Abstract
Butyrate is a histone deacetylase inhibitor (HDACi) with anti-neoplastic properties, which theoretically reactivates epigenetically silenced genes by increasing global histone acetylation. However, recent studies indicate that a similar number or even more genes are down-regulated than up-regulated by this drug. We treated hepatocarcinoma HepG2 cells with butyrate and characterized the levels of acetylation at DNA-bound histones H3 and H4 by ChIP-chip along the ENCODE regions. In contrast to the global increases of histone acetylation, many genomic regions close to transcription start sites were deacetylated after butyrate exposure. In order to validate these findings, we found that both butyrate and trichostatin A treatment resulted in histone deacetylation at selected regions, while nucleosome loss or changes in histone H3 lysine 4 trimethylation (H3K4me3) did not occur in such locations. Furthermore, similar histone deacetylation events were observed when colon adenocarcinoma HT-29 cells were treated with butyrate. In addition, genes with deacetylated promoters were down-regulated by butyrate, and this was mediated at the transcriptional level by affecting RNA polymerase II (POLR2A) initiation/elongation. Finally, the global increase in acetylated histones was preferentially localized to the nuclear periphery, indicating that it might not be associated to euchromatin. Our results are significant for the evaluation of HDACi as anti-tumourogenic drugs, suggesting that previous models of action might need to be revised, and provides an explanation for the frequently observed repression of many genes during HDACi treatment.
Early research efforts on carcinogenesis have focused on genetic and cytogenetic aspects, but recent years have brought a greater understanding of the role of epigenetics (Jones and Baylin 2002). Initial observations showed that DNA hypermethylation at CpG island promoters of tumor-suppressor genes was a major epigenetic event associated with neoplasia (Baylin 2005). However, recent reports have also indicated that the patterns of histone modifications are abnormally altered in tumor cells (Fraga et al. 2005). The N-terminal tails of H3 and H4 are acetylated at several lysine residues and these modifications are associated with the 5′ ends of transcriptionally active genes (Kim et al. 2005b). The acetylation of histones is the result of the net activity of histone acetyl transferases (HATs) and histone deacetylases (HDACs).
A group of drugs generally known as HDACi have been demonstrated to possess anti-tumourogenic properties including growth arrest and/or apoptosis in tumor cells (Lindemann et al. 2004). Molecularly, HDACi block the activity of class I and class II HDACs and increase global histone acetylation, which theoretically should up-regulate many genes. However, HDACi treatment induces significant changes in expression patterns of 5%–10% of human genes, and surprisingly, approximately equal numbers of genes are up- and down-regulated (Daly and Shirazi-Beechey 2006). In addition, it has been reported that an even larger fraction of the transcriptome is affected by HDACi, but most changes in expression are of low magnitude (de Ruijter et al. 2005; Peart et al. 2005), with down-regulation being even more frequent than up-regulation.
Butyrate is similar in structure and effects to other carboxylic acids like phenyl-butyrate or valproic acid, which are already used in the treatment of cancer and epilepsy (Villar-Garea and Esteller 2004). Butyrate has received great attention due to its anti-neoplastic properties and natural occurrence in our bodies (Davie 2003). The molecular mechanisms behind the anti-tumourogenic properties and changes in gene expression exerted by butyrate are not well understood. Typically, activation of tumor-suppressor genes, like CDKN1A (formerly P21/WAF1), is claimed to be the major cause of growth arrest and/or apoptosis, and increased histone acetylation in the promoter of this gene after butyrate treatment has been reported (Richon et al. 2000). However, down-regulation of genes with growth-promoting or antiapoptotic effects, like MYC (Heruth et al. 1993), cyclin D1 (CCND1) (Lallemand et al. 1996), or SRC (Kostyniuk et al. 2002), have also been observed in butyrate-treated cells. This has been associated with the pleiotropic effects of butyrate, which can affect nonhistone (e.g., transcription factors) and/or noneuchromatic targets (e.g., constitutive heterochromatin) (Davie 2003; Taddei et al. 2005). However, most of the experiments to date have focused on a few promoters, typically in genes up-regulated by HDACi (Suzuki et al. 2002), while the common gene-repression events have frequently been neglected.
Using chromatin immunoprecipitation and microarray hybridization (ChIP-chip) we analyzed the acetylation patterns of DNA-bound histone H3 and H4 after treatment of HepG2 cells with butyrate. As expected, global histone acetylation was increased by butyrate treatment. However, by interrogating 1% of the human genome as defined in the ENCODE Project Consortium (2004), we observed histone deacetylation at many promoter regions. The loss of acetylation affected both H3 and H4, was shown to be reversible upon withdrawal of the drug, and did not affect total histone content or H3 lysine 4 trimethylation (H3K4me3). The deacetylation of promoters was also seen after treatment with trichostatin A (TSA) and may be a general effect of HDACi. Furthermore, deacetylation was accompanied by decreased expression, reduced RNA polymerase II (POLR2A) initiation, and/or elongation and diminished transcription-factor binding to regulatory regions. Finally, the global increase in acetylated histones was mainly localized at the nuclear periphery, possibly affecting heterochromatin or free histones. In summary, our study provides a mechanistic explanation for the commonly observed gene repression under HDACi treatment and challenges pre-established models of action.
Results
Bulk histone acetylation increases upon butyrate treatment
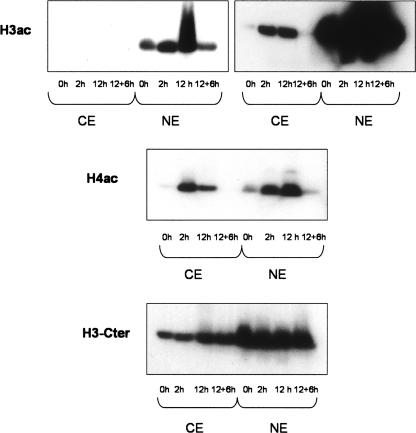
HepG2 cells were treated with butyrate for 12 h, which resulted in a clear increase in the overall acetylation of H3 and H4 upon butyrate treatment (Fig. 1). Removal of the drug for 6 h after a 12-h treatment resulted in acetylation returning to basal levels, indicating that the effects are reversible. Low levels of acetylated histones were also observed in cytoplasmic extracts. This could be due to de novo acetylation of histones in the cytoplasm upon synthesis and inhibition of cytoplasmic HDACs. The specificity of our results is shown by the analysis of the same extracts with an antibody against the C-terminal part of H3, which is not known to be acetylated.
Figure 1.
Butyrate increases the total levels of H3ac and H4ac. Nuclear and cytoplasmic protein extracts were prepared from HepG2 cells that were either untreated (0 h), treated with 5 mM Na-Butyrate for 2 h or 12 h, or treated for 12 h and then kept for 6 h without the drug (12 + 6 h). The same amount of protein was used in all cases. For H3ac, the same Western blot is presented with different exposure times.
Genome-wide changes in acetylation of DNA-bound histones upon butyrate treatment
After reproducing the expected increases in global histone acetylation, we wanted to identify DNA regions associated with altered histone acetylation. We performed chromatin immunoprecipitation using antibodies against H3ac and H4ac in untreated HepG2 cells or in cells treated for 12 h with 5 mM Na-Butyrate. This butyrate concentration is in the same range as naturally occurring levels and was previously used in other studies (Lallemand et al. 1996; Dashwood et al. 2006). As for the duration of the treatment, we were interested in as long a treatment as possible, without inducing apoptosis in many cells (Emanuele et al. 2004). The reason for this is that long treatments better reflect the clinical use of HDACi in cancer therapy, and because changes of higher magnitude were expected.
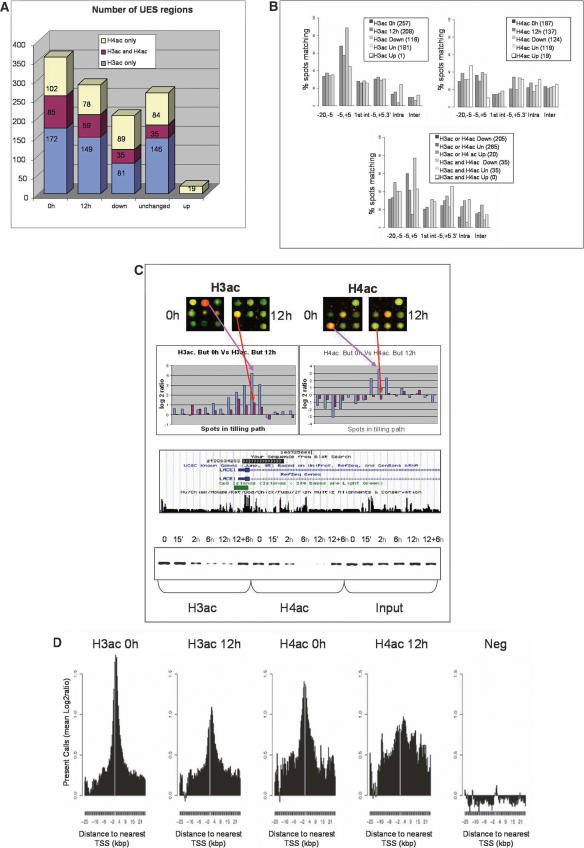
We found, as previously reported (Schubeler et al. 2004), a high correlation between H3ac and H4ac data sets in normal-growing cells (R = 0.667, Supplemental Fig. 1) and enrichment of both modifications at TSSs (Fig. 2B). This localization around TSS was more pronounced for H3ac, while the H4ac signal was also found in more upstream and downstream regions. After a 12-h incubation with butyrate, we observed that the signal at 116 regions for H3ac and 124 regions for H4ac was significantly decreased or lost (down groups), while the 181 and 119 regions remained unchanged, respectively (unchanged groups) (Table 1). In many cases deacetylation affected both H3 and H4 (30% of H3ac down, 28% of H4ac down), while at regions where only one histone was deacetylated, the other was generally not acetylated at 0 h (66% of H3ac down were absent for H4ac, 45% of H4ac down were absent for H3ac). It was more frequent that when both histones were acetylated, H4ac suffered butyrate-mediated deacetylation alone (25% of H4ac down were unchanged for H3ac; 4% of H3ac down were unchanged for H4ac) (Fig. 2A; Table 1). Globally, the ENCODE arrays contain around 400 genes, depending on the various annotations (e.g., 392 unique Entrez Gene IDs). Considering the unchanged and down regions (UES regions) (see Table 1; Supplemental material) described above, we selected those mapping within 5 kb of TSS of known genes. A total of 102/116 regions for H3ac and 47/124 regions for H4ac belonging to down groups were in proximity of TSS, while 80/181 and 42/119 of the unchanged regions for H3ac and H4ac, respectively, were in such location. These numbers indicate that around 25% and 12% of genes in the ENCODE regions could be deacetylated at H3 and H4, respectively, after butyrate treatment.
