Abstract
The comprehensive inventory of functional elements in 44 human genomic regions carried out by the ENCODE Project Consortium enables for the first time a global analysis of the genomic distribution of transcriptional regulatory elements. In this study we developed an intuitive and yet powerful approach to analyze the distribution of regulatory elements found in many different ChIP–chip experiments on a 10∼100-kb scale. First, we focus on the overall chromosomal distribution of regulatory elements in the ENCODE regions and show that it is highly nonuniform. We demonstrate, in fact, that regulatory elements are associated with the location of known genes. Further examination on a local, single-gene scale shows an enrichment of regulatory elements near both transcription start and end sites. Our results indicate that overall these elements are clustered into regulatory rich “islands” and poor “deserts.” Next, we examine how consistent the nonuniform distribution is between different transcription factors. We perform on all the factors a multivariate analysis in the framework of a biplot, which enhances biological signals in the experiments. This groups transcription factors into sequence-specific and sequence-nonspecific clusters. Moreover, with experimental variation carefully controlled, detailed correlations show that the distribution of sites was generally reproducible for a specific factor between different laboratories and microarray platforms. Data sets associated with histone modifications have particularly strong correlations. Finally, we show how the correlations between factors change when only regulatory elements far from the transcription start sites are considered.
Transcription of protein-coding genes is mediated by RNA polymerase II (POLR2A, formerly known as Pol2) and requires a complex set of cis-acting transcriptional control sequences and factors that bind them. POLR2A is dependent on auxiliary general transcription factors (TFs), such as the TBP-associated factors, or TAF proteins, to be fully functional. The complex that they form—known as the basal transcription apparatus (Nikolov and Burley 1997)—recognizes the core promoters located at nucleotide positions from −45 to +40 relative to the transcription initiation site (Butler and Kadonaga 2002) to initiate constitutive gene transcription. Immediately upstream of the core promoter region are the promoter proximal elements, which are typically multiple recognition sites for particular sequence-specific ubiquitous TFs such as SP1, NFI, and NFY that serve to modulate the basal transcription activity of the core promoter (Kadonaga 2004). However, the large size of the mammalian genomes and the general need for more sophisticated control systems to regulate very large numbers of interacting genes require mammalian cells to use rather elaborate control elements to regulate gene transcription. For example, regulation of expression of individual human genes is often controlled by several sets of cis-acting regulatory elements, including promoters, enhancers (Martin 2001), silencers (Pozzoli and Sironi 2005; Boyer et al. 2006), insulators (Bell et al. 2001; Kuhn and Geyer 2003), and response elements (Geserick et al. 2005). In concert with chromatin remodeling, histone modifications such as acetylation and methylation also play an important role in the transcriptional regulatory process (Berger 2002; Turner 2002). In recent years it has become possible to globally map transcriptional regulatory elements (TREs) using high-throughput methods such as chromatin immunoprecipitation coupled with microarray probing (ChIP–chip) (Horak and Snyder 2002) or DNA sequencing of immunoprecipitated fragments (ChIP–PET) (Ng et al. 2005).
Launched in September 2003, The ENCyclopedia of DNA Elements (ENCODE) Project Consortium aims to identify all functional elements in the human genome sequence (The ENCODE Project Consortium 2004). The pilot phase of the project is focused on 14 manually chosen human genomic regions and 30 randomly selected ones, which in total compose 30 mega-bases (∼1%) of the human genome sequence. Of all possible functional elements in the ENCODE regions, epigenetic modifications and cis-regulatory elements, including promoters and TF-binding sites (TFBSs; together referred to as TREs in this report), are a major form of transcriptional regulation in eukaryotes. To identify the complex set of cis-acting transcriptional control sequences and modification sites in the ENCODE regions, a large number of proteins (including POLR2A) that play various roles in transcription and several types of histone modifications were assayed by different participating laboratories.
