenable genetics, we chose T. gondii as our model protozoan for an analysis of virulence genes.
To date, there have been few virulence genes identified in T. gondii. Deletion of the carbamoyl phosphate synthetase II gene (CPSII) completely attenuates the parasite in mice (7). Additionally, avirulence has been associated with the disruption of the surface antigen 3 (SAG3), dense granule protein 2 (GRA2), the catalase TgPrx2, and the microneme adhesin MIC2 (8–11). Finally, genetic mapping has revealed two polymorphic kinases, ROP18 and ROP16, which are secreted by the rhoptry organelles and are major determinants of virulence (12, 13).
With the completion of the T. gondii genome (www.toxodb.org), a high-throughput genetic screen that allows for examination of large panels of mutants would greatly facilitate a global investigation of parasite virulence. One successful method for such analysis is signature-tagged mutagenesis (STM). This technique uses a unique DNA sequence to “tag” a microbe so it can subsequently be identified from a pool. Although STM has been used to study pathogenesis in both bacterial and fungal pathogens (14–18), it has never before been used in a protozoan parasite. We previously reported an adaptation to the conventional STM technique for T. gondii, and showed its usefulness in a cell culture screen to identify mutants resistant to the pro-drug FUDR (19). Here, we describe the expansion of that pilot study to 6,300 insertional mutants, the analysis of these mutants in a mouse model of toxoplasmosis, and the discovery of a regulator of chromosome condensation 1 (RCC1) protein that is involved in parasite virulence [schematized in supporting information (SI) Fig. 6].
Results
Construction and Analysis of T. gondii STM Library.
With the success of the pilot T. gondii STM study in vitro, we expanded the library to include 6,300 insertional mutants for analysis in vivo. Sixty T. gondii clones, each with a stable insertion of a different signature tag, underwent insertional mutagenesis. Approximately 105 clones from each parental tag strain were isolated and arrayed into 96-well plates creating a library of >6,300 clones (20). For the in vivo screen, 60 parasites from each plate were pooled and grown in either cell culture or in mice for 22 days (the time when parasites have established an early chronic infection). This allowed the comparison of parasite growth within a host cell during tissue culture and in vivo conditions, enabling us to identify genes required only for pathogenesis.
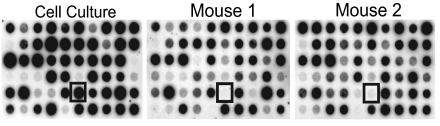
Parasite DNA from cell culture or the brains of infected mice was used as a template to PCR radiolabel each unique tag as a probe for comparative hybridization analysis (Fig. 1). Strains that exhibited a reduced hybridization signal in mice were examined in a clonal infection. For the single clone infections, mice were infected with mutant or wild-type parasites, and the number of cysts per brain was counted at 22 days after infection. Thirty-nine STM clones exhibited a ≈10-fold reduction in cyst counts (Table 1). The genomic DNA adjacent to the insertion site of these less-virulent mutants was isolated and compared with www.toxodb.org. Although we list the annotation for the gene that is interrupted by the mutagenesis plasmid, we have not verified for each of the clones that the insertion is responsible for the avirulent phenotype.
Fig. 1.
Differential hybridization uncovers less virulent parasites. Pools of 60 parasites were grown for 22 days either by serial passage in fibroblasts or i.p. injected into two mice. T. gondii DNA was isolated from tissue culture and mice brains, and then used as template in PCRs to radiolabel the tags from the pools. Identical membranes containing PCR product from all 60 original tags were hybridized with these labeled probes to compare cell culture and the two mice. The boxed spots indicate a potential virulence mutant that produces a strong hybridization signal from tissue culture and is absent from the brains of mice.
Table 1.