Figure 2.
Genome-wide changes in acetylation patterns of histone H3 and H4 in 1% of the genome after butyrate treatment. (A) Number of unique enriched spots (UES, see Supplemental Material and Methods) for H3ac and H4ac, respectively, and overlapping regions at 0 h, 12 h, deacetylated (down), unchanged (Un), and Up groups. (B) Distribution of the different UES respective to their distance to TSS is presented for the 0 h, 12 h, down, unchanged (Un), and up groups either for H3 (left) or H4 (right) alone, or for both histones (bottom). Due to multiple TSSs for unique genes, each UES could map to more than one category, and therefore, total numbers can sum up more than 100%. Distances to TSS or 3′ ends are in Kbp. (1st int) First intron; (Intra) intragenic; (Inter) intergenic. (C) H3ac and H4ac decreased around TSS of LACE1, according to our ChIP-chip experiments (top histograms), which was verified when analyzed by PCR in the same region (bottom). The spots presented in the upper part come from one of the biological replicates in each case, where ChIP DNA was always labeled with Cy5 (red), while input DNA was labeled with Cy3 (green). The log2-ratios in the histograms represent the ratios between the signals for ChIP DNA labeled with Cy5 and the input DNA labeled with Cy3, which have been transformed to log2 values. In such histograms, the blue and red bars correspond to untreated (0 h) and treated (12 h) experiments, respectively. (D) Log2-ratios of acetylated histones and negative control arrays, depicting the spots mapping within 5 kb of Transcription Start Sites (TSS) for UES. For all selected spots a sequence window of ±25 Kb around TSS is presented.
Table 1.
Regions with different histone acetylation patterns before and after butyrate treatment
The numbers represent the overlaps between unique enriched spots (UES) regions as defined in the Supplemental material for the different categories (up, down, unchanged, or absent) of H3ac vs. H4ac. (RS) Remaining spots in array.
The results for a representative gene, LACE1, are presented in Figure 2C. The regions deacetylated upon butyrate treatment were preferentially focused on TSSs compared with the regions that were unchanged after treatment (Fig. 2B), especially for H3ac. By plotting the Log2-ratios against the distance to TSS for both acetylated histones at 0 and 12 h after butyrate treatment, it was evident that at 0 h, there was a peak of acetylation immediately downstream or upstream of TSS for H3ac and H4ac, respectively. Butyrate treatment induced a clear decrease in the intensity of the peaks (Fig. 2D). These results are unexpected, but previous reports have found deacetylation in promoters of genes down-regulated by HDACi (Duan et al. 2005; Reid et al. 2005) and gene repression is commonly observed in HDACi-treated cells. On the other hand, a limited number of the loci we examined displayed increases in acetylation levels (Up group), although of low magnitude and mostly for H4ac. The majority of these changes were located in far upstream or intronic regions (Fig. 2B).
Time-series verification of butyrate-mediated deacetylation and effects of TSA
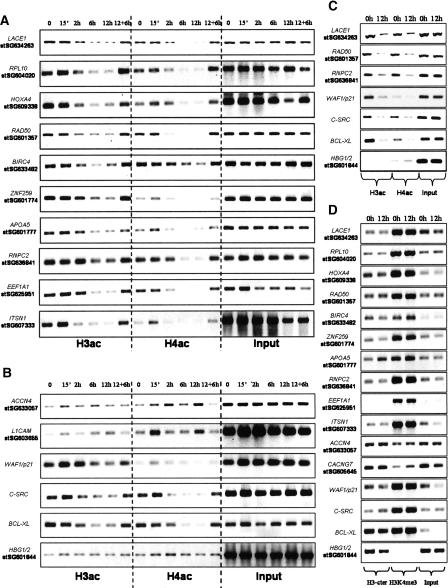
In order to verify the ChIP-chip results, we selected regions with butyrate-mediated deacetylation (10 loci) or increased acetylation (two loci). Since we could not find any spot in the proximity of a TSS with increased acetylation for both histones, we selected two regions with a trend toward hyperacetylation, although below statistical significance. In addition, we analyzed the promoter regions of genes previously shown to be affected by butyrate. CDKN1A is up-regulated by butyrate in HepG2 cells (Hirsch and Bonham 2004), while SRC (Kostyniuk et al. 2002) and BCL2L1 (formerly BCL-XL) (Ruemmele et al. 2003) are inhibited. As a negative control we selected a region close to a TSS without enrichment or change for H3ac or H4ac (Supplemental Table 1).
We generated ChIP material for H3ac and H4ac after 0-h, 15-min, 2-h, 6-h, and 12-h butyrate treatment, and after treatment for 12 h followed by 6 h without butyrate. For the 10 regions where deacetylation was found by ChIP-chip, we concluded that butyrate response was time dependent, with slight initial increase after 15 min at some of the targets, before deacetylation was detected after 2 h and reaching maximum levels already at 6 h. All of the deacetylations were almost completely reversed after removal of butyrate (Fig. 3A).
Figure 3.
Validation of ChIP-chip results and effects on nucleosome occupancy and H3K4me3 of butyrate-mediated histone deacetylation. ChIPs were prepared from cells that were either untreated (0 h) or treated with 5 mM Na-butyrate for 15 min (15’), 2 h, 6 h, or 12 h, and cells were exposed to butyrate for 12 h and then grown for 6 h without (12 + 6 h) using antibodies against H3ac and H4ac. ChIPs were analyzed by amplifying the regions indicated in Supplemental Table 1, which displayed histone deacetylation (A) or histone hyperacetylation (B) in our ChIP-chip analysis. A negative control region (HBG1/2) was analyzed together with some gene promoters from the literature (B). ChIPs were prepared from cells that were either nontreated (0 h) or treated with 100 ng/mL TSA for 12 h, using antibodies against H3ac and H4ac. Some of the selected targets from A and B were analyzed and presented here (C) and in Supplemental Figure 2. ChIPs were also prepared from cells that were either untreated (0 h) or treated with 5 mM Na-butyrate for 12 h using an antibody against H3-cter or against H3K4me3 (D).
In the two regions with increased acetylation by ChIP-chip, the results were less clear. The levels of acetylation were much lower than above, and even after 12 h, the signals compared with input levels were low (Fig. 3B). This was expected, since in our ChIP-chip analysis these regions were below our cut-off values. However, in general, we could detect slight increases in acetylation that were detectable after a 15-min treatment.
After a short treatment, butyrate provoked a clear increase in histone acetylation at the CDKN1A promoter, especially of H3, that reached a maximum after 15 min and remained high after 2 h. However, longer exposure resulted in deacetylation, and the levels after 12 h were lower than those before treatment. In the case of SRC and BCL2L1, the results were similar to the 10 deacetylated targets (Fig. 3B). The specificity of the previous results was shown by the low and stable levels of H3ac and H4ac in the negative control region (HBG1/2) (Fig. 3B). Overall, we can conclude that at very early time points (15 min), there seems to be a common increase in histone acetylation, which, however, is transient. Furthermore, histone deacetylation occurred in a delayed fashion, since it was not clearly evident until 6 h after treatment for most regions.
Similar results were obtained in cells before and after treatment with TSA for 12 h, with H3 and H4 deacetylations observed at the same selected regions, indicating that the deacetylations are not restricted to butyrate (Fig. 3; Supplemental Fig. 2). Furthermore, when treating a different cell line, HT29, derived from a colon adenocarcinoma with butyrate for 12 h, a similar response was observed with clear deacetylations of H3 and H4 at investigated regions (Supplemental Fig. 3). These results are highly relevant, due to the natural production of butyrate in the colon, where it might play a protective role against tumor development. Finally, due to the global increase in histone acetylation provoked by butyrate, it was important to determine that in the treated cells the antibody against acetylated histones was not a limiting factor and the reason for the observed decrease in signal for H3ac and H4ac. ChIP experiments were performed with ∼10 times less cells than in ChIP-chip and either 5 μg or 10 μg of antibody, the latter the amount used in ChIP-chip. When analyzing the previously selected regions, clear histone deacetylation was evident for both H3 and H4, irrespective of amount of antibody (data not shown). In conclusion, there was no indication of the amount of antibody being a limiting factor.
Butyrate neither changes nucleosome density nor H3K4trimetylation
Decreased histone acetylation signal could be the result of nucleosome loss, perhaps due to further induction of nearby genes, as has been observed in yeast, where promoters of active genes have lower nucleosome density (Lee et al. 2004). Using an antibody against the C-terminal end of H3 that served as a measurement of nucleosome occupancy, we found by ChIP no indication of nucleosome loss, at least at the resolution we can achieve, for any of the regions analyzed (Fig. 3D). Furthermore, H3K4me3 levels did not change upon butyrate treatment. Interestingly, most of the regions with high levels of acetylated histones at 0 h displayed high levels of H3K4me3 as well, in concordance with previous studies (Bernstein et al. 2005). The stability of the methylation mark gives a possible explanation for the reversibility of the butyrate-mediated deacetylations, given the presumed cross-talk between these modifications (Dou et al. 2005; Pray-Grant et al. 2005) (see Discussion).
Interaction of transcription factors (TF)s, HATs, and HDACs with deacetylated regions
The acetylation status of histones depends on HDACs and HATs, which do not have histones as their only substrates. Western blotting demonstrated that HDAC1 was not detectable and that SIRT1 was mainly cytoplasmic in HepG2 extracts. Hence, it was not surprising that we could not detect, by ChIP, binding of these proteins at any of the analyzed targets (data not shown). Establishment of HDACs’ role in butyrate-mediated histone deacetylation will require systematic screening of all HDACi-sensitive (e.g., HDAC1) and HDACi-insensitive HDACs (e.g., SIRT1).
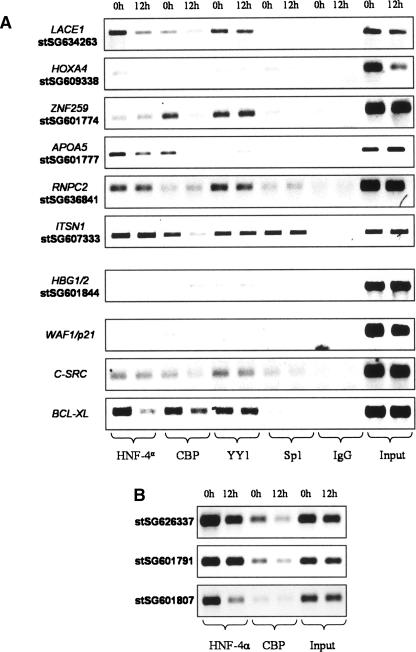
CBP is a HAT whose binding has been reported to be reduced in the BCL2 promoter upon TSA treatment, accompanied by histone deacetylation (Duan et al. 2005). TFs are known to interact with HATs and/or HDACs, and they can also be substrates of such enzymes, e.g., SP1 (Davie 2003), YY1 (Yao et al. 2001), and HNF4A (Soutoglou et al. 2000). Both HNF4A and CBP bindings were reduced by butyrate at 3/10 and 5/10 targets, respectively. Neither YY1 nor SP1 bindings were affected by butyrate (Fig. 4A). HNF4A is known to interact with distal-regulatory elements (Rada-Iglesias et al. 2005), and two of such regions (3 kb downstream from ENPP1 [stSG626337] and 15 kb upstream from APOA5 [stSG601791]), plus a strongly bound promoter (2.5 kb upstream from APOC3 [stSG601807]) were investigated for HNF4A and CBP (Fig. 4B). Some decrease in binding was observed, although the bindings to distal regions seemed less affected by butyrate.