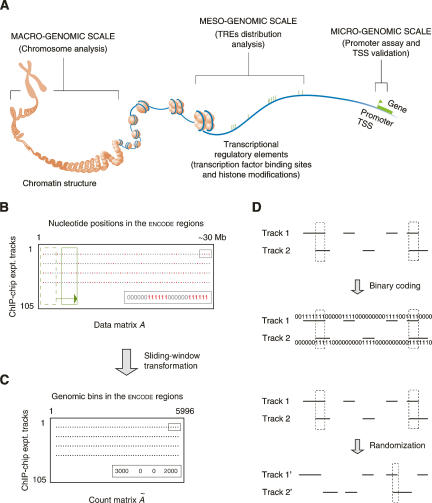
The ENCODE experimental assays of the transcriptional regulation, which collectively represent the first concerted effort to systematically identify TREs in the human genome on a large scale, have generated a large amount of data. With this information available (The ENCODE Project Consortium 2007), it is now possible to conduct detailed surveys of different TFs and their TREs on various genomic levels (Fig. 1A). The promoter assay finds the promoter regions immediately upstream to genes’ transcription start sites (TSSs) on a 100-base-pair (bp) level, and the chromatin structure analysis examines the correspondence between various TREs and aspects of chromatin architecture that implicates mega-bases of DNA. In contrast, our analysis of the genomic distribution of TREs was conducted on an intermediate genomic level, which involves 10∼100 kb of DNA encompassing several genes on average.
Figure 1.
Schematic introduction of the several concepts used in this study. (A) Studies of different transcription factors and their regulatory elements on various genomic levels. (TRE) Transcriptional regulatory element; (TSS) transcription start site. (Modified from The ENCODE Project Consortium 2004 and reprinted with permission from AAAS [www.sciencemag.org] © 2004.) (B) The binary data matrix. Each row is the result track of a ChIP–chip experiment. Red dots are identified transcriptional regulatory elements, in which each nucleotide position is coded as one. (C) The count matrix. A sliding window (the green boxes in B) was used to incorporate contextual information from neighboring positions. Each gray dot represents the number of nucleotide positions in TREs in a sliding window. (D) Correlating two ChIP–chip tracks. The correlation can be done on either two binary vectors or two corresponding count integer vectors (actually used, not shown). Two tracks can also be randomized to generate a background distribution of the correlation.
With such an unprecedented data set, it is now also possible to examine TF coassociation on a large genomic scale. It is highly desirable to present the problem and subsequently analyze the data in a consistent and coherent statistical framework. To do this, we first coded the ChIP–chip experimental results as a binary 105 × ∼30,000,000 data matrix (Fig. 1B) and then transformed it into a 105 × 5669 count matrix using a sliding window to both reduce the matrix size and incorporate contextual information from neighboring nucleotide positions (Fig. 1C). By presenting the data set in the matrix form, many well-studied, mathematically-sound statistical methods and techniques such as the principal component analysis and data randomization (Fig. 1D) can be adopted to tackle the problem.
Below, we evaluate the genomic distribution of the newly identified TREs both by themselves and together with the gene distribution, determine TRE clusters and deserts in the ENCODE regions, and study the relationship among the TFs that have been assayed.
Results
We analyzed 105 lists of regulatory elements of 29 TFs in the ENCODE regions. A list of TREs of a particular TF specifies the location in the genome of the regulatory elements of this factor under certain cellular and experimental conditions. Disregarding overlaps among sites, there are a total of 15,211 TREs identified. The numbers of TREs in each list, ranging from 1–1083 with an average of ∼145 per list, are plotted in Supplemental Figure 1 with lists from the same laboratory grouped together and labeled accordingly. The overall landscape of all 44 ENCODE regions with identified TREs is depicted in Figure 2 and clearly shows a positive correlation of the TRE density with both nonexonic conservation and gene density in a genomic region.
Figure 2.
The landscape of the TREs identified by the 105 ChIP–chip experiments in the ENCODE regions. The green and blue ticks represent TREs of sequence-specific and sequence-nonspecific transcription factors identified by the original experiments respectively. The smaller red ticks mark the locations of genomic elements from the integrated tree-weighted composite list (Trinklein et al. 2007). The placement of 30 randomly picked ENCODE regions (ENr—) in a three-by-three table reflects the stratification in their original selection: The rows are 0%–50%, 50%–80%, and 80%–100% nonexonic conservation from top to bottom, and the columns are 0%–50%, 50%–80%, and 80%–100% gene density from left to right.
