Abstract
We generated high-resolution maps of histone H3 lysine 9/14 acetylation (H3ac), histone H4 lysine 5/8/12/16 acetylation (H4ac), and histone H3 at lysine 4 mono-, di-, and trimethylation (H3K4me1, H3K4me2, H3K4me3, respectively) across the ENCODE regions. Studying each modification in five human cell lines including the ENCODE Consortium common cell lines GM06990 (lymphoblastoid) and HeLa-S3, as well as K562, HFL-1, and MOLT4, we identified clear patterns of histone modification profiles with respect to genomic features. H3K4me3, H3K4me2, and H3ac modifications are tightly associated with the transcriptional start sites (TSSs) of genes, while H3K4me1 and H4ac have more widespread distributions. TSSs reveal characteristic patterns of both types of modification present and the position relative to TSSs. These patterns differ between active and inactive genes and in particular the state of H3K4me3 and H3ac modifications is highly predictive of gene activity. Away from TSSs, modification sites are enriched in H3K4me1 and relatively depleted in H3K4me3 and H3ac. Comparison between cell lines identified differences in the histone modification profiles associated with transcriptional differences between the cell lines. These results provide an overview of the functional relationship among histone modifications and gene expression in human cells.
A comprehensive understanding of the operation of the human genome will require definition of all the functional elements contained within the DNA sequence and a description of their utilization in normal and diseased cells. Progress on definition of the protein coding elements of the genome has been substantial (International Human Genome Sequencing Consortium 2004; Maeda et al. 2006) and we are now beginning to recognize the existence of many non-protein coding genes (Katayama et al. 2005; Mattick and Makunin 2006). However, systematic identification of DNA elements involved in the regulation of gene expression has only recently been the concern of high throughput approaches in mammals (Schübeler et al. 2004; Bernstein et al. 2005; Kim et al. 2005; Roh et al. 2005; Bernstein et al. 2006; Prabhakar et al. 2006). It remains an open question as to how best to identify such elements.
In its native form within the cell the human genome is packaged with histones and other proteins into chromatin. Many studies over recent years in yeast and also in mammals have identified a wide range of post-translational modifications to the N-terminal tails of the histones in chromatin (Jenuwein and Allis 2001; Martin and Zhang 2005). These include a series of methylations and acetylations at defined lysine and arginine residues. A growing literature is defining the mechanisms for addition and removal of the modifications catalyzed by a range of methyl- (Kouzarides 2002; Martin and Zhang 2005) and acetyl-transferases (Roth et al. 2001), deacetylases (Kurdistani and Grunstein 2003), and most recently demethylases (Shi et al. 2004; Cloos et al. 2006; Klose et al. 2006). Lysine acetylation can occur on histones H2A, H2B, H3, and H4 and is generally associated with activation of transcription via the neutralization of the positive charge of the lysine residues, so reducing the affinity of histones for DNA and opening the chromatin (Wolffe and Pruss 1996; Grunstein 1997; Kurdistani et al. 2004). Recent discoveries indicate that histone acetylation correlates with transcriptional activation and that histone deacetylation correlates in many cases with repression in yeast (Robyr et al. 2004). Histone lysine methylation is more complex in that there are 24 known sites of methylation on histones (17 lysine residues and seven arginine residues) with up to three methyl groups, and the methylation state at different residues is associated with opposite effects on transcription. For instance, histone H3 lysine 4 di- (H3K4me2) and tri-methylation (H3K4me3) are associated with positive regulation of transcription and recruitment of chromatin remodeling factors and histone acetyltransferases (Bernstein et al. 2002; Santos-Rosa et al. 2002; Ng et al. 2003). In particular H3K4me3 was found to be associated with the promoter and 5′-coding regions of active genes in yeast (Bernstein et al. 2002; Santos-Rosa et al. 2002) and higher eukaryotes (Schneider et al. 2004; Bernstein et al. 2005), whereas H3K4me2 appeared on active and inactive genes in yeast (Santos-Rosa et al. 2002). Conversely methylation at H3K9 and H3K27 are generally associated with heterochromatin formation associated with HP1 (Lachner et al. 2001) and silencing of transcription through the polycomb group proteins (Cao et al. 2002), respectively. However H3K9me2 and me3 have also recently been shown to be associated with transcription elongation with active genes (Vakoc et al. 2005).
The wide range of histone modifications and the definition of specific effects associated with individual residues has led to the proposal of the histone code hypothesis (Jenuwein and Allis 2001), postulating that the specific pattern of histone post-translational modifications in a locus extends the information conveyed by the genomic code. The ‘‘histone code’’ would be formed by histone modifications, on one or more tails, acting sequentially or in combination. This code would be read by proteins containing specific interacting domains such as chromo- and bromo-domain proteins to initiate biological processes such as transcriptional activation or repression, chromosome condensation, and DNA repair (de la Cruz et al. 2005). In principle the mixture of histone modifications may be the same over large chromatin regions, but the specific occurrence of modifications on nucleosomes could create local structures, leading to functional diversity and thus defining chromatin subdomains. The existence of a code in the strictest sense is still the matter of debate, but it is evident that the modification status at nucleosomes or regions is at least a major factor in the regulation of chromatin structure and transcription. To investigate this further in higher eukaryotes, Bernstein et al. (2005) used genomic tiling oligonucleotide arrays and chromatin immunoprecipitation on microarrays (ChIP-chip) to map H3K4me2 and me3 and lysine 9/14 acetylation across regions of human chromosomes 21 and 22 in human and the orthologous regions in mouse and observed punctate modification sites with H3K4me3 correlating with transcription starts. A later study identified overlapping domains of activating (H3K4me3) and repressive (H3K27me3) histone marks at developmentally important loci in ES cells (Bernstein et al. 2006). Roh et al. (2005) used a genome-wide sequencing-based approach to demonstrate that histone H3 lysine 9/14 acetylation (H3ac) was found in gene rich regions and that hyperacetylation at promoters was correlated with active genes in resting and activated T cells. In Drosophila, Schübeler et al. (2004) found active genes associated with hyperacetylation of H3 and H4 and hypermethylation of lysine 4 and lysine 79 genome-wide, albeit at lower resolution using cDNA expression arrays. In this way, establishing high-resolution maps of the state of histone tail modifications in human cells can provide insights on the regulatory state of chromatin.
The ENCyclopedia of DNA Elements (ENCODE) Project is a community resource project which aims to identify functional elements within 1% (30 Mb) of the human genome through the application of diverse methods (The ENCODE Project Consortium 2004). In order to pilot the generation of high-resolution maps of histone tail modifications which might localize functional elements within the genome, we have applied ChIP-chip using the well studied histone H3 and H4 modifications H3ac, histone H4 acetylation at lysine 5, 8, 12, 16 (H4ac) and histone H3 at lysine 4 mono-, di-, and trimethylation (H3K4me1, H3K4me2, H3K4me3, respectively) across the ENCODE genomic regions in five human cell lines. The resulting maps indicated clear patterns of histone modifications across the genome. H3K4me3, H3K4me2, and H3ac were tightly associated with the transcriptional start sites (TSSs) of genes, while H3K4me1 and H4ac have more broad distributions including some extensive regions. At TSSs there are characteristic patterns of modifications, in relation to both the type of modification and the position relative to the TSS, and these patterns differ between active and inactive genes. In particular, expressed genes had distinct peaks of H3K4me2, H3K4me3, and H3ac modification downstream from the TSS. H3K4me1 signal was low but showed some evidence of enrichment further downstream from the TSS than H3K4me2 and H3K4me3. For non-expressed genes, the pattern was strikingly different with low signals of each of the modifications but with some residual signal for H3K4me2 and H3K4me3 centered on the TSS. Histone modification sites away from TSSs showed enrichment of H3K4me1. The histone modification profiles showed differences between cell lines associated with differences in gene transcription. In this way we demonstrate that maps of histone modifications can be generated with high throughput and used as a generic tool to identify functional elements in the genome.
Results and Discussion
The ENCODE PCR tiling microarray and ChIP-chip
Single-stranded DNA derived from double-stranded PCR products specifically immobilized on microarrays via 5′ aminolinks allows high-sensitivity detection of genome copy number changes (Dhami et al. 2005) and chromatin immunoprecipitation (ChIP) enrichment (P. Dhami, A.W. Bruce, J.H. Jim, S.C. Dillon, A. King, J.L. Cooper, R.M. Andrews, P.D. Ellis, C. Langford, and D. Vetrie, in prep.). In order to identify DNA sequences associated with modified histones by ChIP-chip, we designed and fabricated such a microarray representing the 44 ENCODE regions which in its final version consisted of 24,005 PCR fragments with an average size of 1024 bp (average non-overlapping tile length = 992 bp) (see Supplemental Table 1 and http://www.sanger.ac.uk/PostGenomics/encode/data-access.shtml). The array covers ∼80% of the targeted regions. This array was used to detect sequences enriched by ChIP without DNA amplification in ChIP-chip experiments across five human cell lines. ChIP-chip experiments were performed separately in at least three biological replicates with one or two technical replicates for antibodies specific to H3K4me1, H3K4me2, H3K4me3, H3ac, and H4ac (see Supplemental Fig. S1 for validation of antibody specificity) in five different cell lines (Table 1). These included the ENCODE Consortium common cell lines GM06990 (lymphoblastoid) and HeLa-S3, as well as K562 (erythroleukemic cell line), HFL-1 (fetal lung fibroblast cell line), and MOLT4 (acute lymphoblastic leukemia CD4+ T cell line). Histone modification profiles for each antibody/cell line combination across the ENCODE regions were determined by hybridization of ChIP-enriched DNA to the ENCODE PCR product tiling array as compared to the ChIP input DNA with repeat suppression by Cot1 DNA.
Table 1.
Combinations of antibodies and cell lines with replicate information
B refers to the number of biological replicates, T refers to the number of technical replicates. Technical replicates were spread evenly between biological replicates.
All replicates were submitted to ArrayExpress (ArrayExpress: E-MEXP-269, E-TABM-140, http://www.ebi.ac.uk/arrayexpress/) (Parkinson et al. 2005) and are also available from our website at http://www.sanger.ac.uk/PostGenomics/encode/data-access.shtml. After normalization, replicates were combined and expressed as the median enrichment per PCR product on the microarray. These histone modification profiles are available through the UCSC genome browser at http://genome.ucsc.edu/ENCODE/encode.hg17.html under the “Sanger ChIP” tab.