Figure 4.
Butyrate affects the binding of some transcription factors bound at deacetylated regions. ChIPs were prepared from cells that were either untreated (0 h) or treated with 5 mM Na-butyrate for 12 h, using antibodies against the proteins indicated in the bottom of the pictures. In A, some of the regions from Supplemental Table 1 were analyzed. In B, we analyzed some distal elements previously identified as HNF4A targets. The amplified regions were 3 kb downstream from ENPP1 (stSG626337), 15 kb upstream from APOA5 (stSG601791), and 2.5 kb upstream from APOC3 (stSG601807).
Butyrate-mediated histone deacetylation results in decreased gene expression
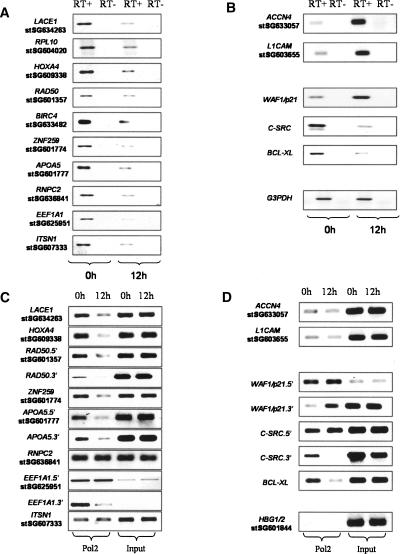
H3ac and H4ac are usually located in the proximity of TSS and are hallmarks of transcriptionally active chromatin, so decreased expression was expected for genes close to deacetylated regions. All of the selected genes with histone deacetylation around the TSS were down-regulated by butyrate (Fig. 5A). Furthermore, EEF1A1 and BIRC4 were previously shown to be down-regulated by butyrate (Goncalves et al. 2005; Kim et al. 2005a), while RBM39 and RPL10 are inhibited under other HDACi treatments (de Ruijter et al. 2005; Gialitakis et al. 2006). For the two genes with increased acetylation close to the TSSs, the overall expression levels were low, but increased expression after treatment was observed (Fig. 5B). The results for our control genes were in concordance with previous reports. CDKN1A expression was increased, while SRC and BCL2L1 levels were diminished. The control GAPDH remained constant after butyrate treatment (Fig. 5B).
Figure 5.
Histone deacetylation at promoter regions results in decreased expression of the associated genes mediated at the transcriptional level. RNAs were prepared from cells that were either untreated (0 h) or treated with 5 mM Na-butyrate for 12 h. Total RNA (1 μg) was used for preparing gene-specific cDNAs using reverse transcriptase (RT), which were analyzed by RT–PCR to investigate the RNA levels of genes displaying histone deacetylation (A), while genes with increased acetylation, genes from the literature, and a loading control (GAPDH) are presented in B. Similar results were obtained in a biological replicate or when cDNAs were obtained using oligo(dT). ChIPs were prepared from cells that were either nontreated (0 h) or treated with 5 mM Na-butyrate for 12 h using an antibody against the C-terminal domain of POLR2A. The analyzed regions included most from Supplemental Table 1, divided into those with histone deacetylation (C) and with histone hyperacetylation from previous literature or serving as negative control (HBG1/2) (D). For some genes, two different genomic regions were analyzed, one close to the TSS (Supplemental Table 1) is indicated by the gene name, followed by 5′, while an intragenic region was analyzed using the same primers used in A and B and is indicated by the gene name, followed by 3′. Similar results were obtained in two other biological replicates.
The decrease in expression occurs at the transcriptional level: Butyrate effects on POLR2A occupancy
It has been previously suggested that histone acetylation around TSS might facilitate POLR2A preinitiation, initiation, and/or elongation steps (Struhl 1998; Kristjuhan and Svejstrup 2004; Zhao et al. 2005). We investigated POLR2A occupancy by ChIP of untreated cells and cells treated with butyrate for 12 h and analyzed regions close to TSS (Supplemental Table 1) and some downstream intragenic regions. We analyzed eight of 10 deacetylated regions from Supplemental Table 1 and targets at some distance upstream from TSS showed no changes in POLR2A occupancy (e.g., RBM39, EEF1A1.5′, ITSN1), while regions very close to TSS or intragenic showed decreased POLR2A binding (e.g., LACE1, HOXA4, RAD50, etc). In all downstream intragenic regions, decreased POLR2A occupancy was observed, including EEF1A1.3′ and SRC.3′, where no changes in the 5′regions were found (Fig. 5C).
For the two genes with increased acetylation and expression, very low levels of POLR2A were detected and no detectable changes in POLR2A occupancy were induced by butyrate (Fig. 5D). CDKN1A showed increased POLR2A occupancy close to TSS, which was more evident further downstream in the gene. For SRC, the deacetylated region was located immediately upstream of a tissue-specific TSS, but more than 1 Kb upstream from a second ubiquitous TSS (Kostyniuk et al. 2002). The POLR2A occupancy was not affected in the upstream region, but when analyzing an intragenic area, reduction in POLR2A binding was mediated by butyrate. POLR2A binding at the BCL2L1 gene was reduced when analyzing an intragenic region close to the TSS (Fig. 5D).
Functional and structural similarities among genes displaying histone deacetylation
We investigated whether histone deacetylation around TSS could be the result of some sequence and/or architecture similarities at the proximal promoters of affected genes and whether they might be functionally related. We studied the Gene Ontology (GO) profiles of genes where histone deacetylation was noted within 5 kb of TSSs and compared them with the genes with no changes, using all of the genes contained in the ENCODE regions as background (Supplemental material). Genes with increased acetylation were few and not investigated. Among deacetylated genes, significantly over-represented categories included several related with cell cycle and proliferation (e.g., M-phase, Meiosis, purine biosynthesis, etc.) and genes that in general promote cell proliferation (e.g., YWHAH) (Wanzel et al. 2005), HCFC1 (Piluso et al. 2002). The other major group of deacetylated genes was mainly involved in protein biosynthesis, including translation elongation factors (e.g., EEF1A1, ITGB4BP) and ribosomal proteins (e.g., RPL10, MRPL23). Inhibition of translation factors and ribosomal proteins seems to be a general observation in cells under stress and slowing of their growth rate (Murray et al. 2004).
We compared the number of CpG islands within 5 kb of deacetylated regions with that in regions remaining acetylated after butyrate treatment. CpG islands were significantly more common in deacetylated areas, especially regarding H3ac (79% for Down vs. 44% for Unchanged, P = 1.3 × 10−9) (Supplemental Table 2). Finally, we evaluated whether the genes responding differently to butyrate might differ in their core promoter elements. Our data indicates that only initiator and initiator-like sequences are frequent and slightly more abundant among deacetylated promoters, while the other analyzed motifs are either rare or equally common in unchanged ones (Supplemental Table 3).
Butyrate-mediated increase in bulk histone acetylation is preferentially located at the nuclear periphery
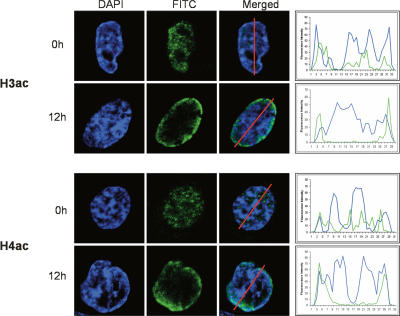
In our ChIP-chip analysis we have interrogated 1% of the human genome as selected in the ENCODE project, which can be considered as part of euchromatin and facultative heterochromatin. Constitutive heterochromatin is primarily located around telomeres and centromeres and excluded from this 1% of the genome. Since the analysis of the changes in histone acetylation by Western-blot and ChIP-chip generated contradictory results, we evaluated possible explanations. We performed immunostaining of HepG2 cells untreated or treated with butyrate for 12 h using antibodies against H3ac and H4ac. Both modifications were mainly nuclear, with a foci-like distribution, and largely excluded from nucleoli and heterochromatic regions at 0 h, as indicated by high-DAPI intensity areas (Fig. 6; Supplemental Fig. 3). Butyrate treatment resulted in a clear increase in acetylation signal, mainly localized at the nuclear periphery and without a foci-like appearance. Immunostaining using the antibody against H3-Cter showed no changes in the staining pattern of total histone H3 (Supplemental Fig. 3).
Figure 6.
Butyrate-mediated increase in total levels of acetylated histones preferentially occurs at the nuclear periphery. HepG2 cells were either untreated (0 h) or treated with 5 mM Na-butyrate for 12 h, and immunostaining assays were performed under both conditions using antibodies against H3ac (top) and H4ac (bottom) and secondary antibody coupled to FITC. From left to right are presented the staining patterns observed for DAPI in blue, FITC in green, and when both stainings are merged. The graphs to the right represent the signal quantification obtained for both DAPI and FITC along the red line drawn in the merged picture. The images represent the overall picture emerging from two biological replicates (Supplemental Fig. 3).
Discussion
Butyrate frequently provokes histone deacetylation around the TSS of active genes
Butyrate and other HDACi are under clinical trials to evaluate their potential as anti-tumourogenic drugs (Villar-Garea and Esteller 2004). However, this evaluation is done without a complete understanding of their molecular mechanisms of action. We have performed the first large-scale characterization of the acetylation status of DNA-bound histones after treating HepG2 cells with butyrate. By interrogating 1% of the human genome by ChIP-chip, we found that many acetylated regions were deacetylated after treatment, frequently affecting both H3 and H4 and mapping close to TSSs. The same effect was seen when HepG2 cells were treated with TSA, indicating that the effect may be general to HDACi. Similar deacetylation was seen in the colon cancer cell line HT29 after butyrate treatment, so the effect may be seen in many cell lines. We then demonstrated that the loss of histone acetylation cannot be explained in terms of nucleosome density (Lee et al. 2004). Furthermore, H3K4me3, which is also a hallmark of transcriptionally active genes (Pokholok et al. 2005), was unaffected by butyrate, as previously observed under TSA treatment (Hazzalin and Mahadevan 2005). This observation could be of importance, since it suggests that the promoters conserve some of their transcriptionally active “landscape” and might explain why butyrate-mediated deacetylation was reversible. Conservation of H3K4me3 and perhaps of other “active” marks, could help in the recruitment of HATs and in the restoration of histone acetylation of these promoters after butyrate withdrawal. Moreover, these stable active marks could render promoter regions in a sufficiently accessible state, allowing Preinitiation complex (PIC)/POLR2A and some TFs to remain bound at core promoters.