TREs are nonrandomly distributed in the ENCODE regions with local enrichment and depletion
Combined regulatory elements of 29 TFs examined in this study are distributed throughout 44 ENCODE regions with an uneven density (Fig. 2). To assess the statistical significance of this density heterogeneity in the TRE genomic distribution, we compared the actual distribution with a randomized one (the null model). Since there are several groups of similar TFs, the actual TRE genomic distribution may be distorted by the repeated measurement of some identical regulatory elements. To minimize this distortion, we used an integrated composite list of 828 genomic elements and performed a χ2 goodness-of-fit test to assess the nature of genomic TRE distribution. The χ2 test rejected the null hypothesis that TREs are randomly distributed in the ENCODE regions (χ2 = 708.68, d.f. = 226, P < 2.2 × 10−16, using 150-kb genomic partitions) and thus confirmed the perception that the TREs are not evenly distributed throughout the ENCODE regions.
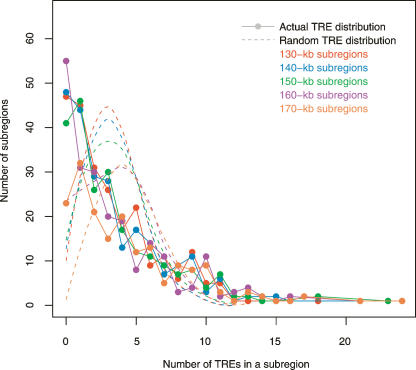
Figure 3 shows the significant difference between the actual TRE distribution and the randomized one (combined from 10 times of genomic permutations of TREs). The distribution of randomly dispersed TREs is a right-skewed, monotonic distribution, which, with 150-kb genomic subregions, peaks at approximately three TREs per bin and then quickly decreases as the number of TREs per subregion deviates further from the average. It resembles a Poisson distribution due to its intrinsically random component but deviates from it as the random dispersion of TREs was restricted to only the nonrepetitive ENCODE sequences. Unlike the “Poissonesque”-null distribution, the actual TRE distribution shows many genomic subregions with extreme numbers of TREs. For example, with 150-kb subregions, there are 87 subregions with zero or one TRE and 16 with >10 TREs.
Figure 3.
TRE distribution in ENCODE regions. Colors signify different genomic subregion sizes. The dots in the same color represent the actual TRE distribution with a particular subregion size. Given the number of TREs in a genomic bin, each dot marks how many such bins are in the ENCODE regions. The null distributions of randomized TREs are represented by dashed curves. Notice both the actual and the null distributions change only slightly when the genomic bin size varies.
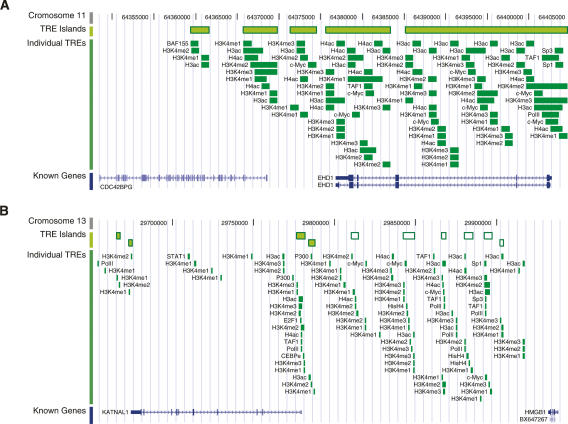
By mapping the full set of TREs onto the human genome sequence, we identified 583 genomic subregions with TRE enrichment and 726 subregions with TRE depletion (the TRE “islands” and “deserts,” respectively) in the ENCODE regions. The longest TRE island is composed of 68 various transcriptional regulatory sites and covers a 35-kb region from HOXA9 to HOXA11 in the HOXA cluster on chromosome 7. High-ranking TRE islands also show that the genomic sequence of EHD, the testilin gene that is highly expressed in testis, is saturated by various histone modification and TFBSs (Fig. 4A). Although most TRE islands are spatially close to known genes, we noticed some of them are located in the intergenic regions in the genome. For example, six small TRE islands are found in a 100-kb intergenic region between KATNAL1 and HMGB1 on chromosome 13 (Fig. 4B).
Figure 4.
Samples of TRE islands. (A) TRE islands that cover the entirety of EHD on chromosome 11. (B) TRE islands in the 100-kb intergenic region between KATNAL1 and HMBG1 on chromosome 13. TRE islands, individual component TREs, and known genes are shown on three different tracks. In B, the distal intergenic TRE islands are shown as white boxes with green borders. The genomic coordinates of both chromosome 11 and 13 are based on the NCBI build 35 of the human genome assembly.