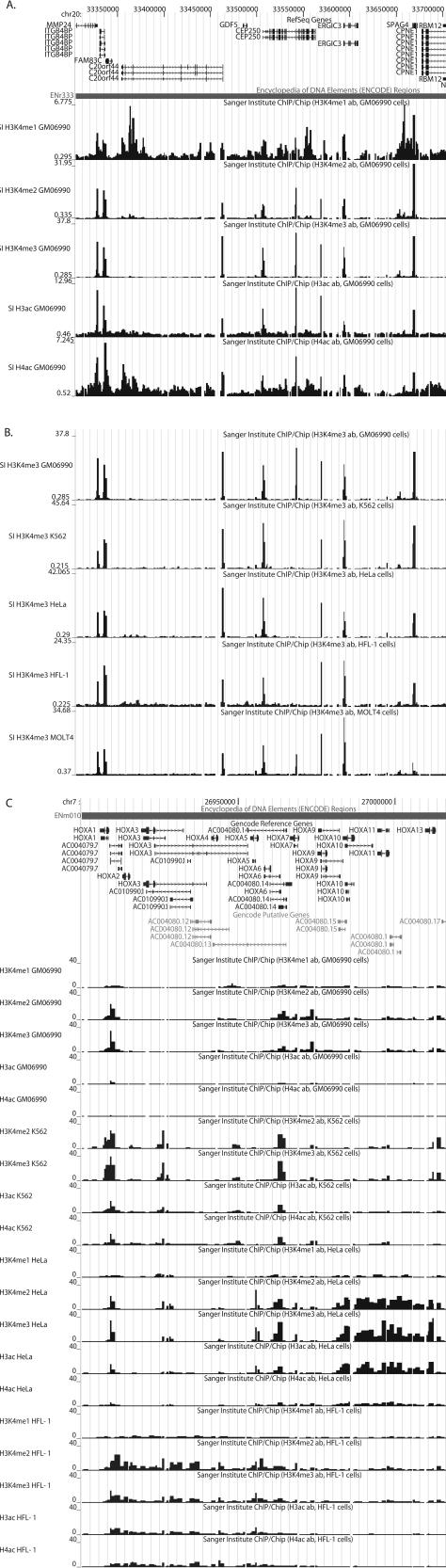
Typical data for the full set of antibodies across one ENCODE region is shown in Figure 1A while a comparison between the five cell lines for H3K4me3 is shown in Figure 1B.
Figure 1.
Example of histone modification profiles across ENCODE regions. (A) Screenshot from the UCSC genome browser (Hinrichs et al. 2006) of ENCODE region ENr333 (human chromosome 20: 33,304,929–33,804,928 bp, NCBI 35) showing ChIP-chip data for the lymphoblastoid cell line, GM06990, using five antibodies for the histone modifications H3K4me1, H3K4me2, H3K4me3, H3ac, and H4ac. The scale in base pairs is indicated by the vertical ticks at the top. The top track shows the UCSC known genes (Hsu et al. 2006) with transcriptional orientation and exons indicated by arrows and vertical ticks, respectively. Below is a track indicating the extent of the ENCODE region ENr333. ChIP-chip data are displayed in the five subsequent tracks as the median value of the ratio of normalized ChIP-chip sample fluorescence to input DNA fluorescence. Each black vertical bar is the enrichment measured at a single amplicon on the ENCODE PCR product microarray with the enrichment represented by the height of the bar. Five tracks represent the data for the five antibodies used in ChIP-chip as indicated by the label at the left of each track. Note that each track is dynamically scaled according to the data displayed, and hence comparison between tracks must take into account the enrichment scale at the left of each data track. (B) Screenshot as in A of ENCODE region ENr333 for ChIP-chip data using the antibody for H3K4me3 with the five cell lines as indicated at the left of the data tracks. The screenshot is aligned to the scale in panel A. Note that the GM06990 data are the same as are displayed in the third data track in panel A. (C) Screenshot of ChIP-chip data for ENCODE region ENm010 (the HOXA cluster on human chromosome 7: 26,730,761–27,230,760bp, NCBI35). At the top is the GENCODE reference gene annotation (Harrow et al. 2006). Data tracks are shown as in panels A and B for all ChIP-chip data on cell lines GM06990, K562, HeLa-S3, and HFL-1. Note the browser is zoomed in to show the HOXA cluster and does not show the full extent of ENm010.
In order to validate the ChIP-chip data, we identified and developed working assays for 74 enriched regions and 27 background signal regions for testing by quantitative PCR (qPCR) on anti-H3K4me3 ChIP material from the GM06990 cell line. Regions for testing were chosen arbitrarily but so as to sample enrichments between 1–2, 2–3, 3–4, 4–5 and >5 standard deviations (s.d.) away from the median signal in the ChIP-chip profile. The enriched sample was found to be significantly above the background sample (Mann-Whitney test, P < 0.00001) as were all the individual s.d. intervals (Mann-Whitney test, P < 0.005). In addition, by designing qPCR assays tiling through six representative ChIP-chip enrichment peaks, we were able to reproduce qualitatively the enrichment profiles observed on the microarray (data not shown).
Roh et al. (2005) have previously determined H3ac profiles across the whole genome using a tag sequencing approach in active and resting T cells. As a further validation step we compared our H3ac data set from the MOLT4 acute lymphoblastic leukemia CD4+ T cell line with these H3ac resting and activating T cell data sets (data obtained from the authors by request) within the ENCODE regions. Using a simple threshold of 2 s.d. from the mean to determine enriched signals in each data set and merging the Roh et al. data onto our microarray coordinates, we found that 62.9% and 61.7% of the enriched fraction of the MOLT4 data overlapped with the Roh et al. resting and activated T cell enrichments, respectively. Thus, despite the use of different cells, there is substantial concordance of the H3ac modification patterns between the two methods.
Histone modification profiles in the lymphoblastoid cell line GM06990
Initially we examined the histone modification profile data from a single ENCODE Consortium common cell line, the lymphoblastoid line GM06990. All observations were consistent across the other cell lines. Examination of the data in the genome browser (Fig. 1) shows that substantial enrichments for H3K4me2, H3K4me3, and H3ac are predominantly confined to sharp peaks and that many of these lie at the TSSs of annotated genes. Enrichments for H4ac and H3K4me1 can also be found at these peaks but in addition have a more widespread distribution with respect to genes. In order to investigate fully the distribution of the histone modification signals, a Hidden Markov Model (HMM) algorithm specifically designed for PCR tiling arrays with dual color hybridizations, multiple replicates, and histone modifications (P. Flicek, C.M. Koch, I. Dunham, and E. Birney, in prep.) was used to identify regions of significant enrichment within the profiles. This two-state HMM partitions the data into locations that are either consistent or inconsistent with antibody binding, and outputs a set of histone modification sites and centers with their probabilities. The advantages of using the HMM are that it incorporates across replicates simultaneously by complex state models and utilizes the sequential nature of the data on genomic tiling arrays so that adjacent tiles are expected to have related enrichments depending on binding site location, i.e., the binding sites have width. Thus the HMM can provide higher sensitivity than a simple cutoff approach.
Analysis of the GM06990 ChIP-chip data with the HMM identified numerous sites of histone modification across the ENCODE regions as shown in Table 2. The HMM identified sites with a median size of 5.2 kb with H4ac having a slightly larger median size at 6.0 kb. In addition there were a small number of larger regions where the HMM identified enriched histone modification over several tens of kilobases, particularly for H3K4me1. For instance, the largest region called for H3K4me1 was over 85 kb and located on chromosome 7 (chr7:116223757–116309020). Inspection of this region showed that there was a generally raised H3K4me1 signal, with some finer peak structure that the HMM in its current implementation does not deconvolute. This phenomenon was also noticeable in the other cell lines. In the HeLa-S3 profiles, there was a large region highly enriched for H3K4me2, H3K4me3, and H3ac across ∼27 kb covering the HOXA10, HOXA11, and HOXA13 genes (Fig. 1C). Others (Bernstein et al. 2005; Guenther et al. 2005) have also found large, cell type-specific Lys4 methylated regions that overlay multiple HOX genes and have suggested that these represent active chromatin domains involved in maintaining HOX gene expression. However, it is noticeable that, in three of the cell lines we examined (K562, GM06990, MOLT4), the modification profiles have more discrete peaks similar to other regions of the genome. These are associated with minimal expression of all the HOXA genes in these cells as judged by Affymetrix U133 plus 2.0 expression arrays. The relevance of these more specific peaks of modification in the HOXA gene cluster remains to be established, but an intriguing feature of the GENCODE annotation (Harrow et al. 2006) in this region is the presence of numerous non-coding transcripts, some of which are antisense to the HOXA genes.
Table 2.
Descriptive statistics for HMM calls for GM06990 ChIP-chip data
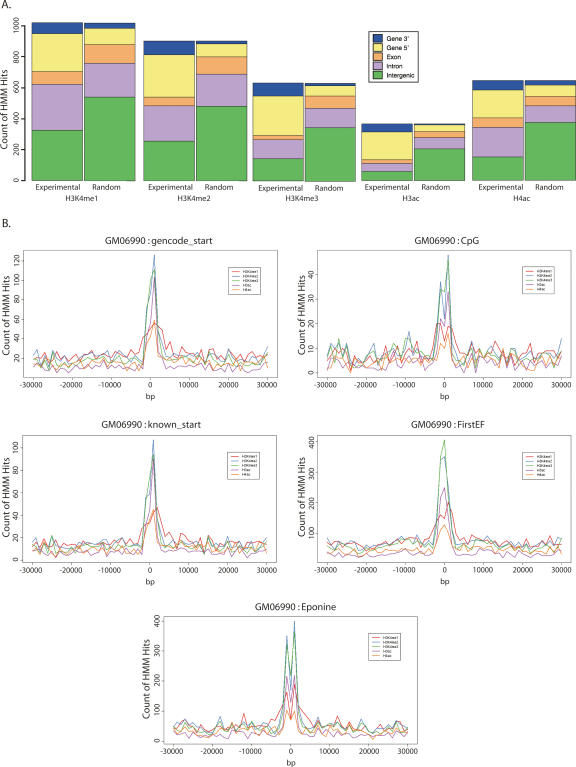
To investigate the utility of the histone modification map in identifying functional elements, we next analyzed the distribution of the histone modification sites as identified by the HMM relative to gene features by examining the overlap of each site with exons, introns, 5′ ends, 3′ ends, and intergenic regions using a window approach (Fig. 2A; Supplemental Table S2). Analyzing a ±2-kb window around the 5′ end of a gene, we found that sites for all antibodies are enriched at gene starts as compared to the simulated random distribution (Supplemental Table S2). Similarly, enrichments of HMM sites was also found at 3′ ends, while a depletion of HMM sites from intergenic regions was observed. This is consistent with previous observations of concordance of Lys4-methylated and acetylated histone sites with TSSs (Liang et al. 2004; Bernstein et al. 2005; Kim et al. 2005).
Figure 2.
Distribution of histone modification sites in the lymphoblastoid cell line, GM06990. (A) The plot shows the number of histone modification sites identified by the HMM which overlap with an exon (orange), intron (mauve), gene 5′ end (yellow), gene 3′ exon (blue), or intergenic sequence (green) for the lymphoblastoid cell line, GM06990, in the ChIP-chip data (left panel) or in random simulated data (right panel). Random data were simulated by generating sites of the same size distribution as the experimental data and placing them at random among the ENCODE regions represented by the PCR product microarray. This was repeated 100 times, and the mean frequencies of overlap were plotted. (B) Distribution of histone modification sites with respect to gene starts. The distance to the nearest gene start for histone modification sites identified by the HMM relative to GENCODE annotation (gencode_start), UCSC known gene starts (known_start), CpG islands (CpG), FirstEF gene start predictions (FirstEF), and Eponine predictions (Eponine) was determined. The plot shows the frequency of distances to the nearest gene start for H3K4me1 (red), H3K4me2 (blue), H3K4me3 (green), H3ac (mauve), and H4ac (orange) in 1-kb windows over ±30 kb from a start.