Insertion site of the mutagenesis plasmid for the attenuated mutants
| Mutant | %WT (no. of mice) | Chromosome | Location | Draft 3 annotation |
|---|---|---|---|---|
| 71C2 | 6 (3) | Ia | 411900 | 83.m01222 |
| 9xG4 | 7 (7) | Ia | 599455 | 83.m01242 |
| 101F9 | 5 (2) | III | 1202200 | 52.m01612 ribosomal protein L15 putative |
| 57G3 | 9 (2) | IV | 2003950 | 27.m00827 |
| 26F7 | 5 (2) | IV | 1407661 | 641.m03156 |
| 73F9 | 7 (6) | V | 1367207 | 31.m00904 regulator of chromosome condensation related |
| 88E8 | 5 (11) | V | 1009640 | 76.m01622 |
| 9B5 | 10 (2) | VI | 178451 | 49.m03099 |
| 19C3 | 4 (2) | VI | 178451 | 49.m03099 |
| 29C3 | 3 (2) | VI | 66771 | 49.m00018 emp24/gp25L/p24 family domain-containing transmembrane protein |
| 98F7 | 4 (2) | VI | 2577040 | 49.m03343 PX domain-containing protein |
| 12F7 | 7 (6) | VIIa | 1175720 | 20.m03930 20k cyclophilin (20.m03931 lysyl-tRNA synthetase) |
| 38C3 | 3 (3) | VIIa | 2463700 | 20.m03798 |
| KMME6 | 5 (12) | VIIb | 1785558 | 55.m04928 (55.m04927) |
| 13E10 | 9 (2) | VIIb | 3944360 | 55.m04701 synaptobrevin-like protein, (55.m04702 L-type amino acid transporter-related) |
| 68C6 | 4 (4) | VIII | 5874600 | 59.m07756 |
| 64F2 | 14 (1) | IX | 1352099 | 57.m01780 acyl-CoA dehydrogenase, short/branched chain specific, mitochondrial (precursor) |
| 90B2 | 33 (2) | IX | 1352051 | 57.m01780 acyl-CoA dehydrogenase, short/branched chain specific, mitochondrial (precursor) |
| 91G8 | 3 (2) | IX | 3220845 | No annotation |
| PRD4 | 3 (12) | IX | 5321038 | 541.m01157 patatin-like phospholipase domain containing protein |
| 49E10 | 7 (4) | X | 3362809 | 42.m00007 elongation factor G putative |
| 56C2 | 7 (2) | X | 1882762 | 42.m03461 |
| 44C6 | 5 (2) | X | 1666204 | 42.m03489 |
| 97F9 | 10 (2) | X | 5856020 | 46.m02916 (46.m01755 ImpB/MucB/SamB family domain-containing protein) |
| 41E2 | 0.2 (4) | XI | 2191215 | 583.m05443 |
| 40E4 | 6 (4) | XII | 5779451 | 65.m02536 |
| 95C5 | 8 (2) | XII | 5331130 | 50.m03390 |
| 42F7 | 11 (2) | XII | 109850 | 145.m00342 staphylococcal nuclease homologue domain-containing protein |
| 24F9 | 8 (2) | XII | 4733853 | 50.m05663 |
| 39B2 | 6 (2) | XII | 78139 | 40.m00346 small nuclear ribonucleoprotein (snRNP) |
| 40F3 | 3 (2) | XII | 327168 | 145.m00321 |
| 40B2 | 6 (2) | XII | 327168 | 145.m00321 |
| 18C5 | 6 (3) | TGG994746 | 45323 | 125.m00072 SPX domain-containing protein |
| 7xC11 | 5 (9) | TGG994767 | 15632 | 177.m00020 |
| 26G5 | 8 (4) | Not identified | ||
| 37B11 | 7 (3) | Not identified | ||
| 87D10 | 1 (2) | Not identified | ||
| 37B2 | 5 (2) | Not identified | ||
| 91E4 | 1 (2) | Not identified |
The 39 mutants with a ≈10-fold reduction in the number of cysts per brain compared with infections with wild-type parasites are listed in the first column. The second column contains the percentage of cysts per brain for each mutant compared with wild type (%WT), along with the number of mice infected with each mutant in parentheses. Also listed is the chromosome number, location within the chromosome, and closest draft 3 annotation to the insertion site for 34 of the 39 less-virulent STM clones. In some cases, no annotation is given because there was no draft 3 prediction within 2 kb of the insertion site. However, other gene predictions or ESTs that are not listed may be directly disrupted by the insertion. Two annotations are given if an insert is between two predicted genes. Although the percentage of cysts compared with wild type is higher for 64F2 and 90B2, they are included because they represent two independent insertion events within the same predicted gene. For the 24F9 mutant, the mice infected with wild-type parasites died during acute infection for that experiment; thus, the number of cysts per brain for wild type was estimated at 10,000 (typical for wild-type infections). It is important to note that each of these disrupted genes has not yet been confirmed by knockout or complementation to be responsible for the less virulent phenotype.
Although the assay readout was the number of cysts per brain, we have isolated mutants that are theoretically defective in any step of the pathogenesis pathway. Mutants with the lowest cyst burden were also examined in a mouse model of acute infection. Several mutants were not lethal in mice at a dose that is 4-fold above the normal LD50, suggesting that they are attenuated during acute infection.
Identification and Characterization of TgRCC1.