Histone deacetylation occurs around TSS of genes down-regulated by butyrate
The observed histone deacetylation around TSSs was accompanied by decreased RNA levels at nearby genes. Furthermore, POLR2A binding was generally diminished both close to TSS and further downstream, while it was rather stable where the analyzed regions were upstream from TSSs. This suggests that histone deacetylation affects transcription initiation and/or elongation, rather than PIC formation (Kristjuhan and Svejstrup 2004; Zhao et al. 2005). In the COX2 and MYC genes in particular, butyrate was reported to suppress elongation, since POLR2A levels were not affected at exon 1, but clearly diminished in downstream exons (Tong et al. 2005).
Overall, our data provides a mechanistic explanation for the frequently observed repression of genes under butyrate and other HDACi treatments. Among the 10 genes identified in the ChIP-chip screen, where we validated the butyrate-mediated histone deacetylation and gene repression, four of them were previously shown to be down-regulated by butyrate or other HDACi (de Ruijter et al. 2005; Goncalves et al. 2005; Kim et al. 2005a; Gialitakis et al. 2006). Furthermore, other genes like STAG2 and MECP2, where we observed histone deacetylation around TSS, were also previously found to be down-regulated under HDACi treatment (de Ruijter et al. 2005; Daly and Shirazi-Beechey 2006). Finally, our data concerning SRC and BCL2L1 is also in agreement with previous data (Kostyniuk et al. 2002; Ruemmele et al. 2003), demonstrating that gene repression mediated by HDACi treatment is a common event and that our data can link to changes in histone acetylation. However, histone deacetylation could be either the cause or a consequence of genes being silenced. In comparison to the early and constant increase in global histone acetylation, as well as the early and transient hyperacetylation of some promoters (e.g., CDKN1A), histone deacetylation seems to occur in a delayed fashion. This could indicate that histone deacetylation is a secondary effect due to a “stress-response” derived from the early events provoked directly by butyrate. In this respect, butyrate is known to induce growth arrest and/or apoptosis in HepG2 cells (Donadel et al. 1991; Emanuele et al. 2004), and it has been previously shown that histone deacetylation occurs at numerous gene promoters in quiescent cells (Rayman et al. 2002; Balciunaite et al. 2005) and cells undergoing terminal growth arrest and acquiring a senescence-like phenotype (Jackson and Pereira-Smith 2006). In both cases, histone deacetylation is accompanied by silencing of the corresponding genes, and E2F/pocket protein complexes bound to their promoters mediate histone deacetylation through recruitment of HDACs. According to our GO analysis of genes with deacetylated promoters, there was a clear over-representation of genes promoting cell proliferation and protein biosynthesis. Silencing of genes belonging to these functional categories could be the signature of a “stress-response” of HepG2 cells to butyrate, with subsequent growth arrest and apoptosis if butyrate exposure is prolonged.
Butyrate-mediated promoter hyperacetylation is an early and transient event
We detected a few examples of increased acetylation in our ChIP-chip analysis. Most were restricted to H4 of low magnitude and commonly located in far upstream and intronic regions. This localization might reflect, in some cases, uncoupled HDAC activity at coding regions, which has been suggested to inhibit spurious transcription-initiation events in connection with H3K36 methylation (Lieb and Clarke 2005). In the case of CDKN1A, it has previously been suggested that elevated mRNA levels in HepG2 cells after butyrate treatment are mainly due to post-transcriptional events (Hirsch and Bonham 2004) and mechanisms not related to histone acetylation (Han et al. 2001). This can be in agreement with our observations, as butyrate provokes an initial increase in histone acetylation, followed by increased transcription that at longer times may be accompanied by post-transcriptional mechanisms once histone acetylation decreases to basal. The importance of post-transcriptional events in HDACi-mediated gene activation is well illustrated by the inhibition of miRNAs by these drugs (81% of detectable miRNAs were down-regulated), since it may lead to mRNA up-regulation (Scott et al. 2006). Richon et al. (2000) found that CDKN1A steady-state mRNA levels, transcription rate, and H4ac rapidly increased after SAHA treatment. However, mRNA levels decreased after 24 h and H4ac went back to basal levels. In summary, the combination of early and transient increases in histone acetylation together with post-transcriptional mechanisms might explain the up-regulation of certain genes (e.g., CDKN1A), without an evident increase in histone acetylation after a 12-h butyrate treatment, as investigated in our ChIP-chip study. Similarly, Hazzalin and Mahadevin (2005) found that upon TSA treatment, H3ac around TSS of the inducible proto-oncogenes Fos and Jun rapidly increased, peaking at 15 min to 1 h after treatment. However, after a 2-h treatment, the levels went back to almost basal and after 4 h, histone deacetylation compared with controls could be observed. This is in agreement with Peart et al. (2005), who observed that HDACi treatment resulted in an early response (1–4 h) involving mainly gene activation, while gene repression steadily increased over time, and at 16 h they found more genes being repressed than activated. Furthermore, in this study it is suggested that a large fraction of genes (40%) are affected by HDACi treatment, although many changes are of low magnitude. Considering that they found more genes being down-regulated than up-regulated, the fraction of known genes displaying histone deacetylation close to their TSS in our study (25% for H3ac, 12% for H4ac) and expected to be down-regulated, is in that same range. In conclusion, we and others have observed that histone hyperacetylation at gene promoters after HDACi treatment is an early and transient event, with important implications for a better understanding of the kinetics of histone modifications (see Hazzalin and Mahadevan 2005 for a detailed study of these early events). However, HDACi cancer treatment is long term, with patients taking the drug for days and weeks. Therefore, we consider the long-term effects of HDACi treatment more relevant from a clinical perspective.
Butyrate-mediated histone deacetylation affects functionally and structurally related genes
CpG islands were more abundant in deacetylated regions than in unchanged ones. Similarly, it was recently reported that CpG island content in promoters positive for TAF1 binding was significantly higher than previous estimates (Kim et al. 2005b). Furthermore, initiator sequences recognized by TAF1(Smale and Kadonaga 2003), were slightly more abundant in deacetylated core promoters than in unchanged ones. Interestingly, HDACi-mediated down-regulation of SRC is TAF1 dependent (Dehm et al. 2004). TAF1 is a general transcription factor with HAT activity specific for histones H3 and H4 in vitro (Mizzen et al. 1996). Disruption of TAF1 HAT activity in mammalian cells results in G1 arrest (Dunphy et al. 2000) and down-regulation of many genes (O’Brien and Tjian 2000). The effects on cell-cycle progression and gene expression induced by disrupting TAF1 HAT activity resemble those observed during butyrate treatment. Therefore, we speculate that direct or indirect disruption of TAF1 HAT activity by butyrate could partly explain the loss of histone acetylation at some active promoters, even though inhibition of other HATs cannot be ruled out. Hazzalin and Mahadevan (2005) found that prolonged TSA treatment (4 h) results in H3 deacetylation around TSS of Fos and Jun, and they suggested that it could be mediated by HDACi-insensitive HDAC or by histone replacement. Finally, as previously mentioned, HDAC recruitment to E2F/pocket protein-target genes under HDACi-mediated growth arrest represents another intriguing possibility requiring further experimental investigation.
The increase in total histone acetylation mediated by butyrate is preferentially localized to the nuclear periphery
Finally, a major paradox is how the increase in global histone acetylation level induced by butyrate can correlate with the observed deacetylation in our ChIP-chip analysis of 1% of the human genome. Heterochromatin consists of facultative heterochromatin, which includes euchromatic regions silenced in a particular cellular context, and constitutive heterochromatin, which are large genomic areas near centromeres and telomeres, mostly formed by repetitive sequences silenced in all cell types (Dimitri et al. 2005). TSA treatment of human cells lead to accumulation of H4K5ac at the nuclear periphery (Taddei et al. 2001). This occurred after 2 h of treatment, but up to 5 d were needed in order to disrupt the pericentric heterochromatin. Similarly, long TSA treatment resulted in accumulation of H3K9ac at pericentric heterochromatic minor satellites (Maison et al. 2002) and, also, butyrate treatment resulted in increased levels of H3K9ac at the nuclear periphery (Bartova et al. 2005). In agreement with these studies, we have found that treatment of HepG2 cells with Na-butyrate for 12 h resulted in increased levels of H3ac and H4ac at the nuclear periphery. Perinuclear localization of chromatin coincides with transcriptional silencing (Andrulis et al. 1998) and acetylated histones are absent close to the nuclear envelope (Sadoni et al. 1999), where centromeric regions are preferentially found in mouse and humans (Weierich et al. 2003). Thus, accumulation of histone acetylation after HDACi treatment could occur at constitutive heterochromatin. Our preliminary analysis of two heterochromatic repeats (i.e., SAT2 and NBL2; data not shown) showed no indication of increased H3ac or H4ac upon butyrate treatment, suggesting that either longer treatment is needed in order to accumulate acetylated histones at heterochromatin or that accumulation occurs at other heterochromatic repeats. Another alternative is that the increased acetylation preferentially affects nuclear histones not bound to DNA, but located at the nuclear periphery. In this respect, H3/H4 interact in vitro with HP1 and LBR, a component of the inner nuclear membrane (Polioudaki et al. 2001). Moreover, HDAC3 contacts with another member of the nuclear inner envelope, LAP2B (Somech et al. 2005). In summary, current data indicate that most of the increase in histone acetylation caused by HDACi treatment localizes to the nuclear periphery, and heterochromatin disruption seems to depend on duration of treatment. Perhaps free histones are acetylated at early times and targeted to the nuclear periphery before they are incorporated into heterochromatin in a cell-cycle dependent manner (Taddei et al. 2005).
Conclusion
Butyrate, in particular, and HDACi, in general, are promising anti-tumourogenic agents that are already used or in clinical trials for treatment of cancer. In spite of their medical interest, the molecular mechanisms of this diverse group of drugs are poorly understood. The work presented here establishes the first genomic characterization of the effects of one of these drugs, Na-Butyrate, on the acetylation status of DNA-bound histones. The major effect in euchromatin is a histone deacetylation centered on TSS, which could be mediated by a pleiotropic effect on HATs and/or HDACs, suggesting that preassumed models for the actions of these drugs might need to be revised. Finally, treatment with TSA resulted in similar histone deacetylation in a number of selected regions, which suggests that such response could be general to HDACi treatment and offers an explanation for the commonly observed down-regulation of genes under these treatments.