TREs have a similar genomic distribution as know genes and are enriched at both ends of genes
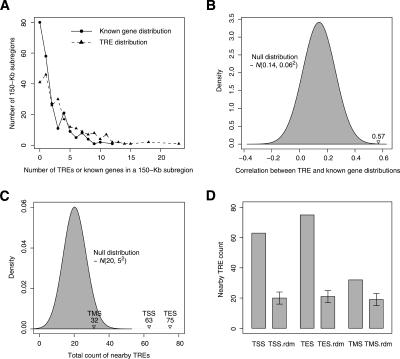
As cis-acting DNA elements through which TFs regulate gene expression, TREs are intimately linked to certain genes or genes in general. To study the spatial relationship between these two types of genomic entities, we first compared the genomic distribution of TREs with that of known genes in ENCODE regions. As Figure 5A shows, there is an overall similarity between these two distributions, which was measured by the correlation between the numbers of TREs and known genes in a series of isometric (150-kb), nonoverlapping partitions of the ENCODE regions. The normality test shows that the correlation coefficient of the null model, which assumes randomly dispersed TREs in the nonrepetitive sequences of the ENCODE regions, is distributed as N(0.14, 0.062). The actual correlation, 0.57, between the occurrence of TREs and known genes is highly significant when it is compared with the null distribution (Fig. 5B).
Figure 5.
Relationship between the TRE and the known gene distributions in ENCODE regions. (A) The distributions of TREs and known genes in ENCODE regions; 150-kb genomic subregions were used. (B) The correlation between the numbers of TREs and known genes in each 150-kb subregion compared with its null distribution, N(0.14, 0.062). (C) The counts of actual TREs within a 1-kb window of gene TSSs, TESs, and TMSs, compared with their corresponding null distributions. All three null distributions are Gaussian-like and almost identical to N(20, 52), which is shown in the plot. (D) Comparison of the actual counts with their corresponding random background, which is depicted as the mean and ±1 SD of each null distribution.
Although this comparison proved that the occurrences of TREs and (known) genes in the genome are highly correlated, it does not explain how TREs are distributed locally relative to the gene transcription sites. To address this problem, we studied the distribution of TREs on a finer scale by examining the enrichment (or the lack of it) of TREs at TSSs, transcription end sites (TESs), and transcription middle sites (TMSs, the genomic middle point between TSSs and TESs) of known genes in ENCODE regions. The comparison between the actual count number, c, of TREs near one type of these sites and its corresponding null distribution, Φ, constitutes an implicit test of the null hypothesis that TREs are not enriched in the vicinity of this particular type of transcription sites.
With 44 ENCODE regions combined, the test rejected the null hypothesis with regard to TSSs as there are 63 TREs near (within 500 bp of) TSSs in all ENCODE regions while the null distribution is normal with 20 as its mean (μΦ) and five as its standard deviation (σΦ) (Fig. 5C). The null distributions of the numbers of TREs near TSSs, TESs, and TMSs (counting was done after the permutation of TRE genomic locations) are all empirically normal and almost identical to N(20, 52). The fold enrichment of TREs near the vicinity of them over the random background (c/μΦ) is 3.2, 3.6, and 1.6, respectively (Fig. 5D). Although there is a slight enrichment of TREs near the middle point of gene transcripts, it is much weaker than that of TREs near the start sites and the end sites of gene transcription.
Multivariate analysis enables biological signal detection despite systematic variation and noise
The result of each ChIP–chip experiment is affected by numerous factors, including systematic experimental design, materials, data analysis methods, and random noise. Since all the experimental data were analyzed by the same false discovery rate method (Efron 2004), a significant portion of this system variability can be explicitly captured by four categorical variables: the TF, the cell line, the microarray platform, and the laboratory. Pairwise correlation of the 105 ChIP–chip experimental results under consideration generated a 105 × 105 symmetric correlation matrix. Although it fully describes the relationships between the experiments in the data set, this correlation matrix is difficult to analyze as the complex experimental factors (the four aforementioned categorical variables) are compounded together.