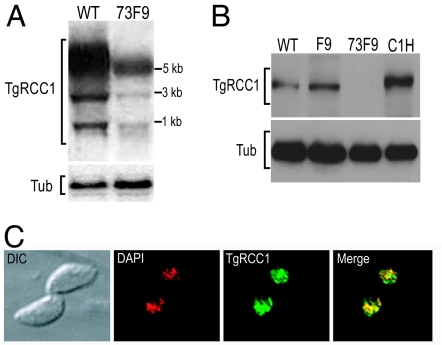
One such acute infection mutant, termed “73F9,” contains an insertion 642 bp upstream of an EST (EST no. 100116788) from the annotated gene 31.m0090. To confirm the disruption of gene 31.m0090, we compared mRNA levels from wild-type and 73F9 parasites by Northern blot analysis, probing with the EST amplified from cDNA. The message of gene 31.m00904 is greatly reduced, but not eliminated in the mutant strain (Fig. 2A). Moreover, this probe (which is specific to the disrupted locus, as confirmed by Southern blot) (data not shown) hybridizes to two smaller products, indicative of alternative splicing of the transcript. PCR from cDNA confirmed multiple transcripts resulting from alternative splicing (SI Fig. 7). We determined the transcription start site for gene 31.m00904 to be 126 bp downstream from the insertion site, consistent with the Northern blot results indicating that we disrupted the promoter. The 5′-UTR is 1,320 bp with a 176-bp intron that is differentially spliced. Mapping of the cDNA uncovered a large ORF of 3.5 kb (with eight exons) and a 400-bp 3′-UTR, corresponding to the 5.2-kb size of the major transcript on the Northern blot. Western blot analysis with antisera generated against the predicted protein shows a 124-kDa protein dramatically reduced in abundance in the 73F9 mutant (Fig. 2B).
Fig. 2.
TgRCC1 is reduced in the 73F9 mutant and nuclear-localized. (A) Total RNA from wild-type Prugniaud (WT) and 73F9 parasites was probed with a 1.6-kb cDNA fragment corresponding to the 5′ region of TgRCC1. The blot was stripped and reprobed for α-tubulin (Tub) as a loading control. (B) Total protein lysates from 5 × 106 parasites per well from Prugniaud (WT), F9 parental (F9), 73F9 mutant, and 73F9 complemented (C1H) strains. The Western blot was probed with a rat polyclonal antibody against TgRCC1. The blot was stripped and reprobed with a rabbit anti-β-tubulin antibody (Tub) as a loading control. (C) HFF cells were infected with the functional complement strain C1H that contains a C-terminal HA-tag for 24 h. Cells were incubated with anti-HA antibody to visualize the location of TgRCC1 (green) and mounted in DAPI to stain the nucleus (red).
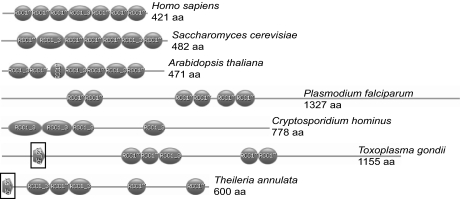
Gene 31.m00904 encodes a large protein of 1,155 aa related to the nuclear protein, RCC1; thus we have designated this gene as “TgRCC1.” RCC1 acts as a guanine exchange factor for Ran GTPase and is critical for maintaining nuclear transport (21–23). In other organisms including yeast, plants, and humans, RCC1 is characterized by seven tandem repeats of 50–60 aa (termed RCC1 repeats) (Fig. 3), which form a propeller protein structure that docks to chromatin and interacts with Ran (refs. 24, 25; reviewed in ref. 26). TgRCC1 contains five nontandem RCC1 repeats, nuclear localization signals (NLSs), and is predicted to fold into a typical RCC1 propeller structure (Fig. 3 and SI Fig. 8). Given that RCC1 proteins must be in the nucleus to function, we examined the intracellular location of TgRCC1 with a C-terminal hemagglutinin (HA) epitope tag expressed from the α-tubulin promoter. The tagged protein localizes exclusively to the parasite nucleus (Fig. 2C). Nuclear localization was also observed with TgRCC1-HA expressed from its native promoter or with an N-terminal FLAG epitope tag (SI Fig. 9).
Fig. 3.
Protozoans contain atypical RCC1 proteins. Shown are domain profiles (www.expasy.org/prosite) illustrating the RCC1 repeats present in RCC1 orthologs from various organisms. Later branching eukaryotes (shown are Homo sapiens, Saccharomyces cerevisiae, and Arabidopsis thaliana) contain the typical RCC1 structure with seven tandem repeats of 50–60 aa (shown as ovals). Apicomplexan parasites display a dispersed arrangement of the RCC1 domains within the protein. In Plasmodium falciparum, there are six repeats within the 1,327-aa protein. The Cryptosporidium hominus ortholog contains four RCC1 repeats. The T. annulata and T. gondii ortholog contain a zinc finger motif (boxed). The human RCC1 protein was used to BLASTp search the National Center for Biotechnology Information database of sequenced protozoa. The proteins with the highest similarity and predicted to be localized to the nucleus are shown.
TgRCC1 Is a Member of Divergent Protozoan RCC1 Proteins.