To refine the analysis of the propensity for histone modification sites to occur at TSSs, we examined the distance between each HMM called histone modification site and 5′ ends of genes as identified by the GENCODE annotation group (Harrow et al. 2006), RefSeq (Pruitt et al. 2005), or Ensembl (Birney et al. 2006). In addition we examined the distance from CpG islands and promoter predictions by First EF (Davuluri et al. 2001) and Eponine (Down and Hubbard 2002). Figure 2B shows that there was a strong enrichment for all the activating histone marks at the 5′ ends of genes. Specifically, considering the GENCODE gene annotation start sites, there is an enrichment for H3K4me2, H3K4me3, and H3ac signals just downstream from the TSS. The positive predictive values with respect to TSSs of these modifications are considerable although at low sensitivity because of the frequency of other HMM sites for the modifications outside of TSSs (see Supplemental Table S3). H4 signals were less enriched but with the same tendency, while H3K4me1 signals spread further around both sides of the TSS although still with a marked polarity toward the downstream side. Similar distributions were seen for RefSeq genes and FirstEF predictions. The distributions for CpG islands and Eponine were slightly different with a biphasic signal around the TSS due to the lack of strand specificity relative to the true TSS in these predictions. However, this biphasic distribution actually serves to emphasize the downstream location of the modification signals and relative depletion directly at the TSS (see below).
Analysis of the combinatorial relationship between histone modification profiles
We next analyzed the overlap of the individual histone modification sites in the GM06990 data by identifying cases of overlap between sites detected by the HMM for each antibody (Fig. 3). Using a 2.5-kb window around each modification site, we examined whether each site overlapped with any other modification site. Although this approach is relatively simplistic, it served to reveal interesting features of the distribution of histone modification sites in the first instance. For the GM06990 data the most frequent combinatorial site found (15.7%) contains modification by all five antibodies (code 11111, Fig. 3A), with sites involving only H3K4me1 (code 10000) being the next most frequent (10.9%). The observation of many H3K4me1-only sites is intriguing and further analysis (see below) indicates that many of these sites lie away from TSSs. It is also apparent that H3K4me3-only sites (code 00100) occur rarely and that H3K4me3 is usually associated with H3K4me2 as has been found previously (Bernstein et al. 2005).
Figure 3.
The coincidences of histone modification sites in the lymphoblastoid cell line, GM06990. (A) The frequency of occurrences of overlapping histone modification sites. The histogram shows the distribution of each type of overlapping histone modification site expressed as a percentage. Histone sites were defined as overlapping if they occurred within a 5-kb window centered on the site. Combinations are indicated as a five-digit binary code where 1 and 0 represent the presence or absence of each modification at a site, respectively, in the order H3K4me1:H3K4me2:H3K4me3:H3ac:H4ac. Combinations for which no sites were found are not shown. (B) Pie chart of occurrences of overlapping histone H3K4 methylation sites. (C) Box plot showing the distribution of distances to the nearest transcriptional start site (TSS) for the main combinations of histone H3K4 methylation sites. The box and horizontal line show the interquartile range and median of the data, while the whiskers extend to 1.5× the interquartile range of the distribution with open circles being the outliers of the distribution of distances from the TSS for each modification pattern.
Further investigation of the specific patterns of H3K4 methylation showed that the most common site shares histones modified at lysine 4 in mono-, di-, and trimethylated forms (Fig. 3B). The next most common forms are jointly mono- and dimethylated sites, monomethylated only, and jointly di- and trimethylated sites. Similarly sites with only di- or only trimethylated H3K4 occur rarely, and mono- and trimethylation sites without dimethylation are almost negligible. This is consistent with a mechanism of sequential enzymatic addition (and removal) of methyl groups at H3K4 (Bernstein et al. 2002; Santos-Rosa et al. 2002; Wysocka et al. 2005). Although these analyses are intriguing, given the size of our microarray tiles and the fact that we used sheared chromatin, each of the sites detected will contain at least several nucleosomes and represents an average across a population of cells, so these combinations of modifications cannot be viewed as occurring at specific single locations. The analysis also does not take into account the quantitative nature of the enrichments at each site, and so treats all levels of enrichment equally, providing they satisfy the parameters of the HMM. We address this further with a quantitative analysis across sites below.
Differences in the histone modification profiles at transcription start sites and other locations
In order to gain insight into the detailed relationships of the histone marks studied, we further analyzed their positions with respect to genomics features. Concentrating initially on H3K4 methylation, we repeated the genomic location analysis with respect to genic features as above. This analysis indicated that the sites with mono-, di-, and trimethylation at H3K4 and those with di- and trimethylation were specifically enriched at gene 5′ ends (data not shown). Analysis of the distance from the nearest TSS for each subclass of lysine methylation sites identified that sites including H3K4me3 signal were significantly (1-sided t-test at 99% confidence: P-value = 0.03252) closer to TSSs than those sites that did not contain H3K4me3 (Fig. 3C). Thus it seems that there are at least two classes of composite histone modification site characterized by histone lysine methylation, one class close (within ±10 kb) to TSSs with H3K4me3 signal and the other class more distal without H3K4me3 signal.
This was confirmed by splitting the HMM-identified sites into those proximal to TSSs and others and plotting the average of the Z-scored histone modification profile at the HMM center for a ±10-kb window around all sites (Fig. 4A). At TSSs the histones have strongly enriched signals for H3K4me2, H3K4me3, and H3ac with slightly lower signals for H3K4me1 and H4ac. Away from TSSs an alternative modification pattern is seen with a relatively strong signal for H3K4me1 and intermediate levels of H3K4me2 and H4ac. These patterns are consistent with the known histone modification patterns at TSSs, but also confirm the presence of an additional set of distal sites. It is worth noting that these profiles are composites of many sites and it remains possible that further refinement of sites included may reveal additional complexity. It is also possible that studying more antibodies to other histone modifications might further refine these elements, for instance as has been seen for combinations of H3K4me3 and H3K27me3 in embryonic stem cells (Bernstein et al. 2006). A separate analysis which clustered the HMM sites by their composite histone modification profile revealed a similar division of sites (see Supplemental Fig. S3).
Figure 4.
The histone modification profile at TSSs and other sites. (A) The average Z-scored histone modification profile for ±10 kb surrounding each HMM-identified histone modification site split into sites at TSSs (within 5 kb) and sites >5 kb from a TSS. Histone modification signals are plotted as lines for the average Z-score over all HMM sites in each cluster with H3K4me1 (red), H3K4me2 (green), H3K4me3 (blue), H3ac (mauve), and H4ac (orange). (B) Heatmaps representing the histone modification enrichment signal over ±10 kb surrounding all TSSs across the ENCODE regions. Histone modification signal is scored on a red (not enriched) to yellow (highest enrichment) scale. Each horizontal line of the heatmap is the histone modification profile in 1-kb windows for a single gene. TSS and TSSs are ordered according to the level of gene expression determined by analysis of the Affymetrix U133 plus 2.0 data from the GM06990 cell line using the gcRMA package of Bioconductor. The scale is the distance from the TSS in bp using 1000-bp windows. A heatmap is presented for each of the five antibodies used as indicated above the panels.
In addition we plotted heatmaps representing the intensity of enrichment for each modification across TSSs for each antibody (Fig. 4B). Examination of the heatmaps for TSSs in Figure 4B reveals further substructure within the histone modification sites. For each of H3K4me2, H3K4me3, and H3ac, there is a clear peak of enriched signal (yellow) extending up to 1–2 kb downstream from the TSS particularly for the highly expressed genes which are placed toward the bottom of the heatmap. The signals for H3K4me1 and H4ac are more diffuse, although, at least for H4ac, they appear to be more directly centered over the TSS. This directionality to the enrichment signal was also implied in the site distribution plot (Fig. 2B) and is reminiscent of the gradient patterns seen in yeast for the same modifications at the 5′ ends of genes (Liu et al. 2005; Pokholok et al. 2005). These patterns were not evident from Figure 4A because of the centering of that plot on the HMM-identified site.
The demonstration that there is a characteristic pattern of histone modification at TSSs raises the possibility of utilizing this profile to identify additional unannotated TSSs. Although we have not yet applied this systematically to these data, visual inspection does identify sites which are associated with rare transcripts. For instance, we were able to identify a TSS histone modification site associated with a lymphoid cell specific antisense transcript in the CEP2 gene (Supplemental Fig. S4).
Histone modification profiles and transcription
To study the association between these histone modifications and gene transcription, we examined the histone modification profiles at TSSs in relation to the transcriptional activity of genes determined in each cell line by Affymetrix U133 plus 2.0 gene expression microarray analysis (see Methods). The heatmaps of enrichments for each modification across TSSs in Figure 4B are ordered by increasing level of transcription of each gene from top to bottom and show two points. First as the relative gene activity increases, the structure of the histone modification signal at the TSS becomes more discrete. Thus the H3K4me2, H3K4me3, H3ac, and H4ac signals were stronger (more yellow) from top to bottom within the heatmap and less dispersed. Second, for genes with higher expression levels, the H3K4me2, H3K4me3, and H3ac signals shifted downstream from the TSS. For genes that were not expressed or were expressed at low levels, the signal for these modifications was weaker but more centered on the TSS.
We then divided the TSSs into sets associated with expressed or non-expressed genes and plotted the average histone modification profile for all sites in each set expressed as the Z-score in Figure 5A. Expressed genes had distinct peaks of H3K4me2, H3K4me3, and H3ac modification 1–2 kb downstream from the TSS. H4ac signal was localized at the TSS while H3K4me1 signal was low but showed some evidence of enrichment further downstream from the TSS than H3K4me2 and H3K4me3. For non-expressed genes, the pattern was strikingly different with low signals of each of the modifications but with some residual signal centered on the TSS. Examination of the levels of histone H3 and H2B (Figure 5B) showed that expressed genes were partly depleted for nucleosomes over the TSS compared to non-expressed genes. Thus active genes in these human cell lines have a reduced nucleosome density at TSS with downstream H3K4me3 and H3ac modifications, H3K4me2 modification slightly further downstream, and finally a slightly raised level of H3K4me1 extending further into the transcript consistent with previous observations for the promoters of yeast genes (Liu et al. 2005). Inactive genes have no nucleosome depletion but appear to have residual H3K4 lysine methylation directly over the TSS. This result could be due to averaging over many genes, some of which may have low, or temporally restricted, levels of transcription or incorrect gene expression assignment so that the inactive TSS signal is contaminated with some active genes. However it also raises the possibility that non-expressed genes have a low-level mark remaining from prior transcription or indicating that they are “primed” for transcription, and, as genes are activated, the histone modification becomes increased by the activity of enzymes such as Set1 associated with RNA polII and moved downstream from the TSS, concomitant with depletion of nucleosomes directly at the TSS. Histone acetylation is then also increased by recruitment of histone acetyltransferases. Assessment of the strength of the association between histone modifications profiles at TSSs and gene expression by estimating the predictive power showed that the histone modification profiles at the TSS were highly predictive of the transcript levels (Supplemental Table S4; Supplemental Fig. S5).
Figure 5.
The histone modification profile at TSSs for active and inactive genes. (A) The average Z-scored histone modification profile for ±10 kb surrounding each TSS of a gene split according to the expression level of the gene as determined by the GeneSpring MAS5 present (Expressed) and absent (Non-expressed) analysis of Affymetrix U133 plus 2.0 gene expression data from the GM06990 cell line. Histone modification signals are plotted as lines for the average Z-score over all HMM sites in each class with lines color-coded as in Figure 4A. (B) For comparison this plot shows the mean log2 value of the enrichment for ChIP-chip using antibodies to histone H3 and H2B over all ENCODE TSSs, split according to the expression level of the gene as determined by the GeneSpring MAS5 present (Expressed) and absent (Non-expressed) analysis of Affymetrix U133 plus 2.0 gene expression data for the K562 cell line.