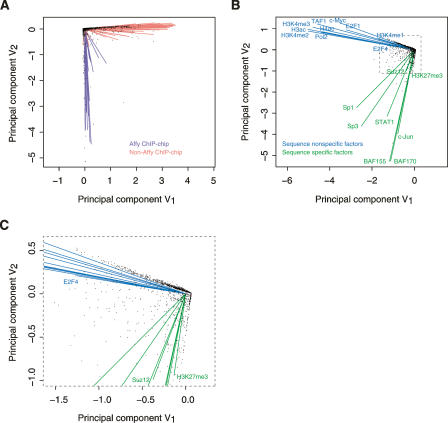
Instead, we use the biplot to explore the relationship among these 105 ChIP–chip experimental results and subsequently among the TFs that were assayed. We obtained a two-dimensional representation of the observations by plotting the first two principal components. By using only the top two principal components, we were able to discard noise but keep main biological signals in the data. Biplot was also used to show TFs and genomic bins together, in a way that represents graphically their joint interrelationship in one plot. It graphs TFs as lines and genomic bins as points together within a common space. Thus we can examine three different relationships—TF to TF, genomic bin to genomic bin, and TF to genomic bin—all in one (bi)plot at the same time.
Formally speaking, a biplot is a graphical representation of the data, in which observations (genomic bins) and variables (experiments or TFs) are plotted in a low dimensional space as points and lines, respectively (Gabriel 1971). Correlations among the experiments are inversely proportional to the angles between the lines. Positive, zero, and negative correlations are represented by acute, right, and obtuse angles, respectively. The distances between the points correspond to the similarities between the profiles of genomic bins: Two bins relatively similar across all the experiments are depicted as points that fall relatively close to each other within the graphic space (see Methods and the Biplot subsection in the Supplemental Material).
As mentioned above, a list of TREs of a particular TF specifies the location of its regulatory elements in the genome under certain cellular and experimental conditions. By partitioning the ENCODE regions into isometric genomic bins, we can quantify the distribution of TREs of a TF by counting the number of nucleotides that its TREs cover in each bin. In essence, this procedure quantitatively describes the relationship between two types of entities—a TRE list and a series of consecutive genomic bins. Given 105 ChIP–chip experiments and 5996 5-kb nonoverlapping genomic bins, this generates a 5996 × 105 data matrix. If the ChIP–chip experiments are treated as 105 random variables and the genomic bins as 5996 observations of them, the relationships between the ChIP–chip experiments and the genomic bins can be studied using a biplot.
The biplot in Figure 6A, generated from our 5996 × 105 matrix, reveals an interesting structure hidden in this data set: The 105 experiments, represented as lines in the figure, can be divided into two highly distinct clusters. One of the clusters is mainly composed of 41 Affy ChIP–chip experiments; the other combines 64 non-Affy ones. The nearly perpendicular orientation of these two line clusters indicates that Affy and non-Affy data are virtually uncorrelated. Affy ChIP–chip experiments were carried out using Affy tiling arrays and the HL-60 cell line, neither of which was used by other laboratories. Either array platform or cell line, or both, contributed to the observed noncorrelation. However, given the currently available data sets, it is not possible to ascertain which factors contributed to the observed noncorrelation and by how much. Based on this observation and the fact that non-Affy data form a larger data set, we focused our following data analysis in this section on non-Affy data only.
Figure 6.
Interrelationship of ChIP–chip experiments and transcription factors with genomic bins. (A) Biplot of 105 original ChIP–chip experiments with 5996 5-kb nonoverlapping genomic bins. Lines represent ChIP–chip experiments, and points indicate genomic bins. (B) Biplot of 18 transcription factors with the same set of genomic bins as in A. Lines represent transcription factors, and points indicate genomic bins. The TREs of each of these 18 transcription factors were merged from the 64 non-Affy ChIP–chip experimental results on a factor basis by taking the union of all TRE lists of each factor. (C) Details of the point-dense region of B inside the box with dashed border. The signs of both coordinates of a point (or the end point of a line) in a biplot are somewhat arbitrary because the data matrix is column-normalized prior to the construction of the plot. Since the scale only reflects the magnitude of the original data, it is not significant in terms of interpreting a biplot either.