Genome analysis of T. gondii yielded the unusual finding that there are no predicted nuclear proteins that contain the conserved seven tandem RCC1 repeats. In addition to TgRCC1, there are other predicted proteins in T. gondii that have RCC1 domains. These other RCC1 repeat-containing proteins are predicted to be localized outside of the nucleus (http://psort.hgc.jp/form2.html) and contain other domains that suggest different functions. TgRCC1 is the only T. gondii protein that contains multiple RCC1 domains, localizes in the nucleus, and is predicted to fold into a typical RCC1 structure. Furthermore, TgRCC1 and a predicted RCC1 protein from the apicomplexan, Theileria annulata, both contain a C2C2 zinc finger motif (Fig. 3) known to bind Ran that is present in the nucleoporins Nup358 (RanBP2) and Nup153 (27–29). This may facilitate TgRCC1 interactions with Ran in the absence of the characteristic seven tandem RCC1 repeats.
We investigated whether this atypical RCC1 phenomenon was shared in other protozoan parasites. We searched the National Center for Biotechnology Information sequence database from Cryptosprodium spp., Plasmodium spp., Leishmania major, Trypanasoma spp., Theileria parva, Dictyostelium discoideum, E. histolytica, and Giardia lamblia. Interestingly, there are no predicted proteins in any of the sequenced protozoan genomes that resemble a typical RCC1 (shown in Fig. 3 are the proteins most similar to human RCC1).
TgRCC1 Is Able to Restore Virulence to 73F9 Mutant.
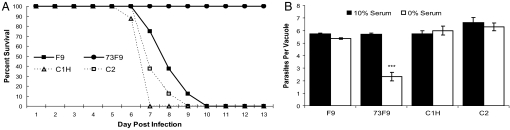
The 73F9 mutant is attenuated in vivo. Mice infected with a normally lethal dose of mutant parasites are able to survive, whereas F9 parental parasites are lethal to mice (Fig. 4A and SI Fig. 6). To determine whether this attenuation was due to the reduction of TgRCC1, we engineered mutant parasites to express the TgRCC1 ORF from the α-tubulin promoter. The parental F9, mutant 73F9, and complemented clones were compared in an acute model of toxoplasmosis. Mutant parasites expressing TgRCC1 were as virulent as the F9 parental strain (Fig. 4A and SI Fig. 10). All mice inoculated with 2 × 105 of either F9 or complemented clones succumbed to infection by day 10, whereas 73F9 parasites were not lethal at this dose. Clone C1H, containing an HA epitope tag, restored virulence similar to nontagged clones, thereby confirming that the HA-tag does not disrupt protein function and validating its use in the localization studies (Fig. 2C).
Fig. 4.
Genetic complementation of 73F9 restores virulence and growth in nutrient-depleted media. (A) F9 parental parasites were compared with 73F9 and complemented strains by i.p. injection of 2 × 105 into four mice each. Morbidity and mortality were evaluated during the acute stage of infection. Mutant parasites expressing TgRCC1 ORF from the α-tubulin promoter are labeled C1H (HA-epitope tag) and C2. The experiment was repeated twice for a total of eight mice. (B) Growth of F9 parental, 73F9, and complemented strains was compared in host cells grown in either 10% or 0% serum. Growth was measured at 36 h after infection. The number of parasites in at least 50 vacuoles was determined by immunofluorescence. Shown is a representative experiment of three. The growth difference of the mutant with and without serum is statistically significant, using an unpaired Student's t test with a value of P < 0.0001.
The TgRCC1 Mutant Is Defective in Growth During Nutrient Limitation.
Our STM screen compared parasite growth in tissue culture versus mice; thus, mutants that are growth impaired during standard cell culture conditions would be eliminated from the pool during 22 days of serial passage. As expected, the 73F9 mutant grows similar to the F9 parental strain in normal tissue culture conditions (10% serum) (Fig. 4B). To examine whether the avirulent phenotype of the 73F9 mutant in mice was due to an overall defect in stress response, various in vivo stresses were mimicked in cell culture, including cyst formation, survival in macrophages, and growth during nutrient limitation. Similar to the F9 parental strain, 73F9 was able to effectively convert to cysts under alkaline conditions, and survived in activated macrophages (data not shown). To examine parasite growth under nutrient limitation in vitro, the fibroblast host cells were serum-starved for ≈12 h before infection with F9, 73F9, and complemented parasites. Interestingly, mutant parasites replicate at a reduced level compared with parental F9 and complemented strains under nutrient-limiting conditions (Fig. 4B). This suggests a mechanism of attenuation for the 73F9 strain, with parasites being unable to replicate as efficiently in a nutrient-deprived host environment.
Nuclear Trafficking Is Defective in the TgRCC1 Mutant.