Comparison of histone modification profiles between cell lines
Having created maps of histone modification profiles across the ENCODE regions for five cell lines, it is pertinent to ask how these maps compare between cell lines. Simple correlation analysis indicated that for the histone modifications with relatively well defined locations (H3K4me2, H3K4me3, and H3ac) there are modest to strong correlations between the data for different cell lines (see Supplemental Table S4). Inspection of the histone modification profiles within the genome browser (see Fig. 1B) showed that the positions of the peaks of enrichment tend to coincide across cell lines for H3K4me2, H3K4me3, and H3ac. The picture with the other modifications was less clear although the broad patterns of modification were similar. However we also identified signals which differentiate between cell lines. For instance, the LSP1 gene in the ENm011 region is a lymphocyte-specific gene expressed in the lymphoblastoid cell line GM06990 and had histone modification signal for H3K4me2, H3K4me3, and H3ac at the upstream TSS (data not shown). In K562 and HeLa-S3, where the gene was not expressed, this signal was greatly diminished. Similar differences can be found at other loci, e.g., the PHEMX gene, which is expressed in K562 cells but not in the other cell lines (data not shown). To identify further cell line-specific differences in histone modification of this sort, we used the gene expression profiles for each cell line to identify genes with differential expression in at least one cell line. We then extracted the histone modification profiles for ±10 kb at the TSSs of these genes and plotted these side by side for each cell line expressed as the Z-score. We then searched for cases where the characteristic histone modification profiles of active and inactive TSSs (Fig. 5) corresponded with the gene expression calls. In total we identified 85 cases of genes with differential gene expression (Supplemental Table S6). The Z-scored modification profiles of the cell lines for each gene were inspected and the concordance or non-concordance with the gene expression present/absence calls was determined. In 10 cases, insufficient data could be extracted from the ChIP-chip profiles in order to make a comparison. In 26 cases, the histone profiles at the TSSs were judged to be concordant with the gene expression profiles, either because the patterns were similar to those identified in Figure 5 or there was a change in the normalized level of one or more of the histone modifications, usually H3K4me3 or H3ac, e.g., the STEAP gene. Some examples of concordant patterns of histone modifications are shown in Supplemental Figure S6. In 23 further cases, the profiles were concordant except for one cell line. Thus inspection of the cell line ChIP-chip data can reveal tissue-specific promoter use. In the remaining cases the patterns could not be reconciled directly with the gene expression calls. This could be because of errors in the gene expression calling where genes are erroneously called as P or A, or it could be because there is additional complexity to the active and inactive histone profiles at some individual TSSs, which is not captured in the averaging approach we took. Alternatively distal sites away from the TSS such as non-promoter regulatory elements which currently we cannot identify could be responsible for tissue-specific differences.
Identification and characterization of cell line specificity of histone modifications
In order to study the cell line specificity of histone modifications more generally, we analyzed ChIP enrichment over background for all the replicates in four cell lines with a complete set of antibody data (not K562) and considered only enrichments common to all replicates from all experiments (see Supplemental Table S6). Enrichments were converted to Z-scores in order to allow comparison between cell lines. A multiclass Significance Analysis of Microarrays (SAM) statistic (Tusher et al. 2001) was used to identify enrichments at PCR products that show significant differences in their histone modification levels across the four cell lines. Enrichments that were differentially modified at a specific false discovery level (FDR) were further filtered to remove low but variable signals based on their average level of modification in each of the cell lines. Figure 6A shows the data for H3ac and the discrimination of PCR products showing enrichments that are cell line specific. The PCR products that do not show cell line-specific enrichment (green spheres) tended to align along the main diagonal while the products that are enriched in a cell line-specific manner (red spheres) had higher mean Z-scores in at least one of the four cell lines. Since many of the cell line-specific enrichments were close to each other, we clustered all the enrichments that were <200 bp apart into regions. Figure 6B shows the number of cell line-specific regions with respect to each of the five histone modifications after filtering. Interestingly, H3K4me1 and H4ac had the largest number of enriched regions that were cell line-specific at all FDR levels. Since H3K4me1 and H4ac are characteristic sites of modification distant from gene TSSs, it is tempting to speculate that these modifications identify non-promoter elements, some of which are cell line-specific. Conversely in these data although there are individual examples of modifications at TSS which are cell line-specific, there is less overall cell line specificity associated with the modifications associated with the TSSs of genes. Out of 13,407 tiles that were analyzed, about 14% (1890) were strongly cell line-specific for at least one modification (FDR = 0.0001, average Z-score in at least one cell line ≥1.5). Figure 6C shows the cell line specificity profile for these 1890 enrichment regions. Nearly 69% of the enrichment regions showed cell line specificity for only a single modification (40% expected by chance) while 20% had two modifications (34% expected by chance). Thus a sizeable proportion of the cell line-specific enrichments are unique to one modification. For each histone modification, Figure 6C shows the contribution of the cell lines to the specificity of the modifications. Surprisingly, for four of the modifications, most of the enrichments were cell line-specific based on the GM06990 cell line. This is probably because overall there were more sites identified in the data for this cell line. For H3K4me3, MOLT4 contributed specificity to nearly half of the enrichments.
Figure 6.
Identifying cell line specificity within histone modification profiles. (A) Four-dimensional plot showing raw data for H3ac for 13,407 PCR products on the microarray with data common to all experiments. Each sphere denotes a PCR product used in the analysis (13,407 spheres in this plot). The three axes denote the mean Z-score of H3ac modification in GM06990, HeLa-S3, and HFL-1 and the sphere size is proportional to the mean Z-score in the fourth cell line (MOLT4). Red spheres are PCR products that are called cell line-specific (FDR = 0.0001), Green are PCR products that are not cell line-specific. Green spheres tend to line up along the main diagonal while the red spheres are biased toward one or more axis. (B) Number of cell line-specific regions at different stringencies. For each FDR, all the PCR products that were cell line-specific were further filtered so that the mean Z-score is >1.5 in at least one of the four cell lines. Products that were closer than 200 bp were merged to define cell line-specific regions. (C) Cell line specificity profiles for 1890 PCR products that are cell line-specific for at least one histone modification. The FDR level was set to 0.01% and the same filtering was applied. The five main columns show the specificity for each of the histone modifications (green: not cell line-specific, red: cell line-specific). Four additional columns next to each of these five columns indicate the contribution of each cell line to the cell line specificity. For each cell line, a cell is colored only if the mean of the replicates for that cell line is significantly higher than the mean of all the replicates (blue: GM06990, gray: HeLa-S3, black: HFL-1, pink: MOLT4). (D) The distributions of the distances from the nearest GENCODE TSS of cell line-specific PCR products for H3K4me2. From left to right, box plots representing: cell line-specific tiles (FDR = 0.01%, filtered as in A), significantly modified (Z-score above the 95th percentile) but not cell line-specific PCR products in GM06990, HFL-1, HeLa-S3, and MOLT4. The width of each box is proportional to the square root of the number of PCR products in each group. Cell line-specific PCR products are significantly farther from TSS compared to highly modified but not cell line-specific tiles (see P-values in Table 3).
We next analyzed the cell line specificity of the histone modifications with respect to their distance from the closest GENCODE TSS. In general, the distribution of distances for the cell line-specific modifications reflects the overall distribution of the histone modification sites (see above). Cell line-specific regions for H3K4me1 tend to be considerably farther from TSSs than the other modifications, whereas H3K4me2 and H4ac are on average slightly closer to TSS compared to H3K4me1 and the cell line-specific regions for H3K4me3 and H3ac are much closer to TSS. Since H3K4me1 and H4ac are the two modifications with the largest numbers of cell line-specific regions, these results suggest that cell line-specific histone modifications tend to be observed more often in regions away from TSSs consistent with the hypothesis that the modifications are marking cell line-specific non-promoter elements. We examined these results in more detail by comparing the distributions of cell line-specific and non-cell line-specific regions of histone modification for each modification type and testing whether the distances to TSS differ. A remarkable shift in distance away from TSSs was found for the H4ac, H3K4me1, and H3K4me2 cell line-specific regions (except in GM06990 for H3K4me1), indicating that the cell line specificity tended to lie further from TSSs than would be expected by chance (Fig. 6D; Table 3).
Table 3.
One-tail Wilcoxon test P-values testing the hypothesis that the cell line-specific regions are farther from TSSs compared to significantly modified but not cell line-specific region
Taken together these data indicate that the histone modification profiles can detect biologically relevant cell-type-specific differences at both the promoter and at distal sites. Future developments of histone modification profiling and the HMM algorithm will attempt to exploit these features to explore the power of this approach.
The ENCODE pilot project
The regions identified for analysis in the ENCODE pilot project were chosen according to two criteria. The manually picked (ENm) regions were identified as regions that had been well studied already or had some specific biological interest. In general these regions have histone modification profiles showing numerous enrichments and examples of cell line-specific differences. The randomly selected (ENr) regions were identified using a stratified random-sampling strategy based on gene density and the level of non-exonic sequence conservation (see http://www.genome.gov/10506161 for details). It is noteworthy that for the regions with very low gene density (e.g., ENr112, ENr113, ENr114, ENr211, ENr213, 311, ENr312, and ENr313) there was little or no histone modification signal for the activating modifications we studied here regardless of the level of non-exonic sequence conservation. In the cases where signal was seen this is associated with the presence of a gene in the region. In fact the amount of histone modification signal present in each region (by % coverage) correlated strongly with both the GC content and gene density of the region but not with the non-exonic sequence conservation (Table 4). As expected for sites that are preferentially associated with gene TSSs H3K4me3 and H3ac have strong correlation with both GC content and gene density, while the other modifications have much stronger correlation with GC content than with gene density. For H4ac and H3K4me1, which preferentially mark sites distant from genes, the GC content of the region as a whole appears to be influential on the distribution of sites. If these modifications are marking non-promoter elements, then these elements appear to be enriched in GC-rich regions of the genome.
Table 4.
Pearson correlation coefficients for histone modification sites with gene density, non-exonic sequence conservation, and GC content by ENCODE region
Concluding remarks
We have constructed histone modification maps over 1% of the human genome using antibodies to histone modifications associated with gene activation in ChIP-chip. Analysis of these maps indicated that TSSs of active genes have a characteristic signature of histone modification signals that was highly distinct from inactive genes and this signal could be used to identify cell line-specific differences in gene expression. This signature consists of H3K4me3 and H3ac signal just downstream from the TSS (which is depleted in nucleosomes) followed by H3K4me2 and then low levels of H3K4me1 further downstream. The histone modification profile at TSSs is qualitatively similar to the signature seen at yeast promoters (Liu et al. 2005). TSSs at inactive genes do not have this signature but may have low-level enrichments of H3K4me3 and H3ac. The existence of the active TSS profile will allow improvement of gene annotation by identification of previously unidentified active promoters when applied throughout the genome. An example of this was given at the THOC2 gene where ChIP-chip enrichment occurred at a promoter which was annotated in the very detailed manual GENCODE annotation but was not previously present in the RefSeq annotation adding support to the annotation. Whole genome profiling with the histone modifications we have used here should provide many more examples and perhaps identify new genes. In addition we have identified a further signature of the histone modification profile which was associated with histone modification sites distant from TSSs comprising H4K3me1 and H4ac signals. This signal may be associated with non-promoter elements and is a major constituent of the cell line-specific differences in histone modification profiles. It is notable that, when examining other data from the ENCODE Consortium, a strong correlation between the histone modification profiles and DNase 1 hypersensitive sites is found both at TSSs and at distal sites. In fact the DNase 1 hypersensitive sites distant from TSSs have H3K4me1 signals, implying that the H3K4me1 sites we have identified might be regulatory elements since enhancers and other regulatory elements are known to exhibit DNase 1 hypersensitivity (The ENCODE Project Consortium 2007). Thus histone modification profiling using multiple different cell lines is a valuable tool to identify active TSSs and to explain expression differences in cell lines. Further functional analysis will reveal whether the additional sites that are identified are also regulatory elements.