Two distinct clusters of TFs emerges from the multivariate analysis
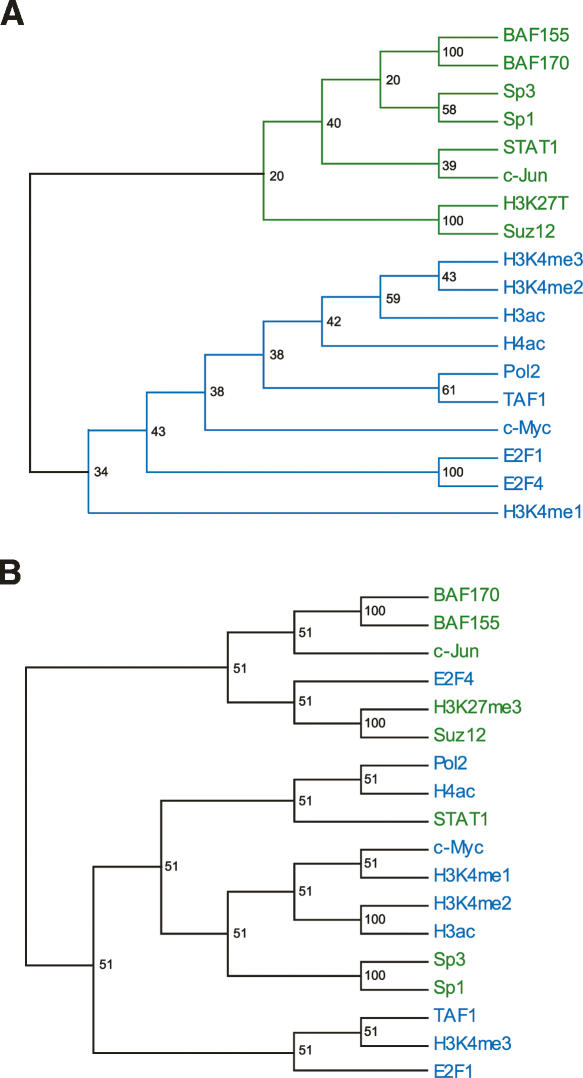
To study the overall relationships between TFs (not their relationships in a particular cell line), we merged TRE lists by taking the union of all TRE lists of each factor. This procedure produced 18 combined lists, one for each of 18 TFs. A second biplot (Fig. 6B) was generated from this non-Affy data set. Again, the biplot reveals an interesting structure hidden in the data. The line orientations show that the TFs can be divided into two relatively distinct clusters. One is composed of POLR2A, TAF1, H3ac, H4ac, H3K4me1, H3K4me2, H3K4me3, E2F1, E2F4, and MYC; the other combines SP1, SP3, STAT1, JUN, SMARCC1, SMARCC2, H3K27me3, and SUZ12. It is interesting to note that the hierarchical clustering of these 18 combined lists reveals very similar cluster structure, both at coarse and fine levels (Fig. 7A). However, hierarchical clustering does nothing about the data noise and shows only one-third of the interrelationship; i.e., the relationship among TFs only. The identities of these two clusters are readily revealed by an examination of their participating factors. In fact, they can be interpreted as clusters of sequence-specific and sequence-nonspecific TFs, respectively.
Figure 7.
Interrelationship of transcription factors. (A) Consensus correlation dendrogram of the same 18 transcription factors as shown in Figure 6B. Bootstrap values are shown at the branching points. Notice the very similar relationship among these 18 transcription factors revealed by these two different methods. (B) Hierarchical clustering of the same 18 transcription factors as in A, but with only TREs >2 kb away from GENCODE-annotated transcription start sites. BAF155 and BAF170 currently known as SMARCC1 and SMARCC2, respectively.
TREs are positively correlated for TFs that fall within the respective clusters; this is indicated by the small angles between lines that represent TFs within the two clusters. One good example is SMARCC1 and SMARCC2 in the second cluster. This high correlation is expected as they are two subunits in the same SWI/SNF chromatin remodeling complex (Wang et al. 1996a, b). Many other pronounced examples can be found in the first cluster, whose compactness indicates high correlation among its constituent TFs, such as POLR2A, TAF1, and histone modification sites. SP1, SP3, and STAT1, the sequence-specific factors, in the second cluster are also correlated with each other, although to a lesser extent. This is expected as they recognize different DNA sequence motifs (see above), which may or may not locate near to each other in the genome. SP1, SP3, and STAT1 in the second cluster also correlate with factors in the first cluster. In contrast, SMARCC1, SMARCC2, H3K27me3, and JUN are virtually uncorrelated with factors in the first cluster.