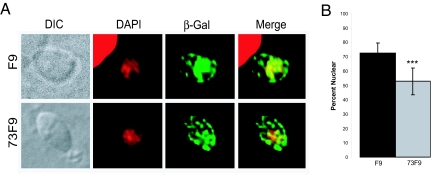
In response to nutrient limitation, many organisms traffic mRNA and regulatory proteins such as transcription factors, kinases, and RNA binding proteins into and out of the nucleus (30–33). Given that the 73F9 mutant has a reduced amount of TgRCC1 protein and RCC1 proteins are known to play critical roles in maintaining the proper gradients essential for nucleocytoplasmic transport, we examined nuclear trafficking in the 73F9 mutant. We used β-galactosidase containing an NLS (β-gal-NLS) (34) as our reporter of nuclear transport in mutant and parental parasites. Twelve hours after transient transfection with β-gal-NLS, the percentage of β-galactosidase within the nucleus was quantified and compared with that within the entire cell by immunofluorescence (Fig. 5). There is a significant decrease in nuclear abundance of β-galactosidase in the 73F9 mutant compared with parental parasites (Fig. 5B). This defect in nuclear trafficking of the 73F9 mutant is likely to impair its growth when it is switched from the nutrient-rich conditions of cell culture to nutrient-limiting conditions in vivo. We hypothesize that key regulatory factors are not able to be efficiently transported between the cytosol and nucleus, which causes an inadequate stress response.
Fig. 5.
73F9 parasites are defective in nuclear trafficking. (A) FLAG-tagged β-gal-NLS was transfected into either F9 or 73F9 parasites. Twelve hours after transfection, cells were fixed and incubated with anti-FLAG antibody to visualize the location of the reporter (green), and then mounted in DAPI to stain the nucleus (red). (B) The amount of β-galactosidase in the nucleus was divided by the total β-gal-NLS per parasite, producing the percentage of nuclear β-galactosidase (% nuclear). Measurements were taken from 25 parasites each of mutant and WT from two independent experiments. Results were statistically significant, using an unpaired Student's t test with a value of P < 0.0001.
Discussion
Identification of 39 Mutants Attenuated in Mice.
We report a large-scale forward genetics approach to identify virulence determinants in the protozoan parasite T. gondii. From screening an insertional library of 6,300 strains, we have identified 39 mutants which show a ≈10-fold decrease in cyst burden compared with wild-type Pru parasites. Unlike any other STM screen, we simultaneously passaged the pools of parasites in tissue culture throughout the entire mouse infection (22 days; schematized in SI Fig. 6). This comparison has allowed us to isolate genes essential only during growth in vivo. Mutants with growth defects in tissue culture would be outcompeted in the pool, eliminating previously identified virulence genes of T. gondii, such as CPSII or MIC2, which are unable to proliferate in standard cell culture conditions when disrupted (7, 11).
Within the 39 mutants we have identified, many are disrupted in genes that encode for proteins with predicted functions in other organisms, but they have not previously been linked to virulence. Further analysis of these proteins will expand our understanding of their known function and define their role in pathogenesis. Interestingly, approximately one-half of the disrupted genes encode proteins without predicted function. Most of these hypothetical proteins are conserved within other organisms. Defining their mechanism of virulence in T. gondii will provide new insight for other pathogens. Investigations of T. gondii pathogenesis will prove useful for the study of other intracellular pathogens, which may use similar mechanisms to maintain their specific niches. Additionally, it is likely that these early branching eukaryotes possess many unique virulence pathways that are divergent from bacterial pathogens that have been more extensively studied.
Identification of Unique RCC1 Proteins in Protozoan Parasites.
Although mutations of RCC1 have been associated with decreased mRNA export and processing, mating defects, and developmental abnormalities (26, 35), this study provides evidence of its role in pathogenesis. It is intriguing that protozoan parasites do not contain the well conserved RCC1 proteins. Because RCC1 proteins have been found to be essential in all organisms examined (26), it is likely that these atypical protozoan RCC1 proteins represent more rudimentary forms. This suggests that these protozoan precursors underwent domain compaction and duplication to evolve into the more condensed version that exists in later branching eukaryotes. With such large differences in protein structure between human and protozoan RCC1, these divergent orthologs may serve as unique drug targets.
RCC1 is so far the only characterized guanine exchange factor for the cellular GTPase Ran (36). We were initially surprised to find a disruption in TgRCC1 because these proteins are essential in other organisms. One possibility is that T. gondii contains multiple proteins capable of exchange activity for Ran. However, functional redundancy does not appear likely. As stated, all other T. gondii proteins that include RCC1 domains are not predicted to be nuclear localized and contain other domains indicative of alternative functions. The 73F9 avirulent mutant is disrupted in the TgRCC1 promoter, resulting in a drastic reduction but not complete loss of protein. Several unsuccessful attempts to create a TgRCC1 null mutant suggest that it cannot be compensated for. This low level of TgRCC1 is sufficient for growth in standard cell culture conditions, but is unable to support growth under nutrient limitation.