The identification of these characteristic signatures associated with active regulatory elements, particularly promoters, suggests that there is a strong correlation between the histone modifications and biological activity. This lends support to the idea that there is a histone code associated with the activity of these elements, although perhaps not a completely deterministic one. Further analysis on the cell lines and regions we have studied here with additional antibodies to further histone modifications may give the opportunity to further refine the code, particularly for non-promoter elements and for silencing modifications. It would also be beneficial to examine additional cell lines, including ES cell lines where interesting signatures of histone modifications have already been identified (Bernstein et al. 2006).
In our approach we have used an HMM to identify sites of modification. Although this approach has advantages as mentioned above, there are also areas of improvement that can be incorporated into new versions of the algorithm. For instance it is possible to run the HMM on the data from multiple antibodies and it would be possible to incorporate multiple states which reflect the histone modification signatures we have identified above. In this way the output for the HMM could ultimately be predictive of active or inactive regulatory elements. In a similar way the data across cell lines could be examined to identify cell line-specific differences. It is also the case that the insight we gained above on the histone modification profiles came from using the HMM to guide us to interesting sites of histone modification and then analyzing the overall raw enrichment signal in the region. It is possible that we may be missing some features of the signal through this approach, for instance depletions or low-level enrichments spread over large regions. An alternative approach for data such as histone modifications which has a continuous nature (rather than discrete such as transcription binding sites) would be to analyze all the data by hierarchical clustering, which might allow us to identify such features.
Methods
Design and fabrication of ENCODE PCR product tiling microarray
The final ENCODE array spanned 23.8 Mb and contained 24,005 array elements (average size 992 bp). Primer pairs used to amplify PCR products for the arrays were designed using primer 3 including repetitive elements where possible. (The primer sequences for amplicons used as array elements are available at ftp://ftp.sanger.ac.uk/pub/encode/microarrays/). In order to generate arrays containing single-stranded array elements, all amplicons used in this study were prepared and printed on arrays as described (Dhami et al. 2005 and www.sanger.ac.uk/Projects/Microarrays/arraylab/methods.shtml). All PCR products were prepared as follows. A 5′-(C6) amino-link was added to all forward primers. The primer pairs (final concentration 0.5 μM) were used to amplify PCR products in a 60-μL final volume PCR containing 50 mM KCl, 5 mM Tris HCl (pH 8.5), 2.5 mM MgCl2, 10 mM dNTPs (Pharmacia), 0.625 U Taq polymerase (Perkin Elmer), and 50 ng of human genomic DNA (Roche). The PCR products were amplified with the following program: 1 × 5 min 95°C, 35 × 95°C 1.5 min, 65°C 1.5 min (−0.2°C per cycle), 72°C 3 min, 1 × 72°C 5 min. For arraying of PCR products, spotting buffer was added at final concentrations of 0.25 M sodium phosphate buffer pH 8.5 and 0.00025% sodium sarkosyl (BDH). The PCR products were filtered through multiscreen-GV 96-well filter plates (Millipore), aliquotted into 384-well plates (Genetix), and were arrayed onto Codelink slides (GE) in a 48-block format using a Microgrid II arrayer (Biorobotics/Genomic Solutions). Slides were processed to generate single-stranded array elements, as described at http://www.sanger.ac.uk/Projects/Microarrays/, and were stored at room temperature until hybridized.
Chromatin immunoprecipitation (ChIP)
Human cell line K562 (Lozzio and Lozzio 1979) was cultured in DMEM, 10% fetal calf serum, 1% penicillin-streptomycin, and 2 mM L-glutamine. Human cell line GM06990 (CEPH/UTAH PEDIGREE 1331) was cultured in RPMI1640, 15% fetal calf serum, 1% penicillin-streptomycin, and 2 mM L-glutamine. Human cell line HeLa-S3 was cultured in Joklic’s DMEM, 5% newborn bovine serum by the National Cell Culture Center Minneapolis, USA. Human cell line MOLT4 was cultured in RPMI640, 10% fetal calf serum, 1% penicillin-streptomycin, and 2 mM L-glutamine. Human cell line HFL-1 was cultured in Ham’s F12, 10% fetal calf serum, 1% penicillin-streptomycin, 2 mM L-glutamine, and 1% nonessential amino acids. Human cell line IMR90 was cultured in DMEM, 10% fetal calf serum, 1% penicillin-streptomycin, 2 mM L-glutamine, and 1% nonessential amino acids. 108 cells of suspension cultures (K562, GM06990, HeLa-S3, MOLT4) were collected by centrifugation and resuspended in 50 mL prewarmed serum free media in a glass flask. Formaldehyde (BDH) was added to final concentrations of 0.37 or 1%. The media of 80% confluent plates of adherent cultures (HFL-1, IMR90) was replaced with prewarmed serum free media containing 1% formaldehyde. After incubating the cells for 10 min with gentle agitation at room temperature, glycine (Sigma) was added to a final concentration of 0.125 M and incubated for 5 min at RT with agitation. Suspension cells were resuspended in 1.5 mL ice-cold PBS and centrifuged at 2000 rpm for 5 min at 4°C (Sorval Heraeus). Adherent cells were washed with PBS, scraped off the plates, collected at 4°C, resuspended in 1.5 mL ice-cold PBS, and centrifuged at 2000 rpm for 5 min at 4°C (Sorval Heraeus). The cell pellets were resuspended in ∼1.5× pellet volume of cell lysis buffer (10 mM Tris-HCl pH 8.0, 10 mM NaCl, 0.2% Igepal CA-630, 10 mM sodium butyrate, 50 μg/mL PMSF, 1 μg/mL leupeptin) and incubated for 10 min on ice. The cell nuclei were collected by centrifugation at 2500 rpm for 5 min at 4°C. The nuclei were resuspended in 1.2 mL of nuclear lysis buffer (NLB 50 mM Tris-HCl pH 8.1, 10 mM EDTA, 1% SDS, 10 mM sodium butyrate, 50 μg/mL PMSF, 1 μg/mL leupeptin) and incubated on ice for 10 min. After adding 0.72 mL of immunoprecipitation dilution buffer (IPDB 20 mM Tris-HCl pH 8.0, 150 mM NaCl, 2 mM EDTA 1% Triton X-100, 0.01% SDS, 10 mM sodium butyrate, 50 μg/mL PMSF, 1 μg/mL leupeptin) the chromatin was transferred to a 5-mL tube (Falcon) and sheared to a fragment size of ∼500 bp by sonication (Branson 450 digital sonifier using settings of time: 8 min, amplitude: 16%, pulse on 0.5 sec, pulse off 2.0 sec). During sonication, samples were cooled in an ice water bath. Debris was removed from the sheared chromatin by centrifugation in a cooled bench centrifuge (Eppendorf) at 20800g for 5 min at 4°C. The supernatant was diluted with 4.1 mL of IPDB to a final ratio of NLB:IPDB of 1:4. The chromatin was precleared by adding 100 μL of normal rabbit IgG (Upstate) and incubating for 1 h at 4°C on a rotating wheel. Two-hundred microliters of homogeneous protein G-agarose suspension was added (Roche) and incubation continued for 3 h to overnight at 4°C on a rotating wheel. The protein G-agarose was spun down at 955g for two minutes at 4°C; 1.35 mL of supernatant (chromatin) was used to set up each ChIP assay while 270 μL were used as input control. Ten micrograms of antibody was used in each ChIP assay. Antibodies used were di-acetylated histone H3 (06–599, Upstate), tetra-acetylated histone H4 (06–866, Upstate), histone H3 mono-methyl lysine 4 (ab8895, Abcam), histone H3 di-methyl lysine 4 (ab7766, Abcam), histone H3 tri-methyl lysine 4 (ab8580, Abcam), histone H2B (ab1791 Abcam), histone H3 (ab1790 Abcam). Antibody specificity was confirmed by Western blot of crude cell extracts in combination with competition by the specific peptide epitope (see Supplemental Fig. S1). The chromatin and antibody were incubated on a rotation wheel overnight at 4°C, then 100 μL of homogeneous protein G-agarose suspension was added (Roche) and incubation continued for 3 h. The protein G-agarose was spun down and the pellet washed twice with 750 μL of IP wash buffer 1 (20 mM Tris-HCl pH 8.1, 50 mM NaCl, 2 mM EDTA, 1% Triton X-100, 0.01% SDS), once with 750 μL of IP wash buffer 2 (10 mM Tris-HCl pH 8.1, 250 mM LiCl, 1 mM EDTA, 1% IGEPAL CA630, 1% deoxycholic acid) and twice with 10 mM Tris-HCl 1 mM EDTA pH 8.0. The immune complexes were twice eluted from the beads by adding 225 μL of IP elution buffer (100 mM NaHCO3, 0.1% SDS). After adding 0.2 μL of RNase A (10 mg/mL, ICN) and 27 μL of 5 M NaCl to the combined elutions and adding 0.1 μL of RNAse A and 16.2 μL of 5 M NaCl to the input sample, the samples were incubated at 65°C for 6 h. Then 9 μL of proteinase K (10 mg/mL, Invitrogen) was added and the samples incubated at 45°C overnight. Immediately before the DNA was recovered using phenol chloroform extraction, 2 μL tRNA (5 mg/mL stock, Invitrogen) was added. The aqueous layer was extracted once over chloroform. Then 5 μg of glycogen (Roche), 1 μL of tRNA (5 mg/mL, Invitrogen), 50 μL of 3 M sodium acetate pH 5.2, and 1.25 mL of ice-cold ethanol were added to precipitate the DNA at −20°C overnight. The DNA pellets were washed with 70% ethanol, air-dried, and resuspended in 100 μL of water for input samples and 50 μL of water for ChIP samples.