When only the TREs that are >2 kb away from annotated TSSs were used to cluster TFs, a different cluster pattern emerged (Fig. 7B). Four TFs changed their cluster membership: SP1, SP3, and STAT1 are now grouped with sequence-nonspecific factors, while E2F4 joins the sequence-specific ones. The two SMARCCs, JUN, SUZ12, and H3K27me3 behave as before. This change of pattern indicates that there may be novel promoters or alternative promoters that are bound by SP1, SP3, and STAT1. Sequence-specific factors can be classified into classes depending upon the genomic distributions of their TREs. Some are heavily involved in the general transcriptional machinery, while many are only functional in certain cell lines or under a specific condition, and these latter factors tend to bind to distinct regions of the genome.
Points in a biplot represent observations and in this case 5996 5-kb genomic bins. Figure 6C shows that some points are distributed along the sequence-specific cluster edge of the right angle spanned by these two clusters, more points along the sequence-nonspecific cluster edge, and the rest are scattered inside of the right angle. This distribution pattern reflects the fact that some genomic bins are bound mainly by sequence-specific TFs, more bins are bound mainly by sequence-nonspecific ones, and the rest are bound by both to a comparable degree. Thus the two distinct clusters of genomic bins represented by the points along the two edges of the right angle can be regarded as the genomic “markers” for sequence-specific and sequence-nonspecific TFs, respectively.
ChIP–chip experimental results are generally reproducible between different laboratories and microarray platforms
As mentioned earlier, four categorical variables—the TF, the cell line, the microarray platform, and the laboratory—capture a significant portion of the variability in the ChIP–chip experiments analyzed here. Although not every possible combination of these variables was assayed, the whole data set can be summarized as follows: The binding of 29 TFs to the ENCODE regions was assayed in eight cell lines on three microarray platforms by seven different laboratories (see Methods). As a result, the 105 ChIP–chip experiments can be classified and selected according to a different combination of those four categorical variables.
The analysis of the relationship between the experimental results of these 105 ChIP–chip experiments is very important, since it can provide an assessment of the quality of the experiments as well as lead to biological knowledge discovery. Due to the difficulty of using the whole correlation matrix, we calculated the correlation between subsets of these ChIP–chip experimental results where some experimental variables were kept fixed. This procedure explicitly controls experimental variations. By keeping the TF and the cell line identical, correlations between a set of ChIP–chip experimental results measure the overall data reproducibility between different laboratories (on the same platform) or between microarray platforms (by the same laboratory).
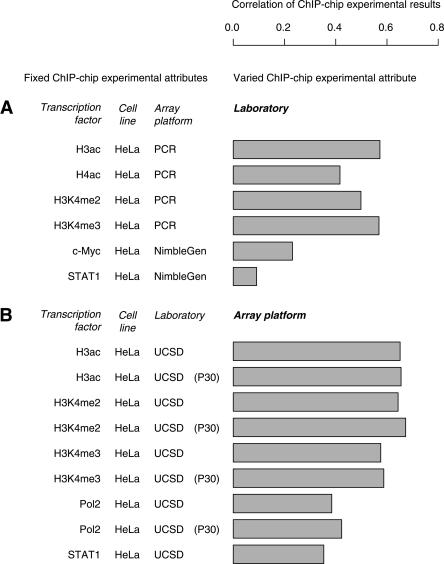
The goal of the pilot phase of the ENCODE Project is not only to find biological novelties but also to standardize the experimental protocols for the next phase of the Project. To assess data reproducibility of ChIP–chip experiments conducted by different laboratories, we selected pairs of experiments that assayed the same TF in the same cell line on the same microarray platform but were conducted by two different laboratories. Six such experimental pairs are present in the data set, and the correlation of the results of each pair of experiments was calculated (Fig. 8A). It shows that the laboratories (not necessarily the same pair for two comparisons) gave much more comparable results for TFs H2ac, H3ac, H3K4me2, and H3K4me3 than for MYC and STAT1. Since the former experiments were all carried out using PCR arrays and the latter high-density tiling arrays, the low agreement on both MYC and STAT1 ChIP–chip experiments by different laboratories may be due to the type of microarray used to assay these two TFs. However, the discrepancy may also be explained by the sequence specificity of these TFs since, coincidentally, H2ac, H3ac, H3K4me2, and H3K4me3 are all sequence-nonspecific histone modifications and MYC and STAT1 are the sequence-specific TFs, which are more sensitive to the noise of the experimental process.