Efficient Nuclear Transport Is Critical During Nutrient Limitation in Vitro and in Vivo.
RCC1 is the guanine exchange factor for Ran that is critical for maintaining high levels of RanGTP in the nucleus. In the cytoplasm, the RanGTPase-activating protein (RanGAP) hydrolyses RanGTP to create a high concentration of RanGDP. The distribution of RanGTP/RanGDP determines loading and unloading of cargo transported either to or from the nucleus (37). We have shown here that the TgRCC1 mutant is defective in nuclear trafficking. Many model organisms rapidly transport regulatory molecules into and out of the nucleus during nutrient limitation (30–33). Our data suggest that T. gondii needs to efficiently traffic molecules into and out of the nucleus under nutrient deprivation, and that serum starvation in cell culture mimics at least some of the conditions that T. gondii encounters in vivo. Further studies should include an investigation of the molecules whose nuclear trafficking is critical during nutrient limitation in vitro and in vivo. Whether the trafficking of one or more specific proteins is required for pathogenesis, or whether the reduction in the overall rate of nuclear transport causes T. gondii to be vulnerable during host stress will need to be explored.
Further sequence analysis of protozoa shows that, even though most components of nuclear trafficking are conserved, the Ran network appears to be unique. Although there is a Ran ortholog, no protozoa encode for a protein with sequence similarity to characterized RanGAPs. This, in conjunction with the differences in RCC1, highlights the importance of investigating the protistan parasites for insights into the evolution of nuclear trafficking.
Materials and Methods
STM in Vivo Screen.
A total of 105 plates each containing ≈60 mutants was pooled and either i.p. injected into two CBA/J mice (JAX, Bar Harbor, ME) at a dose of 2 × 104 parasites per mouse, or grown in human foreskin fibroblast (HFF) cells by serial passage. Twenty-two days after infection, brains were harvested and ground by a tissue homogenizer. DNA was prepared from the infected brains and the parasites passed in HFFs by the TELT method (38) and compared by radiolabeled PCR and dot blot (19). Mutants that had a reduced hybridization signal in the two mice compared with growth in HFFs were selected as potential avirulent mutants and examined as clones in mice.
Acute and Chronic Mouse Infections.
For chronic infections, 8-week-old CBA/J mice received, i.p., 2 × 104 PrugniaudΔHPT, parental STM tag parasites, or potential avirulent mutants. Plaque assays were performed immediately after inoculation to ensure counting accuracy and extracellular stability of the mutants. Only experiments where the plaque counts for the mutants were within 2-fold of wild type were analyzed. Twenty-two days after inoculation (early chronic infection), the mice were killed. Their brains were stained and counted as described in ref. 20. For acute mouse infections, a range between 2 × 105 and 1 × 106 parasites was i.p. injected into CBA/J mice (National Cancer Institute, Charles River Laboratories, Frederick, MD). Plaque assays were performed immediately after the inoculation to confirm the number of viable parasites that were injected. Time of death was determined over 22 days. When mice were moribund (severely hunched and not moving), they were killed. For all mouse experiments, four mice were infected per strain and repeated at least two times for each concentration.
Measurement of T. gondii Growth in HFFs.
Lysed parasites were infected in triplicate in confluent HFF monolayers seeded on glass coverslips in a 24-well plate. For nutrient-limiting conditions, confluent HFF monolayers were grown for ≈12 h in DMEM without FBS before parasites were added. At 12, 24, and 36 h after parasite infection, monolayers were washed in PBS and fixed in 3% formaldehyde. Coverslips were permeabilized and blocked in 0.2% Triton X-100, 3% BSA in PBS. Antisera from chronically infected mice was used at a 1:500 dilution followed by 488-Alexa Fluor (Molecular Probes, Eugene, OR) goat anti-mouse secondary antibody at a 1:1,000 dilution and visualized by using a Zeiss (Oberkochen, Germany) inverted Axiovert 100 microscope. At least 50 vacuoles were counted and the number of parasites per vacuole 36 h after infection is shown in Fig. 4. Experiments were repeated at least three times.
Nuclear Transport Assay.