Fluorescent DNA labeling, microarray hybridization, and data analysis
Fluorescently labeled DNA samples were prepared using a modified Bioprime labeling kit (Invitrogen) in 150 μL reaction volumes containing 450 ng Input DNA or 40% of ChIP DNA, dNTPs (0.2 mM dATP, 0.2 mM dTTP, 0.2 mM dGTP, and 0.1 mM dCTP), 0.01 mM Cy5/Cy3 dCTP (GE Healthcare), 60 μL 2.5× random primer solution (750 μg/mL, Invitrogen), and 3 μL of Klenow fragment (Invitrogen). Input DNA samples were labeled with Cy5, and ChIP DNA samples were labeled with Cy3 overnight at 37°C. Labeling reactions were purified using Micro-spin G50 columns (Pharmacia-Amersham) in accordance with the manufacturer's instructions. Input and ChIP sample were combined and precipitated with 3 M sodium acetate (pH 5.2) in 2.5 volumes of ethanol with 135 μg human Cot DNA (Invitrogen). The DNA pellet was resuspended in 80 μL hybridization buffer containing 50% deionized formamide (Sigma), 10 mM Tris-HCl (pH 7.4), 5% dextran sulphate, 2× SSC, 0.1% Tween-20. Two combined labeling reactions were denatured for 10 min at 100°C, snap-frozen on ice, and used for one microarray hybridization. Microarrays were hybridized on an automatic hybridization station (HS4800, Tecan) for 45 h at 37°C with medium agitation, washed 10× for 1 min with PBS 0.05% Tween-20 (BDH) at 37°C, 5× for 1 min with 0.1× SSC at 52°C, 10× for 1 min with PBS 0.05% Tween-20 at 23°C, followed by a final wash with HPLC-grade water (BDH) at 23°C and drying under nitrogen flow for 4 min. Microarrays were scanned using a ScanArray 4000 confocal laser-based scanner (Perkin Elmer). Mean spot intensities from images were quantified using ScanArray Express (Perkin Elmer) with background subtraction. Spots affected by dust were manually flagged as “not found” and subsequently excluded from the analysis.
Data processing for analysis
The ratios of the background corrected ChIP signal divided by the background corrected input signal, both globally normalized, were used for the HMM analysis. Ratios of duplicated spots were averaged. Ratios of spots defined as “not found” and ratios with a value below zero were excluded from the analysis and also excluded from the final composite median data. Technical and biological replicates were automatically affiliated as the median value with an individual R script (i.e., ftp://ftp.sanger.ac.uk/pub/encode/H3K4me3_GM06990_2/H3K4me3_GM06990_2.R) which combines only positive values of technical replicates not classified as “not found”.
Hit regions and peak centers for Sanger ChIP/Chip data, as identified by Hidden Markov Model (HMM) analysis
A two-state HMM (Rabiner 1989) was used to analyze the Sanger ChIP-chip data. The states of the HMM represent regions of the tile path corresponding to locations either consistent or inconsistent with antibody binding. The emission probabilities of the states are derived from the probability that a point is part of a normal distribution fitted from the 45% of the data with the lowest enrichment values. The fitted distribution is calculated separately for each of the ENCODE regions using the Levenberg-Marquart curve-fitting technique (Gill et al. 1981).
The optimal state sequence for the observed data was calculated from the HMM using the Viterbi algorithm. The resulting list of tiles assigned to the state consistent with antibody binding was post-processed to develop a final hit list, which combined positive tiles within 1000 bp of each other into “hit regions.” The score of each hit region was determined by taking the summation of the median enrichment values of the tiles in the contiguous portions (i.e., the area under the peak). The center position of the PCR tile with the highest enrichment value in the hit region was deemed the center of the peak.
Real-time PCR
PCR primer pairs were designed to amplify 80–150 bp fragments from selected genomic regions (Supplemental Data). Real-time PCR reactions were carried out using SYBR green Mastermix plus (Eurogentec) in Applied Biosystems 7000 and 7500 instruments according to manufacturers' instructions. The enrichment (relative copy number) was determined in real-time PCR reactions using either 25 ng ChIP DNA or 25 ng non-enriched DNA (Input) as template. Each sample was evaluated in triplicate by real-time PCR. The efficiency of primer pairs were calculated using human genomic DNA (Roche) and the standard curve method. Only primer pairs with efficiencies between 0.7 and 1.3 were used. The enrichment was calculated with the primer efficiency corrected 2-ΔΔCT method (Livak and Schmittgen 2001) using the median of the ChIP triplicate and the Input triplicate.
Gene expression analysis
To determine transcriptional activity, we prepared total RNA from each of the five cell lines studied by ChIP-chip in at least two biological replicates and hybridized the labeled samples to the Affymetrix U133 plus 2.0 gene expression microarray. 0.5–1 × 107 asynchronously growing cells were pelleted and washed twice with cold PBS. The cell pellet was resuspended in 1 mL Trizol (Invitrogen) and allowed to stand for 5 min at room temperature. The cell suspension was extracted with chloroform and the aqueous layer precipitated with isopropanol. The resulting pellet was washed in 70% ethanol, air-dried, and resuspended in TE. Hybridization samples were prepared according to the Affymetrix GeneChip Expression Analysis Manual (Affymetrix) using 5 μg of total RNA and hybridized to Affymetrix human genome U133 plus 2.0 arrays. Normalized MAS5 (Affymetrix) data were used for present, marginal, and absent calling. Calls of multiple replicates were combined to the majority call. Array data is available at http://www.sanger.ac.uk/PostGenomics/encode/data-access.shtml. Expression levels were computed using the GCRMA (http://www.bioconductor.org/repository/devel/vignette/gcrma.pdf), limma (Smyth 2005), and affy packages in bioconductor (Gentleman et al. 2004), using the default parameters.
Analysis of histone enrichments and genomic features
Raw enrichment data were normalized and transformed using scripts written in the R programming language (http://www.r-project.org/). Scripts are available (http://www.sanger.ac.uk/PostGenomics/encode/data-access.shtml). All data used in the analysis were searched and archived in an Ensembl database (http://www.ensembl.org/) with an additional table for storing Affymetrix microarray expression data. The Ensembl database and companion API permits the retrieval of data objects in the context of sequences and features, such as gene start sites. Customization enabled the integration of microarray expression data. All data were stored and analyzed on NCBI genome build 35 (hg17) and genome features were from EnsEMBL homo_sapiens_ core_33_35f now archived at http://sep2005.archive.ensembl.org/index.html. GENCODE annotations were downloaded from GENCODE genes version 02.2 from ftp://genome.imim.es/pub/projects/gencode/data/havana-encode/current/44regions/44regions_CHR_coord.gtf. Principal Component Analysis and hierarchical clustering were performed using the R programming language (http://www.r-project.org/). Randomization strategy for Figure 2A: Each experimental HHM-center data set was randomized 100 times to generate 100 random HMM center data sets within the ENCODE region, conserving the number of centers. Parameters were generated for the overlap of HMM centers with gene features for the 100 randomized data sets. The corresponding value in the parent experimental dataset was compared to the population of randomized values using a two-sided t-test.
Evolutionary sequence conservation analysis
HMM sites were analyzed using phastOdds to compute log-odds scores that represent how likely a given feature fits a “conserved” model versus a “neutral” model. The neutral model was calculated from fourfold degenerate coding sites, and the conserved model was scaled accordingly to represent more slowly evolving sequence. phastOdds is closely related to phastCons but scores genomics intervals given as input (Siepel et al. 2005).
Acknowledgments
We thank Janine Thiele for assistance in setting up the ChIP-chip pipeline, Kate Rosenbloom and the staff at the UCSC Genome Browser for assistance in data curation for submission, and Wolfgang Huber for advice on the R programming language. This work was supported by NHGRI grant U01HG003168 to I.D., D.V., and N.P.C. and the Wellcome Trust, and partly supported by NHGRI grant R01HG03110 to Z.W.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article is online at http://www.genome.org/cgi/doi/10.1101/gr.5704207
References
- Bernstein B.E., Humphrey E.L., Erlich R.L., Schneider R., Bouman P., Liu J.S., Kouzarides T., Schreiber S.L., Humphrey E.L., Erlich R.L., Schneider R., Bouman P., Liu J.S., Kouzarides T., Schreiber S.L., Erlich R.L., Schneider R., Bouman P., Liu J.S., Kouzarides T., Schreiber S.L., Schneider R., Bouman P., Liu J.S., Kouzarides T., Schreiber S.L., Bouman P., Liu J.S., Kouzarides T., Schreiber S.L., Liu J.S., Kouzarides T., Schreiber S.L., Kouzarides T., Schreiber S.L., Schreiber S.L. Methylation of histone H3 Lys 4 in coding regions of active genes. Proc. Natl. Acad. Sci. 2002;99:8695–8700. doi: 10.1073/pnas.082249499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein B.E., Kamal M., Lindblad-Toh K., Bekiranov S., Bailey D.K., Huebert D.J., McMahon S., Karlsson E.K., Kulbokas E.J., III, Gingeras T.R., Kamal M., Lindblad-Toh K., Bekiranov S., Bailey D.K., Huebert D.J., McMahon S., Karlsson E.K., Kulbokas E.J., III, Gingeras T.R., Lindblad-Toh K., Bekiranov S., Bailey D.K., Huebert D.J., McMahon S., Karlsson E.K., Kulbokas E.J., III, Gingeras T.R., Bekiranov S., Bailey D.K., Huebert D.J., McMahon S., Karlsson E.K., Kulbokas E.J., III, Gingeras T.R., Bailey D.K., Huebert D.J., McMahon S., Karlsson E.K., Kulbokas E.J., III, Gingeras T.R., Huebert D.J., McMahon S., Karlsson E.K., Kulbokas E.J., III, Gingeras T.R., McMahon S., Karlsson E.K., Kulbokas E.J., III, Gingeras T.R., Karlsson E.K., Kulbokas E.J., III, Gingeras T.R., Kulbokas E.J., III, Gingeras T.R., Gingeras T.R., et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Bernstein B.E., Mikkelsen T.S., Xie X., Kamal M., Huebert D.J., Cuff J., Fry B., Meissner A., Wernig M., Plath K., Mikkelsen T.S., Xie X., Kamal M., Huebert D.J., Cuff J., Fry B., Meissner A., Wernig M., Plath K., Xie X., Kamal M., Huebert D.J., Cuff J., Fry B., Meissner A., Wernig M., Plath