Methods
Cell culture and treatments
HepG2 and HT-29 cells were grown in RPMI-1640 medium (Sigma-Aldrich) supplemented with 10% FBS (GIBCO, Invitrogen), 1% PEST (GIBCO, Invitrogen), and 1% Glutamine (GIBCO, Invitrogen) at 37°C with 5% CO2. When cells were subconfluent, medium was changed for both untreated and treated cells. The treated cells were grown for the indicated period (15 min, 2 h, 6 h, 12 h) in medium supplemented with 5 mM Na-butyrate (Sigma-Aldrich) or 100 ng/mL TSA. Control cells were grown in parallel with the treated cells, typically for 12 h, since the majority of the treatments were performed for those same hours. For the 12 + 6-h treatment, 12 h after butyrate addition, the medium was removed, the cells washed with PBS, and fresh medium without butyrate was added and cells maintained in such conditions for an additional 6 h.
Protein extracts, Western blot, and antibodies
Nuclear and cytoplasmic extracts from HepG2 cells and Western-blot analysis were performed as described (Rada-Iglesias et al. 2005). Anti-Histone H3 acetyl K9/14 (06–599), anti-Histone H4 acetyl K5/8/12/16 (06–866), anti-SIRT1 (05–707), and normal rabbit IgG (12–370) were from Upstate. Anti-Histone H3 trimethyl K4 (ab8580) and anti-Histone H3 (ab1791) were from AbCam. Antibodies against HNF4A (sc-6556), SP1 (sc-17824), CBP (sc-369), HDAC1 (sc-6299), and YY1 (sc-1703) were from Santa Cruz. Antibody against C-terminal domain (CTD) of POLR2A (MMS-126R) was from Covance.
Chromatin immunoprecipiation (ChIPs) and microarray hybridization
ChIPs were performed as previously described (Rada-Iglesias et al. 2005). Briefly, one 175-cm2 cell flask with subconfluent cells was used per ChIP experiment. Cells were cross-linking with 0.37% formaldehyde for 10 min. Chromatin was sonicated to a size of 0.5–2 kb, a fraction was kept as input (∼15%–20% of total chromatin), and the rest was incubated with 10 μg of antibody and protein G-agarose. ProteinG-agarose pellets were washed and DNA–protein complexes eluted, treated with RNaseA (Amersham Bioscience), and cross-links were reversed. Proteins were degraded by Proteinase K (Amersham Bioscience) and DNA was extracted, purified, and resuspended in water. Microarray construction, DNA labeling, and microarray hybridization were exactly as previously described (Rada-Iglesias et al. 2005). Briefly, for each histone modification and butyrate condition, three completely independent biological replicates were performed. The DNA obtained from a single ChIP reaction was labeled with Cy5 and a 1/3 of the corresponding input was labeled with Cy3. For labeling reactions, the Bioprime Labeling system (Invitrogen) was used. Labeled ChIP/Cy5 and Total input/Cy3 DNAs were combined and hybridized to the array.
Microarray data analysis, Gene Ontology (GO) categories, CpG islands content, and Core promoter’s analysis
The computational analysis was divided in various parts, some of them as previously described (Rada-Iglesias et al. 2005). The LCB-DataWareHouse was used for analysis of microarray data and GO categories (Ameur et al. 2006). Extensive description of all the procedures can be found in the Supplemental material.
RNA preparation and RT–PCR
5–10 x 106 HepG2 cells were treated with 5 mM Na-butyrate for 12 h or grown in parallel without treatment for the same time. RNA was extracted using 1 mL TRIzol (Invitrogen). Samples were treated with RNAguard RNAse inhibitor (Amersham) and Turbo DNA-free DNase I (AmBion). Gene-specific or oligo-dT primed RT-PCR was performed using 1 μg of RNA and the M-MLV Reverse Transcriptase kit (Invitrogen). The resulting cDNAs were amplified with specific primers for each gene.
Immunostaining, image acquisition, and analysis
HepG2 cells were grown in glass slides, fixed with 4% paraformaldehyde, and permeabilized. Slides were treated with Image-iT FX Signal enhancer (Molecular Probes) and an additional blocking step was carried out in a solution containing 2% (w/v) BSA and 0.1% (v/v) Triton X-100. Slides were incubated with primary antibodies for 1–2 h, rinsed, and incubated with anti-rabbit FITC (Jackson Immunoresearch Laboratories) for 1 h. The slides were rinsed and mounted in ProLong Gold Antifade (Molecular Probes). Images were acquired using a confocal microscope 510 META (Zeiss), Axiovision LE rel. 4.2 (Axioplan II), and LSM 5 Image browser (510 META). Signal quantification was performed using Image J software by calculating the signal intensity along a line crossing one of the nucleus’ axis.
Acknowledgments
This work was supported by the Swedish Research Council, the Wellcome Trust, The US National Human Genome Research Institute (Grant 5 U01 HG003168), the Markus Borgström Foundation, and the Knut and Alice Wallenberg Foundation.
Footnotes
[Supplemental material is available online at www.genome.org. The microarray data presented in this work have been submitted to ArrayExpress database, with accession number E-MEXP-693. The processed ChIP-chip data can be visualized at www.genome.ucsc.edu using the Human May 2004 (hg17) Assembly.]
Article is online at http://www.genome.org/cgi/doi/10.1101/gr.5540007
References
- Ameur A., Yankovski V., Enroth S., Spjuth O., Komorowski J., Yankovski V., Enroth S., Spjuth O., Komorowski J., Enroth S., Spjuth O., Komorowski J., Spjuth O., Komorowski J., Komorowski J. The LCB DataWareHouse. Bioinformatics. 2006;22:1024–1026. doi: 10.1093/bioinformatics/btl036. [DOI] [PubMed] [Google Scholar]
- Andrulis E.D., Neiman A.M., Zappulla D.C., Sternglanz R., Neiman A.M., Zappulla D.C., Sternglanz R., Zappulla D.C., Sternglanz R., Sternglanz R. Perinuclear localization of chromatin facilitates transcriptional silencing. Nature. 1998;394:592–595. doi: 10.1038/29100. [DOI] [PubMed] [Google Scholar]
- Balciunaite E., Spektor A., Lents N.H., Cam H., Te Riele H., Scime A., Rudnicki M.A., Young R., Dynlacht B.D., Spektor A., Lents N.H., Cam H., Te Riele H., Scime A., Rudnicki M.A., Young R., Dynlacht B.D., Lents N.H., Cam H., Te Riele H., Scime A., Rudnicki M.A., Young R., Dynlacht B.D., Cam H., Te Riele H., Scime A., Rudnicki M.A., Young R., Dynlacht B.D., Te Riele H., Scime A., Rudnicki M.A., Young R., Dynlacht B.D., Scime A., Rudnicki M.A., Young R., Dynlacht B.D., Rudnicki M.A., Young R., Dynlacht B.D., Young R., Dynlacht B.D., Dynlacht B.D. Pocket protein complexes are recruited to distinct targets in quiescent and proliferating cells. Mol. Cell. Biol. 2005;25:8166–8178. doi: 10.1128/MCB.25.18.8166-8178.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartova E., Pachernik J., Harnicarova A., Kovarik A., Kovarikova M., Hofmanova J., Skalnikova M., Kozubek M., Kozubek S., Pachernik J., Harnicarova A., Kovarik A., Kovarikova M., Hofmanova J., Skalnikova M., Kozubek M., Kozubek S., Harnicarova A., Kovarik A., Kovarikova M., Hofmanova J., Skalnikova M., Kozubek M., Kozubek S., Kovarik A., Kovarikova M., Hofmanova J., Skalnikova M., Kozubek M., Kozubek S., Kovarikova M., Hofmanova J., Skalnikova M., Kozubek M., Kozubek S., Hofmanova J., Skalnikova M., Kozubek M., Kozubek S., Skalnikova M., Kozubek M., Kozubek S., Kozubek M., Kozubek S., Kozubek S. Nuclear levels and patterns of histone H3 modification and HP1 proteins after inhibition of histone deacetylases. J. Cell Sci. 2005;118:5035–5046. doi: 10.1242/jcs.02621. [DOI] [PubMed] [Google Scholar]
- Baylin S.B. DNA methylation and gene silencing in cancer. Nat. Clin. Pract. Oncol. 2005;(Suppl. 1) 2:S4–11. doi: 10.1038/ncponc0354. [DOI] [PubMed] [Google Scholar]
- Bernstein B.E., Kamal M., Lindblad-Toh K., Bekiranov S., Bailey D.K., Huebert D.J., McMahon S., Karlsson E.K., Kulbokas E.J., 3rd, Gingeras T.R., Kamal M., Lindblad-Toh K., Bekiranov S., Bailey D.K., Huebert D.J., McMahon S., Karlsson E.K., Kulbokas E.J., 3rd, Gingeras T.R., Lindblad-Toh K., Bekiranov S., Bailey D.K., Huebert D.J., McMahon S., Karlsson E.K., Kulbokas E.J., 3rd, Gingeras T.R., Bekiranov S., Bailey D.K., Huebert D.J., McMahon S., Karlsson E.K., Kulbokas E.J., 3rd, Gingeras T.R., Bailey D.K., Huebert D.J., McMahon S., Karlsson E.K., Kulbokas E.J., 3rd, Gingeras T.R., Huebert D.J., McMahon S., Karlsson E.K., Kulbokas E.J., 3rd, Gingeras T.R., McMahon S., Karlsson E.K., Kulbokas E.J., 3rd, Gingeras T.R., Karlsson E.K., Kulbokas E.J., 3rd, Gingeras T.R., Kulbokas E.J., 3rd, Gingeras T.R., Gingeras T.R., et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Daly K., Shirazi-Beechey S.P., Shirazi-Beechey S.P. Microarray analysis of butyrate regulated genes in colonic epithelial cells. DNA Cell Biol. 2006;25:49–62. doi: 10.1089/dna.2006.25.49. [DOI] [PubMed] [Google Scholar]