73F9 and F9 parasites were transiently transfected with 75 μg of β-gal-NLS (34) and allowed to invade for 4 h before changing the media from 10% to 0% serum supplementation. Twelve hours after transfection, cells were fixed, stained by using the anti-FLAG antibody, and mounted in VectaShield with DAPI (supplemental). Images were captured and pseudocolored (green for β-galactosidase and red for DAPI) as described above, and no alterations (e.g., deconvolution) were performed before quantification. The nucleus was marked by DAPI in the red channel, and then the amount of fluorescence in the green channel was calculated by multiplying the mean pixel intensity by the number of pixels within the selected area. This number represents the total fluorescence intensity of FLAG-tagged β-gal-NLS in the nucleus (39). We then marked the entire parasite and calculated the total β-gal-NLS by identical methods. Background was subtracted from both calculations and the nuclear β-galactosidase was divided by the total intensity to determine the percentage of β-galactosidase within the nucleus. This value was determined for 25 cells each for 73F9 and F9 parasites from two separate transfections.
Mutagenic Insertion Identification, Plasmids, and RNA and Protein Analysis.
Complete details are provided in the SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank William Sullivan (Indiana University School of Medicine, Indianapolis, IN) for providing the β-gal-NLS expression plasmid; David Sibley (Washington University School of Medicine, St. Louis, MO) for the anti-β-tubulin antibody; Jay Bangs for the use of the Zeiss Axioplan IIi; John Boothroyd, Rob Striker, Ned Ruby, and Margaret McFall-Ngai for critical reading of the manuscript; Casey F. Scott-Weathers and Jeremy J. Johnson for excellent technical assistance; and all members of the L.J.K. laboratory for their contributions to Table 1. This research was supported by the Burroughs Wellcome Fund Career Award 992908 and National Institutes of Health Awards A1054603 and AI41014.
Abbreviations
- STM
signature-tagged mutagenesis
- RCC1
regulator of chromosome condensation 1
- NLS
nuclear localization signal
- HA
hemagglutinin
- β-gal-NLS
β-galactosidase containing an NLS
- HFF
human foreskin fibroblast.
Footnotes
This article is a PNAS Direct Submission.
The authors declare no conflict of interest.
Data deposition: The sequence reported in this paper has been deposited in the National Center for Biotechnology Information database (accession no. EF591127).
This article contains supporting information online at www.pnas.org/cgi/content/full/0701893104/DC1.
References
- 1.Mandell GL, Bennett JE, Dolin R. In: Principles and Practice of Infectious Diseases. Hartman T, Dudlick M, editors. Philadelphia: Elsevier; 2005. pp. 3095–3319. [Google Scholar]
- 2.Roos DS, Donald RG, Morrissette NS, Moulton AL. Methods Cell Biol. 1994;45:27–63. doi: 10.1016/s0091-679x(08)61845-2. [DOI] [PubMed] [Google Scholar]
- 3.Kim K, Weiss LM. Int J Parasitol. 2004;34:423–432. doi: 10.1016/j.ijpara.2003.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dubey JP. J Am Vet Med Assoc. 1994;205:1593–1598. [PubMed] [Google Scholar]
- 5.Tenter AM, Heckeroth AR, Weiss LM. Int J Parasitol. 2000;30:1217–1258. doi: 10.1016/s0020-7519(00)00124-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dubey JP. Int J Parasitol. 1998;28:1019–1024. doi: 10.1016/s0020-7519(98)00023-x. [DOI] [PubMed] [Google Scholar]
- 7.Fox BA, Bzik DJ. Nature. 2002;415:926–929. doi: 10.1038/415926a. [DOI] [PubMed] [Google Scholar]
- 8.Dzierszinski F, Mortuaire M, Cesbron-Delauw MF, Tomavo S. Mol Microbiol. 2000;37:574–582. doi: 10.1046/j.1365-2958.2000.02014.x. [DOI] [PubMed] [Google Scholar]
- 9.Mercier C, Howe DK, Mordue D, Lingnau M, Sibley LD. Infect Immun. 1998;66:4176–4182. doi: 10.1128/iai.66.9.4176-4182.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ding M, Kwok LY, Schluter D, Clayton C, Soldati D. Mol Microbiol. 2004;51:47–61. doi: 10.1046/j.1365-2958.2003.03823.x. [DOI] [PubMed] [Google Scholar]