K., Kamal M., Huebert D.J., Cuff J., Fry B., Meissner A., Wernig M., Plath K., Huebert D.J., Cuff J., Fry B., Meissner A., Wernig M., Plath K., Cuff J., Fry B., Meissner A., Wernig M., Plath K., Fry B., Meissner A., Wernig M., Plath K., Meissner A., Wernig M., Plath K., Wernig M., Plath K., Plath K., et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Birney E., Andrews D., Caccamo M., Chen Y., Clarke L., Coates G., Cox T., Cunningham F., Curwen V., Cutts T., Andrews D., Caccamo M., Chen Y., Clarke L., Coates G., Cox T., Cunningham F., Curwen V., Cutts T., Caccamo M., Chen Y., Clarke L., Coates G., Cox T., Cunningham F., Curwen V., Cutts T., Chen Y., Clarke L., Coates G., Cox T., Cunningham F., Curwen V., Cutts T., Clarke L., Coates G., Cox T., Cunningham F., Curwen V., Cutts T., Coates G., Cox T., Cunningham F., Curwen V., Cutts T., Cox T., Cunningham F., Curwen V., Cutts T., Cunningham F., Curwen V., Cutts T., Curwen V., Cutts T., Cutts T., et al. Ensembl 2006. Nucleic Acids Res. 2006;34:D556–D561. doi: 10.1093/nar/gkj133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R., Wang L., Wang H., Xia L., Erdjument-Bromage H., Tempst P., Jones R.S., Zhang Y., Wang L., Wang H., Xia L., Erdjument-Bromage H., Tempst P., Jones R.S., Zhang Y., Wang H., Xia L., Erdjument-Bromage H., Tempst P., Jones R.S., Zhang Y., Xia L., Erdjument-Bromage H., Tempst P., Jones R.S., Zhang Y., Erdjument-Bromage H., Tempst P., Jones R.S., Zhang Y., Tempst P., Jones R.S., Zhang Y., Jones R.S., Zhang Y., Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- Cloos P.A., Christensen J., Agger K., Maiolica A., Rappsilber J., Antal T., Hansen K.H., Helin K., Christensen J., Agger K., Maiolica A., Rappsilber J., Antal T., Hansen K.H., Helin K., Agger K., Maiolica A., Rappsilber J., Antal T., Hansen K.H., Helin K., Maiolica A., Rappsilber J., Antal T., Hansen K.H., Helin K., Rappsilber J., Antal T., Hansen K.H., Helin K., Antal T., Hansen K.H., Helin K., Hansen K.H., Helin K., Helin K. The putative oncogene GASC1 demethylates tri- and dimethylated lysine 9 on histone H3. Nature. 2006;442:307–311. doi: 10.1038/nature04837. [DOI] [PubMed] [Google Scholar]
- Davuluri R.V., Grosse I., Zhang M.Q., Grosse I., Zhang M.Q., Zhang M.Q. Computational identification of promoters and first exons in the human genome. Nat. Genet. 2001;29:412–417. doi: 10.1038/ng780. [DOI] [PubMed] [Google Scholar]
- de la Cruz X., Lois S., Sanchez-Molina S., Martinez-Balbas M.A., Lois S., Sanchez-Molina S., Martinez-Balbas M.A., Sanchez-Molina S., Martinez-Balbas M.A., Martinez-Balbas M.A. Do protein motifs read the histone code? Bioessays. 2005;27:164–175. doi: 10.1002/bies.20176. [DOI] [PubMed] [Google Scholar]
- Dhami P., Coffey A.J., Abbs S., Vermeesch J.R., Dumanski J.P., Woodward K.J., Andrews R.M., Langford C., Vetrie D., Coffey A.J., Abbs S., Vermeesch J.R., Dumanski J.P., Woodward K.J., Andrews R.M., Langford C., Vetrie D., Abbs S., Vermeesch J.R., Dumanski J.P., Woodward K.J., Andrews R.M., Langford C., Vetrie D., Vermeesch J.R., Dumanski J.P., Woodward K.J., Andrews R.M., Langford C., Vetrie D., Dumanski J.P., Woodward K.J., Andrews R.M., Langford C., Vetrie D., Woodward K.J., Andrews R.M., Langford C., Vetrie D., Andrews R.M., Langford C., Vetrie D., Langford C., Vetrie D., Vetrie D. Exon array CGH: Detection of copy-number changes at the resolution of individual exons in the human genome. Am. J. Hum. Genet. 2005;76:750–762. doi: 10.1086/429588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Down T.A., Hubbard T.J., Hubbard T.J. Computational detection and location of transcription start sites in mammalian genomic DNA. Genome Res. 2002;12:458–461. doi: 10.1101/gr.216102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium, The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium, Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007 doi: 10.1038/nature05874. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman R.C., Carey V.J., Bates D.M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., Carey V.J., Bates D.M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., Bates D.M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., Ellis B., Gautier L., Ge Y., Gentry J., Gautier L., Ge Y., Gentry J., Ge Y., Gentry J., Gentry J., et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill P R Murray W., Wright M.H., Wright M.H. The Levenberg-Marquardt Method. In: Gill P.E., et al., editors. Practical Optimization. Academic Press; London: 1981. pp. 136–137. [Google Scholar]
- Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- Guenther M.G., Jenner R.G., Chevalier B., Nakamura T., Croce C.M., Canaani E., Young R.A., Jenner R.G., Chevalier B., Nakamura T., Croce C.M., Canaani E., Young R.A., Chevalier B., Nakamura T., Croce C.M., Canaani E., Young R.A., Nakamura T., Croce C.M., Canaani E., Young R.A., Croce C.M., Canaani E., Young R.A., Canaani E., Young R.A., Young R.A. Global and HOX-specific roles for the MLL1 methyltransferase. Proc. Natl. Acad. Sci. 2005;102:8603–8608. doi: 10.1073/pnas.0503072102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrow J., Denoeud F., Frankish A., Reymond A., Chen C., Chrast J., Lagarde J., Gilbert J.G.R., Storey R., Swarbreck D., Denoeud F., Frankish A., Reymond A., Chen C., Chrast J., Lagarde J., Gilbert J.G.R., Storey R., Swarbreck D., Frankish A., Reymond A., Chen C., Chrast J., Lagarde J., Gilbert J.G.R., Storey R., Swarbreck D., Reymond A., Chen C., Chrast J., Lagarde J., Gilbert J.G.R., Storey R., Swarbreck D., Chen C., Chrast J., Lagarde J., Gilbert J.G.R., Storey R., Swarbreck D., Chrast J., Lagarde J., Gilbert J.G.R., Storey R., Swarbreck D., Lagarde J., Gilbert J.G.R., Storey R., Swarbreck D., Gilbert J.G.R., Storey R., Swarbreck D., Storey R., Swarbreck D., Swarbreck D., et al. GENCODE: Producing a reference annotation for ENCODE. Genome Biol. 2006;7 (Suppl.):S4.1–S4.9. doi: 10.1186/gb-2006-7-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinrichs A.S., Karolchik D., Baertsch R., Barber G.P., Bejerano G., Clawson H., Diekhans M., Furey T.S., Harte R.A., Hsu F., Karolchik D., Baertsch R., Barber G.P., Bejerano G., Clawson H., Diekhans M., Furey T.S., Harte R.A., Hsu F., Baertsch R., Barber G.P., Bejerano G., Clawson H., Diekhans M., Furey T.S., Harte R.A., Hsu F., Barber G.P., Bejerano G., Clawson H., Diekhans M., Furey T.S., Harte R.A., Hsu F., Bejerano G., Clawson H., Diekhans M., Furey T.S., Harte R.A., Hsu F., Clawson H., Diekhans M., Furey T.S., Harte R.A., Hsu F., Diekhans M., Furey T.S., Harte R.A., Hsu F., Furey T.S., Harte R.A., Hsu F., Harte R.A., Hsu F., Hsu F., et al. The UCSC Genome Browser Database: Update 2006. Nucleic Acids Res. 2006;34:D590–D598. doi: 10.1093/nar/gkj144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu F., Kent W.J., Clawson H., Kuhn R.M., Diekhans M., Haussler D., Kent W.J., Clawson H., Kuhn R.M., Diekhans M., Haussler D., Clawson H., Kuhn R.M., Diekhans M., Haussler D., Kuhn R.M., Diekhans M., Haussler D., Diekhans M., Haussler D., Haussler D. The UCSC known genes. Bioinformatics. 2006;22:1036–1046. doi: 10.1093/bioinformatics/btl048. [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium, Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- Jenuwein T., Allis C.D., Allis C.D. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Katayama S., Tomaru Y., Kasukawa T., Waki K., Nakanishi M., Nakamura M., Nishida H., Yap C.C., Suzuki M., Kawai J., Tomaru Y., Kasukawa T., Waki K., Nakanishi M., Nakamura M., Nishida H., Yap C.C., Suzuki M., Kawai J., Kasukawa T., Waki K., Nakanishi M., Nakamura M., Nishida H., Yap C.C., Suzuki M., Kawai J., Waki K., Nakanishi M., Nakamura M., Nishida H., Yap C.C., Suzuki M., Kawai J., Nakanishi M., Nakamura M., Nishida H., Yap C.C., Suzuki M., Kawai J., Nakamura M., Nishida H., Yap C.C., Suzuki M., Kawai J., Nishida H., Yap C.C., Suzuki M., Kawai J., Yap C.C., Suzuki M., Kawai J., Suzuki M., Kawai J., Kawai J., et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- Kim T.H., Barrera L.O., Zheng M., Qu C., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B., Barrera L.O., Zheng M., Qu C., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B., Zheng M., Qu C., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B., Qu C., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B., Richmond T.A., Wu Y., Green R.D., Ren B., Wu Y., Green R.D., Ren B., Green R.D., Ren B., Ren B. A high-resolution map of active promoters in the human genome. Nature. 2005;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose R.J., Yamane K., Bae Y., Zhang D., Erdjument-Bromage H., Tempst P., Wong J., Zhang Y., Yamane K., Bae Y., Zhang D., Erdjument-Bromage H., Tempst P., Wong J., Zhang Y., Bae Y., Zhang D., Erdjument-Bromage H., Tempst P., Wong J., Zhang Y., Zhang D., Erdjument-Bromage H., Tempst P., Wong J., Zhang Y., Erdjument-Bromage H., Tempst P., Wong J., Zhang Y., Tempst P., Wong J., Zhang Y., Wong J., Zhang Y., Zhang Y. The transcriptional repressor JHDM3A demethylates trimethyl histone H3 lysine 9 and lysine 36. Nature. 2006;442:312–316. doi: 10.1038/nature04853. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Histone methylation in transcriptional control. Curr. Opin. Genet. Dev. 2002;12:198–209. doi: 10.1016/s0959-437x(02)00287-3. [DOI] [PubMed] [Google Scholar]
- Kurdistani S.K., Grunstein M., Grunstein M. Histone acetylation and deacetylation in yeast. Nat. Rev. Mol. Cell Biol. 2003;4:276–284. doi: 10.1038/nrm1075. [DOI] [PubMed] [Google Scholar]
- Kurdistani S.K., Tavazoie S., Grunstein M., Tavazoie S., Grunstein M., Grunstein M. Mapping global histone acetylation patterns to gene expression. Cell. 2004;117:721–733. doi: 10.1016/j.cell.2004.05.023. [DOI] [PubMed] [Google Scholar]