- Dashwood R.H., Myzak M.C., Ho E., Myzak M.C., Ho E., Ho E. Dietary HDAC inhibitors: Time to rethink weak ligands in cancer chemoprevention? Carcinogenesis. 2006;27:344–349. doi: 10.1093/carcin/bgi253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davie J.R. Inhibition of histone deacetylase activity by butyrate. J. Nutr. 2003;133:2485S–2493S. doi: 10.1093/jn/133.7.2485S. [DOI] [PubMed] [Google Scholar]
- Dehm S.M., Hilton T.L., Wang E.H., Bonham K., Hilton T.L., Wang E.H., Bonham K., Wang E.H., Bonham K., Bonham K. SRC proximal and core promoter elements dictate TAF1 dependence and transcriptional repression by histone deacetylase inhibitors. Mol. Cell. Biol. 2004;24:2296–2307. doi: 10.1128/MCB.24.6.2296-2307.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Ruijter A.J., Meinsma R.J., Bosma P., Kemp S., Caron H.N., van Kuilenburg A.B., Meinsma R.J., Bosma P., Kemp S., Caron H.N., van Kuilenburg A.B., Bosma P., Kemp S., Caron H.N., van Kuilenburg A.B., Kemp S., Caron H.N., van Kuilenburg A.B., Caron H.N., van Kuilenburg A.B., van Kuilenburg A.B. Gene expression profiling in response to the histone deacetylase inhibitor BL1521 in neuroblastoma. Exp. Cell Res. 2005;309:451–467. doi: 10.1016/j.yexcr.2005.06.024. [DOI] [PubMed] [Google Scholar]
- Dimitri P., Corradini N., Rossi F., Verni F., Corradini N., Rossi F., Verni F., Rossi F., Verni F., Verni F. The paradox of functional heterochromatin. Bioessays. 2005;27:29–41. doi: 10.1002/bies.20158. [DOI] [PubMed] [Google Scholar]
- Donadel G., Garzelli C., Frank R., Gabrielli F., Garzelli C., Frank R., Gabrielli F., Frank R., Gabrielli F., Gabrielli F. Identification of a novel nuclear protein synthesized in growth-arrested human hepatoblastoma HepG2 cells. Eur. J. Biochem. 1991;195:723–729. doi: 10.1111/j.1432-1033.1991.tb15759.x. [DOI] [PubMed] [Google Scholar]
- Dou Y., Milne T.A., Tackett A.J., Smith E.R., Fukuda A., Wysocka J., Allis C.D., Chait B.T., Hess J.L., Roeder R.G., Milne T.A., Tackett A.J., Smith E.R., Fukuda A., Wysocka J., Allis C.D., Chait B.T., Hess J.L., Roeder R.G., Tackett A.J., Smith E.R., Fukuda A., Wysocka J., Allis C.D., Chait B.T., Hess J.L., Roeder R.G., Smith E.R., Fukuda A., Wysocka J., Allis C.D., Chait B.T., Hess J.L., Roeder R.G., Fukuda A., Wysocka J., Allis C.D., Chait B.T., Hess J.L., Roeder R.G., Wysocka J., Allis C.D., Chait B.T., Hess J.L., Roeder R.G., Allis C.D., Chait B.T., Hess J.L., Roeder R.G., Chait B.T., Hess J.L., Roeder R.G., Hess J.L., Roeder R.G., Roeder R.G. Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell. 2005;121:873–885. doi: 10.1016/j.cell.2005.04.031. [DOI] [PubMed] [Google Scholar]
- Duan H., Heckman C.A., Boxer L.M., Heckman C.A., Boxer L.M., Boxer L.M. Histone deacetylase inhibitors down-regulate bcl-2 expression and induce apoptosis in t(14;18) lymphomas. Mol. Cell. Biol. 2005;25:1608–1619. doi: 10.1128/MCB.25.5.1608-1619.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunphy E.L., Johnson T., Auerbach S.S., Wang E.H., Johnson T., Auerbach S.S., Wang E.H., Auerbach S.S., Wang E.H., Wang E.H. Requirement for TAF(II)250 acetyltransferase activity in cell cycle progression. Mol. Cell. Biol. 2000;20:1134–1139. doi: 10.1128/mcb.20.4.1134-1139.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuele S., D’Anneo A., Bellavia G., Vassallo B., Lauricella M., De Blasio A., Vento R., Tesoriere G., D’Anneo A., Bellavia G., Vassallo B., Lauricella M., De Blasio A., Vento R., Tesoriere G., Bellavia G., Vassallo B., Lauricella M., De Blasio A., Vento R., Tesoriere G., Vassallo B., Lauricella M., De Blasio A., Vento R., Tesoriere G., Lauricella M., De Blasio A., Vento R., Tesoriere G., De Blasio A., Vento R., Tesoriere G., Vento R., Tesoriere G., Tesoriere G. Sodium butyrate induces apoptosis in human hepatoma cells by a mitochondria/caspase pathway, associated with degradation of β-catenin, pRb and Bcl-XL. Eur. J. Cancer. 2004;40:1441–1452. doi: 10.1016/j.ejca.2004.01.039. [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium, The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- Fraga M.F., Ballestar E., Villar-Garea A., Boix-Chornet M., Espada J., Schotta G., Bonaldi T., Haydon C., Ropero S., Petrie K., Ballestar E., Villar-Garea A., Boix-Chornet M., Espada J., Schotta G., Bonaldi T., Haydon C., Ropero S., Petrie K., Villar-Garea A., Boix-Chornet M., Espada J., Schotta G., Bonaldi T., Haydon C., Ropero S., Petrie K., Boix-Chornet M., Espada J., Schotta G., Bonaldi T., Haydon C., Ropero S., Petrie K., Espada J., Schotta G., Bonaldi T., Haydon C., Ropero S., Petrie K., Schotta G., Bonaldi T., Haydon C., Ropero S., Petrie K., Bonaldi T., Haydon C., Ropero S., Petrie K., Haydon C., Ropero S., Petrie K., Ropero S., Petrie K., Petrie K., et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat. Genet. 2005;37:391–400. doi: 10.1038/ng1531. [DOI] [PubMed] [Google Scholar]
- Gialitakis M., Kretsovali A., Spilianakis C., Kravariti L., Mages J., Hoffmann R., Hatzopoulos A.K., Papamatheakis J., Kretsovali A., Spilianakis C., Kravariti L., Mages J., Hoffmann R., Hatzopoulos A.K., Papamatheakis J., Spilianakis C., Kravariti L., Mages J., Hoffmann R., Hatzopoulos A.K., Papamatheakis J., Kravariti L., Mages J., Hoffmann R., Hatzopoulos A.K., Papamatheakis J., Mages J., Hoffmann R., Hatzopoulos A.K., Papamatheakis J., Hoffmann R., Hatzopoulos A.K., Papamatheakis J., Hatzopoulos A.K., Papamatheakis J., Papamatheakis J. Coordinated changes of histone modifications and HDAC mobilization regulate the induction of MHC class II genes by Trichostatin A. Nucleic Acids Res. 2006;34:765–772. doi: 10.1093/nar/gkj462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goncalves J., Malta-Vacas J., Louis M., Brault L., Bagrel D., Monteiro C., Brito M., Malta-Vacas J., Louis M., Brault L., Bagrel D., Monteiro C., Brito M., Louis M., Brault L., Bagrel D., Monteiro C., Brito M., Brault L., Bagrel D., Monteiro C., Brito M., Bagrel D., Monteiro C., Brito M., Monteiro C., Brito M., Brito M. Modulation of translation factor's gene expression by histone deacetylase inhibitors in breast cancer cells. Clin. Chem. Lab. Med. 2005;43:151–156. doi: 10.1515/CCLM.2005.025. [DOI] [PubMed] [Google Scholar]
- Han J.W., Ahn S.H., Kim Y.K., Bae G.U., Yoon J.W., Hong S., Lee H.Y., Lee Y.W., Lee H.W., Ahn S.H., Kim Y.K., Bae G.U., Yoon J.W., Hong S., Lee H.Y., Lee Y.W., Lee H.W., Kim Y.K., Bae G.U., Yoon J.W., Hong S., Lee H.Y., Lee Y.W., Lee H.W., Bae G.U., Yoon J.W., Hong S., Lee H.Y., Lee Y.W., Lee H.W., Yoon J.W., Hong S., Lee H.Y., Lee Y.W., Lee H.W., Hong S., Lee H.Y., Lee Y.W., Lee H.W., Lee H.Y., Lee Y.W., Lee H.W., Lee Y.W., Lee H.W., Lee H.W. Activation of p21(WAF1/Cip1) transcription through Sp1 sites by histone deacetylase inhibitor apicidin: Involvement of protein kinase C. J. Biol. Chem. 2001;276:42084–42090. doi: 10.1074/jbc.M106688200. [DOI] [PubMed] [Google Scholar]
- Hazzalin C.A., Mahadevan L.C., Mahadevan L.C. Dynamic acetylation of all lysine 4-methylated histone H3 in the mouse nucleus: Analysis at c-fos and c-jun. PLoS Biol. 2005;3:e393. doi: 10.1371/journal.pbio.0030393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heruth D.P., Zirnstein G.W., Bradley J.F., Rothberg P.G., Zirnstein G.W., Bradley J.F., Rothberg P.G., Bradley J.F., Rothberg P.G., Rothberg P.G. Sodium butyrate causes an increase in the block to transcriptional elongation in the c-myc gene in SW837 rectal carcinoma cells. J. Biol. Chem. 1993;268:20466–20472. [PubMed] [Google Scholar]
- Hirsch C.L., Bonham K., Bonham K. Histone deacetylase inhibitors regulate p21WAF1 gene expression at the post-transcriptional level in HepG2 cells. FEBS Lett. 2004;570:37–40. doi: 10.1016/j.febslet.2004.06.018. [DOI] [PubMed] [Google Scholar]
- Jackson J.G., Pereira-Smith O.M., Pereira-Smith O.M. Primary and compensatory roles for RB family members at cell cycle gene promoters that are deacetylated and downregulated in doxorubicin-induced senescence of breast cancer cells. Mol. Cell. Biol. 2006;26:2501–2510. doi: 10.1128/MCB.26.7.2501-2510.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P.A., Baylin S.B., Baylin S.B. The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- Kim E.H., Kim H.S., Kim S.U., Noh E.J., Lee J.S., Choi K.S., Kim H.S., Kim S.U., Noh E.J., Lee J.S., Choi K.S., Kim S.U., Noh E.J., Lee J.S., Choi K.S., Noh E.J., Lee J.S., Choi K.S., Lee J.S., Choi K.S., Choi K.S. Sodium butyrate sensitizes human glioma cells to TRAIL-mediated apoptosis through inhibition of Cdc2 and the subsequent downregulation of survivin and XIAP. Oncogene. 2005a;24:6877–6889. doi: 10.1038/sj.onc.1208851. [DOI] [PubMed] [Google Scholar]