- 11.Huynh MH, Carruthers VB. PLoS Pathog. 2006;2:e84. doi: 10.1371/journal.ppat.0020084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taylor S, Barragan A, Su C, Fux B, Fentress SJ, Tang K, Beatty WL, Hajj HE, Jerome M, Behnke MS, et al. Science. 2006;314:1776–1780. doi: 10.1126/science.1133643. [DOI] [PubMed] [Google Scholar]
- 13.Saeij JP, Boyle JP, Coller S, Taylor S, Sibley LD, Brooke-Powell ET, Ajioka JW, Boothroyd JC. Science. 2006;314:1780–1783. doi: 10.1126/science.1133690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hensel M, Shea JE, Gleeson C, Jones MD, Dalton E, Holden DW. Science. 1995;269:400–403. doi: 10.1126/science.7618105. [DOI] [PubMed] [Google Scholar]
- 15.Chiang SL, Mekalanos JJ. Mol Microbiol. 1998;27:797–805. doi: 10.1046/j.1365-2958.1998.00726.x. [DOI] [PubMed] [Google Scholar]
- 16.Edelstein PH, Edelstein MA, Higa F, Falkow S. Proc Natl Acad Sci USA. 1999;96:8190–8195. doi: 10.1073/pnas.96.14.8190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cormack BP, Ghori N, Falkow S. Science. 1999;285:578–582. doi: 10.1126/science.285.5427.578. [DOI] [PubMed] [Google Scholar]
- 18.Nelson RT, Hua J, Pryor B, Lodge JK. Genetics. 2001;157:935–947. doi: 10.1093/genetics/157.3.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knoll LJ, Furie GL, Boothroyd JC. Mol Biochem Parasitol. 2001;116:11–16. doi: 10.1016/s0166-6851(01)00295-x. [DOI] [PubMed] [Google Scholar]
- 20.Mordue DG, Scott-Weathers CF, Tobin CM, Knoll LJ. Mol Microbiol. 2007;63:482–496. doi: 10.1111/j.1365-2958.2006.05538.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bischoff FR, Ponstingl H. Nature. 1991;354:80–82. doi: 10.1038/354080a0. [DOI] [PubMed] [Google Scholar]
- 22.Bischoff FR, Ponstingl H. Proc Natl Acad Sci USA. 1991;88:10830–10834. doi: 10.1073/pnas.88.23.10830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tachibana T, Imamoto N, Seino H, Nishimoto T, Yoneda Y. J Biol Chem. 1994;269:24542–24545. [PubMed] [Google Scholar]
- 24.Nemergut ME, Mizzen CA, Stukenberg T, Allis CD, Macara IG. Science. 2001;292:1540–1543. doi: 10.1126/science.292.5521.1540. [DOI] [PubMed] [Google Scholar]
- 25.Renault L, Nassar N, Vetter I, Becker J, Klebe C, Roth M, Wittinghofer A. Nature. 1998;392:97–101. doi: 10.1038/32204. [DOI] [PubMed] [Google Scholar]
- 26.Dasso M. Trends Biochem Sci. 1993;18:96–101. doi: 10.1016/0968-0004(93)90161-f. [DOI] [PubMed] [Google Scholar]
- 27.Wu J, Matunis MJ, Kraemer D, Blobel G, Coutavas E. J Biol Chem. 1995;270:14209–14213. doi: 10.1074/jbc.270.23.14209. [DOI] [PubMed] [Google Scholar]
- 28.Yaseen NR, Blobel G. Proc Natl Acad Sci USA. 1999;96:5516–5521. doi: 10.1073/pnas.96.10.5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakielny S, Shaikh S, Burke B, Dreyfuss G. EMBO J. 1999;18:1982–1995. doi: 10.1093/emboj/18.7.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li H, Tsang CK, Watkins M, Bertram PG, Zheng XF. Nature. 2006;442:1058–1061. doi: 10.1038/nature05020. [DOI] [PubMed] [Google Scholar]
- 31.Ueta R, Fukunaka A, Yamaguchi-Iwai Y. J Biol Chem. 2003;278:50120–50127. doi: 10.1074/jbc.M305046200. [DOI] [PubMed] [Google Scholar]
- 32.Cox KH, Kulkarni A, Tate JJ, Cooper TG. J Biol Chem. 2004;279:10270–10278. doi: 10.1074/jbc.M312023200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qin J, Kang W, Leung B, McLeod M. Mol Cell Biol. 2003;23:3253–3264. doi: 10.1128/MCB.23.9.3253-3264.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bhatti MM, Sullivan WJ., Jr J Biol Chem. 2005;280:5902–5908. doi: 10.1074/jbc.M410656200. [DOI] [PubMed] [Google Scholar]
- 35.Shi WY, Skeath JB. Dev Biol. 2004;270:106–121. doi: 10.1016/j.ydbio.2004.02.011. [DOI] [PubMed] [Google Scholar]
- 36.Bischoff FR, Krebber H, Kempf T, Hermes I, Ponstingl H. Proc Natl Acad Sci USA. 1995;92:1749–1753. doi: 10.1073/pnas.92.5.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Macara IG. Microbiol Mol Biol Rev. 2001;65:570–594. doi: 10.1128/MMBR.65.4.570-594.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Medina-Acosta E, Cross GA. Mol Biochem Parasitol. 1993;59:327–329. doi: 10.1016/0166-6851(93)90231-l. [DOI] [PubMed] [Google Scholar]
- 39.Pu RT, Dasso M. Mol Biol Cell. 1997;8:1955–1970. doi: 10.1091/mbc.8.10.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.