- Lachner M., O'Carroll D., Rea S., Mechtler K., Jenuwein T., O'Carroll D., Rea S., Mechtler K., Jenuwein T., Rea S., Mechtler K., Jenuwein T., Mechtler K., Jenuwein T., Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- Liang G., Lin J.C., Wei V., Yoo C., Cheng J.C., Nguyen C.T., Weisenberger D.J., Egger G., Takai D., Gonzales F.A., Lin J.C., Wei V., Yoo C., Cheng J.C., Nguyen C.T., Weisenberger D.J., Egger G., Takai D., Gonzales F.A., Wei V., Yoo C., Cheng J.C., Nguyen C.T., Weisenberger D.J., Egger G., Takai D., Gonzales F.A., Yoo C., Cheng J.C., Nguyen C.T., Weisenberger D.J., Egger G., Takai D., Gonzales F.A., Cheng J.C., Nguyen C.T., Weisenberger D.J., Egger G., Takai D., Gonzales F.A., Nguyen C.T., Weisenberger D.J., Egger G., Takai D., Gonzales F.A., Weisenberger D.J., Egger G., Takai D., Gonzales F.A., Egger G., Takai D., Gonzales F.A., Takai D., Gonzales F.A., Gonzales F.A., et al. Distinct localization of histone H3 acetylation and H3-K4 methylation to the transcription start sites in the human genome. Proc. Natl. Acad. Sci. 2004;101:7357–7362. doi: 10.1073/pnas.0401866101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C.L., Kaplan T., Kim M., Buratowski S., Schreiber S.L., Friedman N., Rando O.J., Kaplan T., Kim M., Buratowski S., Schreiber S.L., Friedman N., Rando O.J., Kim M., Buratowski S., Schreiber S.L., Friedman N., Rando O.J., Buratowski S., Schreiber S.L., Friedman N., Rando O.J., Schreiber S.L., Friedman N., Rando O.J., Friedman N., Rando O.J., Rando O.J. Single-nucleosome mapping of histone modifications in S. cerevisiae. PLoS Biol. 2005;3:e328. doi: 10.1371/journal.pbio.0030328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lozzio B.B., Lozzio C.B., Lozzio C.B. Properties and usefulness of the original K-562 human myelogenous leukemia cell line. Leuk. Res. 1979;3:363–370. doi: 10.1016/0145-2126(79)90033-x. [DOI] [PubMed] [Google Scholar]
- Maeda N., Kasukawa T., Oyama R., Gough J., Frith M., Engstrom P.G., Lenhard B., Aturaliya R.N., Batalov S., Beisel K.W., Kasukawa T., Oyama R., Gough J., Frith M., Engstrom P.G., Lenhard B., Aturaliya R.N., Batalov S., Beisel K.W., Oyama R., Gough J., Frith M., Engstrom P.G., Lenhard B., Aturaliya R.N., Batalov S., Beisel K.W., Gough J., Frith M., Engstrom P.G., Lenhard B., Aturaliya R.N., Batalov S., Beisel K.W., Frith M., Engstrom P.G., Lenhard B., Aturaliya R.N., Batalov S., Beisel K.W., Engstrom P.G., Lenhard B., Aturaliya R.N., Batalov S., Beisel K.W., Lenhard B., Aturaliya R.N., Batalov S., Beisel K.W., Aturaliya R.N., Batalov S., Beisel K.W., Batalov S., Beisel K.W., Beisel K.W., et al. Transcript annotation in FANTOM3: Mouse gene catalog based on physical cDNAs. PLoS Genet. 2006;2:e62. doi: 10.1371/journal.pgen.0020062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C., Zhang Y., Zhang Y. The diverse functions of histone lysine methylation. Nat. Rev. Mol. Cell Biol. 2005;6:838–849. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- Mattick J.S., Makunin I.V., Makunin I.V. Non-coding RNA. Hum. Mol. Genet. 2006;(Spec No 1) 15:R17–R29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- Ng H.H., Robert F., Young R.A., Struhl K., Robert F., Young R.A., Struhl K., Young R.A., Struhl K., Struhl K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol. Cell. 2003;11:709–719. doi: 10.1016/s1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- Parkinson H., Sarkans U., Shojatalab M., Abeygunawardena N., Contrino S., Coulson R., Farne A., Lara G.G., Holloway E., Kapushesky M., Sarkans U., Shojatalab M., Abeygunawardena N., Contrino S., Coulson R., Farne A., Lara G.G., Holloway E., Kapushesky M., Shojatalab M., Abeygunawardena N., Contrino S., Coulson R., Farne A., Lara G.G., Holloway E., Kapushesky M., Abeygunawardena N., Contrino S., Coulson R., Farne A., Lara G.G., Holloway E., Kapushesky M., Contrino S., Coulson R., Farne A., Lara G.G., Holloway E., Kapushesky M., Coulson R., Farne A., Lara G.G., Holloway E., Kapushesky M., Farne A., Lara G.G., Holloway E., Kapushesky M., Lara G.G., Holloway E., Kapushesky M., Holloway E., Kapushesky M., Kapushesky M., et al. ArrayExpress—A public repository for microarray gene expression data at the EBI. Nucleic Acids Res. 2005;33:D553–D555. doi: 10.1093/nar/gki056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokholok D.K., Harbison C.T., Levine S., Cole M., Hannett N.M., Lee T.I., Bell G.W., Walker K., Rolfe P.A., Herbolsheimer E., Harbison C.T., Levine S., Cole M., Hannett N.M., Lee T.I., Bell G.W., Walker K., Rolfe P.A., Herbolsheimer E., Levine S., Cole M., Hannett N.M., Lee T.I., Bell G.W., Walker K., Rolfe P.A., Herbolsheimer E., Cole M., Hannett N.M., Lee T.I., Bell G.W., Walker K., Rolfe P.A., Herbolsheimer E., Hannett N.M., Lee T.I., Bell G.W., Walker K., Rolfe P.A., Herbolsheimer E., Lee T.I., Bell G.W., Walker K., Rolfe P.A., Herbolsheimer E., Bell G.W., Walker K., Rolfe P.A., Herbolsheimer E., Walker K., Rolfe P.A., Herbolsheimer E., Rolfe P.A., Herbolsheimer E., Herbolsheimer E., et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- Prabhakar S., Poulin F., Shoukry M., Afzal V., Rubin E.M., Couronne O., Pennacchio L.A., Poulin F., Shoukry M., Afzal V., Rubin E.M., Couronne O., Pennacchio L.A., Shoukry M., Afzal V., Rubin E.M., Couronne O., Pennacchio L.A., Afzal V., Rubin E.M., Couronne O., Pennacchio L.A., Rubin E.M., Couronne O., Pennacchio L.A., Couronne O., Pennacchio L.A., Pennacchio L.A. Close sequence comparisons are sufficient to identify human cis-regulatory elements. Genome Res. 2006;16:855–863. doi: 10.1101/gr.4717506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt K.D., Tatusova T., Maglott D.R., Tatusova T., Maglott D.R., Maglott D.R. NCBI Reference Sequence (RefSeq): A curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2005;33:D501–D504. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabiner L.R. A tutorial on hidden Markov-models and selected applications in speech recognition. Proc. IEEE. 1989;77:257–286. [Google Scholar]
- Robyr D., Kurdistani S.K., Grunstein M., Kurdistani S.K., Grunstein M., Grunstein M. Analysis of genome-wide histone acetylation state and enzyme binding using DNA microarrays. Methods Enzymol. 2004;376:289–304. doi: 10.1016/S0076-6879(03)76019-4. [DOI] [PubMed] [Google Scholar]
- Roh T.Y., Cuddapah S., Zhao K., Cuddapah S., Zhao K., Zhao K. Active chromatin domains are defined by acetylation islands revealed by genome-wide mapping. Genes & Dev. 2005;19:542–552. doi: 10.1101/gad.1272505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth S.Y., Denu J.M., Allis C.D., Denu J.M., Allis C.D., Allis C.D. Histone acetyltransferases. Annu. Rev. Biochem. 2001;70:81–120. doi: 10.1146/annurev.biochem.70.1.81. [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H., Schneider R., Bannister A.J., Sherriff J., Bernstein B.E., Emre N.C., Schreiber S.L., Mellor J., Kouzarides T., Schneider R., Bannister A.J., Sherriff J., Bernstein B.E., Emre N.C., Schreiber S.L., Mellor J., Kouzarides T., Bannister A.J., Sherriff J., Bernstein B.E., Emre N.C., Schreiber S.L., Mellor J., Kouzarides T., Sherriff J., Bernstein B.E., Emre N.C., Schreiber S.L., Mellor J., Kouzarides T., Bernstein B.E., Emre N.C., Schreiber S.L., Mellor J., Kouzarides T., Emre N.C., Schreiber S.L., Mellor J., Kouzarides T., Schreiber S.L., Mellor J., Kouzarides T., Mellor J., Kouzarides T., Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- Schneider R., Bannister A.J., Myers F.A., Thorne A.W., Crane-Robinson C., Kouzarides T., Bannister A.J., Myers F.A., Thorne A.W., Crane-Robinson C., Kouzarides T., Myers F.A., Thorne A.W., Crane-Robinson C., Kouzarides T., Thorne A.W., Crane-Robinson C., Kouzarides T., Crane-Robinson C., Kouzarides T., Kouzarides T. Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nat. Cell Biol. 2004;6:73–77. doi: 10.1038/ncb1076. [DOI] [PubMed] [Google Scholar]
- Schübeler D., MacAlpine D.M., Scalzo D., Wirbelauer C., Kooperberg C., van Leeuwen F., Gottschling D.E., O'Neill L.P., Turner B.M., Delrow J., MacAlpine D.M., Scalzo D., Wirbelauer C., Kooperberg C., van Leeuwen F., Gottschling D.E., O'Neill L.P., Turner B.M., Delrow J., Scalzo D., Wirbelauer C., Kooperberg C., van Leeuwen F., Gottschling D.E., O'Neill L.P., Turner B.M., Delrow J., Wirbelauer C., Kooperberg C., van Leeuwen F., Gottschling D.E., O'Neill L.P., Turner B.M., Delrow J., Kooperberg C., van Leeuwen F., Gottschling D.E., O'Neill L.P., Turner B.M., Delrow J., van Leeuwen F., Gottschling D.E., O'Neill L.P., Turner B.M., Delrow J., Gottschling D.E., O'Neill L.P., Turner B.M., Delrow J., O'Neill L.P., Turner B.M., Delrow J., Turner B.M., Delrow J., Delrow J., et al. The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes & Dev. 2004;18:1263–1271. doi: 10.1101/gad.1198204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Lan F., Matson C., Mulligan P., Whetstine J.R., Cole P.A., Casero R.A., Shi Y., Lan F., Matson C., Mulligan P., Whetstine J.R., Cole P.A., Casero R.A., Shi Y., Matson C., Mulligan P., Whetstine J.R., Cole P.A., Casero R.A., Shi Y., Mulligan P., Whetstine J.R., Cole P.A., Casero R.A., Shi Y., Whetstine J.R., Cole P.A., Casero R.A., Shi Y., Cole P.A., Casero R.A., Shi Y., Casero R.A., Shi Y., Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Siepel A., Bejerano G., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., Bejerano G., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., Clawson H., Spieth J., Hillier L.W., Richards S., Spieth J., Hillier L.W., Richards S., Hillier L.W., Richards S., Richards S., et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth G.K. Limma: Linear models for microarray data. In: Gentleman R., et al., editors. Bioinformatics and computational biology solutions using R and bioconductor. Springer; New York: 2005. pp. 397–420. [Google Scholar]
- Tusher V.G., Tibshirani R., Chu G., Tibshirani R., Chu G., Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakoc C.R., Mandat S.A., Olenchock B.A., Blobel G.A., Mandat S.A., Olenchock B.A., Blobel G.A., Olenchock B.A., Blobel G.A., Blobel G.A. Histone H3 lysine 9 methylation and HP1γ are associated with transcription elongation through mammalian chromatin. Mol. Cell. 2005;19:381–391. doi: 10.1016/j.molcel.2005.06.011. [DOI] [PubMed] [Google Scholar]
- Wolffe A.P., Pruss D., Pruss D. Targeting chromatin disruption: Transcription regulators that acetylate histones. Cell. 1996;84:817–819. doi: 10.1016/s0092-8674(00)81059-4. [DOI] [PubMed] [Google Scholar]
- Wysocka J., Swigut T., Milne T.A., Dou Y., Zhang X., Burlingame A.L., Roeder R.G., Brivanlou A.H., Allis C.D., Swigut T., Milne T.A., Dou Y., Zhang X., Burlingame A.L., Roeder R.G., Brivanlou A.H., Allis C.D., Milne T.A., Dou Y., Zhang X., Burlingame A.L., Roeder R.G., Brivanlou A.H., Allis C.D., Dou Y., Zhang X., Burlingame A.L., Roeder R.G., Brivanlou A.H., Allis C.D., Zhang X., Burlingame A.L., Roeder R.G., Brivanlou A.H., Allis C.D., Burlingame A.L., Roeder R.G., Brivanlou A.H., Allis C.D., Roeder R.G., Brivanlou A.H., Allis C.D., Brivanlou A.H., Allis C.D., Allis C.D. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell. 2005;121:859–872. doi: 10.1016/j.cell.2005.03.036. [DOI] [PubMed] [Google Scholar]