- Kim T.H., Barrera L.O., Zheng M., Qu C., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B., Barrera L.O., Zheng M., Qu C., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B., Zheng M., Qu C., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B., Qu C., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B., Richmond T.A., Wu Y., Green R.D., Ren B., Wu Y., Green R.D., Ren B., Green R.D., Ren B., Ren B. A high-resolution map of active promoters in the human genome. Nature. 2005b;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostyniuk C.L., Dehm S.M., Batten D., Bonham K., Dehm S.M., Batten D., Bonham K., Batten D., Bonham K., Bonham K. The ubiquitous and tissue specific promoters of the human SRC gene are repressed by inhibitors of histone deacetylases. Oncogene. 2002;21:6340–6347. doi: 10.1038/sj.onc.1205787. [DOI] [PubMed] [Google Scholar]
- Kristjuhan A., Svejstrup J.Q., Svejstrup J.Q. Evidence for distinct mechanisms facilitating transcript elongation through chromatin in vivo. EMBO J. 2004;23:4243–4252. doi: 10.1038/sj.emboj.7600433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lallemand F., Courilleau D., Sabbah M., Redeuilh G., Mester J., Courilleau D., Sabbah M., Redeuilh G., Mester J., Sabbah M., Redeuilh G., Mester J., Redeuilh G., Mester J., Mester J. Direct inhibition of the expression of cyclin D1 gene by sodium butyrate. Biochem. Biophys. Res. Commun. 1996;229:163–169. doi: 10.1006/bbrc.1996.1774. [DOI] [PubMed] [Google Scholar]
- Lee C.K., Shibata Y., Rao B., Strahl B.D., Lieb J.D., Shibata Y., Rao B., Strahl B.D., Lieb J.D., Rao B., Strahl B.D., Lieb J.D., Strahl B.D., Lieb J.D., Lieb J.D. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat. Genet. 2004;36:900–905. doi: 10.1038/ng1400. [DOI] [PubMed] [Google Scholar]
- Lieb J.D., Clarke N.D., Clarke N.D. Control of transcription through intragenic patterns of nucleosome composition. Cell. 2005;123:1187–1190. doi: 10.1016/j.cell.2005.12.010. [DOI] [PubMed] [Google Scholar]
- Lindemann R.K., Gabrielli B., Johnstone R.W., Gabrielli B., Johnstone R.W., Johnstone R.W. Histone-deacetylase inhibitors for the treatment of cancer. Cell Cycle. 2004;3:779–788. [PubMed] [Google Scholar]
- Maison C., Bailly D., Peters A.H., Quivy J.P., Roche D., Taddei A., Lachner M., Jenuwein T., Almouzni G., Bailly D., Peters A.H., Quivy J.P., Roche D., Taddei A., Lachner M., Jenuwein T., Almouzni G., Peters A.H., Quivy J.P., Roche D., Taddei A., Lachner M., Jenuwein T., Almouzni G., Quivy J.P., Roche D., Taddei A., Lachner M., Jenuwein T., Almouzni G., Roche D., Taddei A., Lachner M., Jenuwein T., Almouzni G., Taddei A., Lachner M., Jenuwein T., Almouzni G., Lachner M., Jenuwein T., Almouzni G., Jenuwein T., Almouzni G., Almouzni G. Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat. Genet. 2002;30:329–334. doi: 10.1038/ng843. [DOI] [PubMed] [Google Scholar]
- Mizzen C.A., Yang X.J., Kokubo T., Brownell J.E., Bannister A.J., Owen-Hughes T., Workman J., Wang L., Berger S.L., Kouzarides T., Yang X.J., Kokubo T., Brownell J.E., Bannister A.J., Owen-Hughes T., Workman J., Wang L., Berger S.L., Kouzarides T., Kokubo T., Brownell J.E., Bannister A.J., Owen-Hughes T., Workman J., Wang L., Berger S.L., Kouzarides T., Brownell J.E., Bannister A.J., Owen-Hughes T., Workman J., Wang L., Berger S.L., Kouzarides T., Bannister A.J., Owen-Hughes T., Workman J., Wang L., Berger S.L., Kouzarides T., Owen-Hughes T., Workman J., Wang L., Berger S.L., Kouzarides T., Workman J., Wang L., Berger S.L., Kouzarides T., Wang L., Berger S.L., Kouzarides T., Berger S.L., Kouzarides T., Kouzarides T., et al. The TAF(II)250 subunit of TFIID has histone acetyltransferase activity. Cell. 1996;87:1261–1270. doi: 10.1016/s0092-8674(00)81821-8. [DOI] [PubMed] [Google Scholar]
- Murray J.I., Whitfield M.L., Trinklein N.D., Myers R.M., Brown P.O., Botstein D., Whitfield M.L., Trinklein N.D., Myers R.M., Brown P.O., Botstein D., Trinklein N.D., Myers R.M., Brown P.O., Botstein D., Myers R.M., Brown P.O., Botstein D., Brown P.O., Botstein D., Botstein D. Diverse and specific gene expression responses to stresses in cultured human cells. Mol. Biol. Cell. 2004;15:2361–2374. doi: 10.1091/mbc.E03-11-0799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien T., Tjian R., Tjian R. Different functional domains of TAFII250 modulate expression of distinct subsets of mammalian genes. Proc. Natl. Acad. Sci. 2000;97:2456–2461. doi: 10.1073/pnas.97.6.2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peart M.J., Smyth G.K., van Laar R.K., Bowtell D.D., Richon V.M., Marks P.A., Holloway A.J., Johnstone R.W., Smyth G.K., van Laar R.K., Bowtell D.D., Richon V.M., Marks P.A., Holloway A.J., Johnstone R.W., van Laar R.K., Bowtell D.D., Richon V.M., Marks P.A., Holloway A.J., Johnstone R.W., Bowtell D.D., Richon V.M., Marks P.A., Holloway A.J., Johnstone R.W., Richon V.M., Marks P.A., Holloway A.J., Johnstone R.W., Marks P.A., Holloway A.J., Johnstone R.W., Holloway A.J., Johnstone R.W., Johnstone R.W. Identification and functional significance of genes regulated by structurally different histone deacetylase inhibitors. Proc. Natl. Acad. Sci. 2005;102:3697–3702. doi: 10.1073/pnas.0500369102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piluso D., Bilan P., Capone J.P., Bilan P., Capone J.P., Capone J.P. Host cell factor-1 interacts with and antagonizes transactivation by the cell cycle regulatory factor Miz-1. J. Biol. Chem. 2002;277:46799–46808. doi: 10.1074/jbc.M206226200. [DOI] [PubMed] [Google Scholar]
- Pokholok D.K., Harbison C.T., Levine S., Cole M., Hannett N.M., Lee T.I., Bell G.W., Walker K., Rolfe P.A., Herbolsheimer E., Harbison C.T., Levine S., Cole M., Hannett N.M., Lee T.I., Bell G.W., Walker K., Rolfe P.A., Herbolsheimer E., Levine S., Cole M., Hannett N.M., Lee T.I., Bell G.W., Walker K., Rolfe P.A., Herbolsheimer E., Cole M., Hannett N.M., Lee T.I., Bell G.W., Walker K., Rolfe P.A., Herbolsheimer E., Hannett N.M., Lee T.I., Bell G.W., Walker K., Rolfe P.A., Herbolsheimer E., Lee T.I., Bell G.W., Walker K., Rolfe P.A., Herbolsheimer E., Bell G.W., Walker K., Rolfe P.A., Herbolsheimer E., Walker K., Rolfe P.A., Herbolsheimer E., Rolfe P.A., Herbolsheimer E., Herbolsheimer E., et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- Polioudaki H., Kourmouli N., Drosou V., Bakou A., Theodoropoulos P.A., Singh P.B., Giannakouros T., Georgatos S.D., Kourmouli N., Drosou V., Bakou A., Theodoropoulos P.A., Singh P.B., Giannakouros T., Georgatos S.D., Drosou V., Bakou A., Theodoropoulos P.A., Singh P.B., Giannakouros T., Georgatos S.D., Bakou A., Theodoropoulos P.A., Singh P.B., Giannakouros T., Georgatos S.D., Theodoropoulos P.A., Singh P.B., Giannakouros T., Georgatos S.D., Singh P.B., Giannakouros T., Georgatos S.D., Giannakouros T., Georgatos S.D., Georgatos S.D. Histones H3/H4 form a tight complex with the inner nuclear membrane protein LBR and heterochromatin protein 1. EMBO Rep. 2001;2:920–925. doi: 10.1093/embo-reports/kve199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pray-Grant M.G., Daniel J.A., Schieltz D., Yates J.R., 3rd, Grant P.A., Daniel J.A., Schieltz D., Yates J.R., 3rd, Grant P.A., Schieltz D., Yates J.R., 3rd, Grant P.A., Yates J.R., 3rd, Grant P.A., Grant P.A. Chd1 chromodomain links histone H3 methylation with SAGA- and SLIK-dependent acetylation. Nature. 2005;433:434–438. doi: 10.1038/nature03242. [DOI] [PubMed] [Google Scholar]
- Rada-Iglesias A., Wallerman O., Koch C., Ameur A., Enroth S., Clelland G., Wester K., Wilcox S., Dovey O.M., Ellis P.D., Wallerman O., Koch C., Ameur A., Enroth S., Clelland G., Wester K., Wilcox S., Dovey O.M., Ellis P.D., Koch C., Ameur A., Enroth S., Clelland G., Wester K., Wilcox S., Dovey O.M., Ellis P.D., Ameur A., Enroth S., Clelland G., Wester K., Wilcox S., Dovey O.M., Ellis P.D., Enroth S., Clelland G., Wester K., Wilcox S., Dovey O.M., Ellis P.D., Clelland G., Wester K., Wilcox S., Dovey O.M., Ellis P.D., Wester K., Wilcox S., Dovey O.M., Ellis P.D., Wilcox S., Dovey O.M., Ellis P.D., Dovey O.M., Ellis P.D., Ellis P.D., et al. Binding sites for metabolic disease related transcription factors inferred at base pair resolution by chromatin immunoprecipitation and genomic microarrays. Hum. Mol. Genet. 2005;14:3435–3447. doi: 10.1093/hmg/ddi378. [DOI] [PubMed] [Google Scholar]
- Rayman J.B., Takahashi Y., Indjeian V.B., Dannenberg J.H., Catchpole S., Watson R.J., te Riele H., Dynlacht B.D., Takahashi Y., Indjeian V.B., Dannenberg J.H., Catchpole S., Watson R.J., te Riele H., Dynlacht B.D., Indjeian V.B., Dannenberg J.H., Catchpole S., Watson R.J., te Riele H., Dynlacht B.D., Dannenberg J.H., Catchpole S., Watson R.J., te Riele H., Dynlacht B.D., Catchpole S., Watson R.J., te Riele H., Dynlacht B.D., Watson R.J., te Riele H., Dynlacht B.D., te Riele H., Dynlacht B.D., Dynlacht B.D. E2F mediates cell cycle-dependent transcriptional repression in vivo by recruitment of an HDAC1/mSin3B corepressor complex. Genes & Dev. 2002;16:933–947. doi: 10.1101/gad.969202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid G., Metivier R., Lin C.Y., Denger S., Ibberson D., Ivacevic T., Brand H., Benes V., Liu E.T., Gannon F., Metivier R., Lin C.Y., Denger S., Ibberson D., Ivacevic T., Brand H., Benes V., Liu E.T., Gannon F., Lin C.Y., Denger S., Ibberson D., Ivacevic T., Brand H., Benes V., Liu E.T., Gannon F., Denger S., Ibberson D., Ivacevic T., Brand H., Benes V., Liu E.T., Gannon F., Ibberson D., Ivacevic T., Brand H., Benes V., Liu E.T., Gannon F., Ivacevic T., Brand H., Benes V., Liu E.T., Gannon F., Brand H., Benes V., Liu E.T., Gannon F., Benes V., Liu E.T., Gannon F., Liu E.T., Gannon F., Gannon F. Multiple mechanisms induce transcriptional silencing of a subset of genes, including oestrogen receptor α, in response to deacetylase inhibition by valproic acid and trichostatin A. Oncogene. 2005;24:4894–4907. doi: 10.1038/sj.onc.1208662. [DOI] [PubMed] [Google Scholar]
- Richon V.M., Sandhoff T.W., Rifkind R.A., Marks P.A., Sandhoff T.W., Rifkind R.A., Marks P.A., Rifkind R.A., Marks P.A., Marks P.A. Histone deacetylase inhibitor selectively induces p21WAF1 expression and gene-associated histone acetylation. Proc. Natl. Acad. Sci. 2000;97:10014–10019. doi: 10.1073/pnas.180316197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruemmele F.M., Schwartz S., Seidman E.G., Dionne S., Levy E., Lentze M.J., Schwartz S., Seidman E.G., Dionne S., Levy E., Lentze M.J., Seidman E.G., Dionne S., Levy E., Lentze M.J., Dionne S., Levy E., Lentze M.J., Levy E., Lentze M.J., Lentze M.J. Butyrate induced Caco-2 cell apoptosis is mediated via the mitochondrial pathway. Gut. 2003;52:94–100. doi: 10.1136/gut.52.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadoni N., Langer S., Fauth C., Bernardi G., Cremer T., Turner B.M., Zink D., Langer S., Fauth C., Bernardi G., Cremer T., Turner B.M., Zink D., Fauth C., Bernardi G., Cremer T., Turner B.M., Zink D., Bernardi G., Cremer T., Turner B.M., Zink D., Cremer T., Turner B.M., Zink D., Turner B.M., Zink D., Zink D. Nuclear organization of mammalian genomes. Polar chromosome territories build up functionally distinct higher order compartments. J. Cell Biol. 1999;146:1211–1226. doi: 10.1083/jcb.146.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubeler D., MacAlpine D.M., Scalzo D., Wirbelauer C., Kooperberg C., van Leeuwen F., Gottschling D.E., O'Neill L.P., Turner B.M., Delrow J., MacAlpine D.M., Scalzo D., Wirbelauer C., Kooperberg C., van Leeuwen F., Gottschling D.E., O'Neill L.P., Turner B.M., Delrow J., Scalzo D., Wirbelauer C., Kooperberg C., van Leeuwen F., Gottschling D.E., O'Neill L.P., Turner B.M., Delrow J., Wirbelauer C., Kooperberg C., van Leeuwen F., Gottschling D.E., O'Neill L.P., Turner B.M., Delrow J., Kooperberg C., van Leeuwen F., Gottschling D.E., O'Neill L.P., Turner B.M., Delrow J., van Leeuwen F., Gottschling D.E., O'Neill L.P., Turner B.M., Delrow J., Gottschling D.E., O'Neill L.P., Turner B.M., Delrow J., O'Neill L.P., Turner B.M., Delrow J., Turner B.M., Delrow J., Delrow J., et al. The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes & Dev. 2004;18:1263–1271. doi: 10.1101/gad.1198204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott G.K., Mattie M.D., Berger C.E., Benz S.C., Benz C.C., Mattie M.D., Berger C.E., Benz S.C., Benz C.C., Berger C.E., Benz S.C., Benz C.C., Benz S.C., Benz C.C., Benz C.C. Rapid alteration of microRNA levels by histone deacetylase inhibition. Cancer Res. 2006;66:1277–1281. doi: 10.1158/0008-5472.CAN-05-3632. [DOI] [PubMed] [Google Scholar]
- Smale S.T., Kadonaga J.T., Kadonaga J.T. The RNA polymerase II core promoter. Annu. Rev. Biochem. 2003;72:449–479. doi: 10.1146/annurev.biochem.72.121801.161520. [DOI] [PubMed] [Google Scholar]
- Somech R., Shaklai S., Geller O., Amariglio N., Simon A.J., Rechavi G., Gal-Yam E.N., Shaklai S., Geller O., Amariglio N., Simon A.J., Rechavi G., Gal-Yam E.N., Geller O., Amariglio N., Simon A.J., Rechavi G., Gal-Yam E.N., Amariglio N., Simon A.J., Rechavi G., Gal-Yam E.N., Simon A.J., Rechavi G., Gal-Yam E.N., Rechavi G., Gal-Yam E.N., Gal-Yam E.N. The nuclear-envelope protein and transcriptional repressor LAP2β interacts with HDAC3 at the nuclear periphery, and induces histone H4 deacetylation. J. Cell Sci. 2005;118:4017–4025. doi: 10.1242/jcs.02521. [DOI] [PubMed] [Google Scholar]
- Soutoglou E., Katrakili N., Talianidis I., Katrakili N., Talianidis I., Talianidis I. Acetylation regulates transcription factor activity at multiple levels. Mol. Cell. 2000;5:745–751. doi: 10.1016/s1097-2765(00)80253-1. [DOI] [PubMed] [Google Scholar]
- Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes & Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- Suzuki H., Gabrielson E., Chen W., Anbazhagan R., van Engeland M., Weijenberg M.P., Herman J.G., Baylin S.B., Gabrielson E., Chen W., Anbazhagan R., van Engeland M., Weijenberg M.P., Herman J.G., Baylin S.B., Chen W., Anbazhagan R., van Engeland M., Weijenberg M.P., Herman J.G., Baylin S.B., Anbazhagan R., van Engeland M., Weijenberg M.P., Herman J.G., Baylin S.B., van Engeland M., Weijenberg M.P., Herman J.G., Baylin S.B., Weijenberg M.P., Herman J.G., Baylin S.B., Herman J.G., Baylin S.B., Baylin S.B. A genomic screen for genes upregulated by demethylation and histone deacetylase inhibition in human colorectal cancer. Nat. Genet. 2002;31:141–149. doi: 10.1038/ng892. [DOI] [PubMed] [Google Scholar]
- Taddei A., Maison C., Roche D., Almouzni G., Maison C., Roche D., Almouzni G., Roche D., Almouzni G., Almouzni G. Reversible disruption of pericentric heterochromatin and centromere function by inhibiting deacetylases. Nat. Cell Biol. 2001;3:114–120. doi: 10.1038/35055010. [DOI] [PubMed] [Google Scholar]
- Taddei A., Roche D., Bickmore W.A., Almouzni G., Roche D., Bickmore W.A., Almouzni G., Bickmore W.A., Almouzni G., Almouzni G. The effects of histone deacetylase inhibitors on heterochromatin: Implications for anticancer therapy? EMBO Rep. 2005;6:520–524. doi: 10.1038/sj.embor.7400441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong X., Yin L., Joshi S., Rosenberg D.W., Giardina C., Yin L., Joshi S., Rosenberg D.W., Giardina C., Joshi S., Rosenberg D.W., Giardina C., Rosenberg D.W., Giardina C., Giardina C. Cyclooxygenase-2 regulation in colon cancer cells: Modulation of RNA polymerase II elongation by histone deacetylase inhibitors. J. Biol. Chem. 2005;280:15503–15509. doi: 10.1074/jbc.M411978200. [DOI] [PubMed] [Google Scholar]
- Villar-Garea A., Esteller M., Esteller M. Histone deacetylase inhibitors: Understanding a new wave of anticancer agents. Int. J. Cancer. 2004;112:171–178. doi: 10.1002/ijc.20372. [DOI] [PubMed] [Google Scholar]
- Wanzel M., Kleine-Kohlbrecher D., Herold S., Hock A., Berns K., Park J., Hemmings B., Eilers M., Kleine-Kohlbrecher D., Herold S., Hock A., Berns K., Park J., Hemmings B., Eilers M., Herold S., Hock A., Berns K., Park J., Hemmings B., Eilers M., Hock A., Berns K., Park J., Hemmings B., Eilers M., Berns K., Park J., Hemmings B., Eilers M., Park J., Hemmings B., Eilers M., Hemmings B., Eilers M., Eilers M. Akt and 14-3-3η regulate Miz1 to control cell-cycle arrest after DNA damage. Nat. Cell Biol. 2005;7:30–41. doi: 10.1038/ncb1202. [DOI] [PubMed] [Google Scholar]
- Weierich C., Brero A., Stein S., von Hase J., Cremer C., Cremer T., Solovei I., Brero A., Stein S., von Hase J., Cremer C., Cremer T., Solovei I., Stein S., von Hase J., Cremer C., Cremer T., Solovei I., von Hase J., Cremer C., Cremer T., Solovei I., Cremer C., Cremer T., Solovei I., Cremer T., Solovei I., Solovei I. Three-dimensional arrangements of centromeres and telomeres in nuclei of human and murine lymphocytes. Chromosome Res. 2003;11:485–502. doi: 10.1023/a:1025016828544. [DOI] [PubMed] [Google Scholar]
- Yao Y.L., Yang W.M., Seto E., Yang W.M., Seto E., Seto E. Regulation of transcription factor YY1 by acetylation and deacetylation. Mol. Cell. Biol. 2001;21:5979–5991. doi: 10.1128/MCB.21.17.5979-5991.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Lu J., Sun H., Chen X., Huang W., Tao D., Huang B., Lu J., Sun H., Chen X., Huang W., Tao D., Huang B., Sun H., Chen X., Huang W., Tao D., Huang B., Chen X., Huang W., Tao D., Huang B., Huang W., Tao D., Huang B., Tao D., Huang B., Huang B. Histone acetylation regulates both transcription initiation and elongation of hsp22 gene in Drosophila. Biochem. Biophys. Res. Commun. 2005;326:811–816. doi: 10.1016/j.bbrc.2004.11.118. [DOI] [PubMed] [Google Scholar]