Figure 8.
Data quality measured by the correlation of two ChIP–chip experiment results. (A) Data reproducibility between two different laboratories. Each bar gives the correlation coefficient between the results of two ChIP–chip experiments performed by two laboratories that assayed the same transcription factor in the same cell line on the same array platform. The pairs of laboratories in all six comparisons are not necessarily the same. (B) Data reproducibility between PCR and NimbleGen tiling microarrays. Each bar gives the correlation coefficient between the results of two ChIP–chip experiments performed by the same laboratory that assayed the same transcription factor in the same cell line on both PCR and NimbleGen tiling microarray platforms.
For platform comparison, pairs of ChIP–chip experiments that assayed the same TF in the same cell line by the same laboratory but using two different array platforms were selected. Nine such experimental pairs, all by UCSD using HeLa cells, were present in the data set, and the correlation of the results of each pair of experiments was calculated. Due to size limitations in the current data set, data reproducibility on different microarray platforms could only be assessed between the NimbleGen high-density tiling array and the traditional PCR array. The result (Fig. 8B) shows that different array platforms gave rather similar results for histone modifications such as H3ac, H3K4me2, and H3K4me3 and gave slightly less similar results for POLR2A and STAT1. It is worth noticing that, on average, the correlation of TREs shows that ChIP–chip data sets generated by the same laboratory on different array platforms agree with each other better than ones generated by different laboratories on the same platform.
Discussion
TRE distribution and clustering in the ENCODE regions
The first comprehensive survey of the regulatory elements enables an assay of the distribution of TREs on a large genomic scale. Such a study could provide insight into the organization of functional elements in the human genome. However, it is not immediately clear what is the most sensible way to carry out this assessment, as three pertinent questions need especially careful consideration. One must decide what is a suitable statistical test for this problem, what subregion size should be used if the ENCODE regions are to be discretized, and how the sequence repeats should be dealt with.
Both the Kolmogorov-Smirnov test and the χ2 goodness-of-fit test can be used to compare two distributions. Given the actual and the randomized genomic locations of TREs, the K-S two-sample test may be used to test whether these two location profiles come from the same distribution. However, because the ENCODE regions are fundamentally discrete entities and a simple concatenation of them makes little biological sense, the K-S test can be applied to each individual ENCODE region separately but not to all the regions combined. By contrast, the χ2 test does not have such limitation and is thus used for this study.
TFs and their TREs can be studied on various genomic levels. Unlike and complementary to the promoter assay on the “micro-genomic” scale and the chromosome analysis on the “macro-genomic” scale, our TRE distribution analysis surveys different TFs and their TREs on an intermediate, “meso-genomic” scale, which involves 100∼200 kb of DNA encompassing several genes on average. Based on 150-kb genomic partitions, the χ2 test of goodness of fit rejects random distribution of TREs in ENCODE regions. Similar observations were made using genomic partitions of different bin sizes (130∼170 kb); thus, the conclusion that TREs are not randomly distributed in ENCODE region (and therefore in the human genome) is not specific to a particular subregion size used in the analysis but is general and truly reflects the underlying TRE distribution.
In the hypothesis test presented above, the alternative to the rejected null hypothesis is that the TREs in the ENCODE regions are not distributed in a random, uniform fashion—i.e., they form clusters in the genome. As Figure 2 reveals, substantial TRE deserts are mainly found in ENCODE regions with low gene density and low nonexonic conservation. Conversely, most of the TRE islands are located in the gene-rich regions in the genome. Indeed, a highly significant association between the regulatory elements and the gene locations has been observed by the comparison of the genomic distribution of TREs with that of known genes in ENCODE regions.
A closer examination of the distribution of TREs around TSSs, TESs, and TMSs of known genes revealed a substantial enrichment of TREs at TSSs and TESs. However, a much weaker enrichment of TREs at TMSs was detected. This observation supports the general belief that the density of TFBSs is much lower in the middle of genes and thus validates the widespread practice of using so-called “deep introns” as negative training samples for machine learning algorithms to predict certain TREs in genomic sequences.
It is an intriguing observation that the enrichment of TREs near TESs is comparable to—in fact slightly higher than—that of TREs near TSSs. This result seems unlikely to be a methodological artifact as it was confirmed independently with a different counting procedure and several composite TRE lists from different origins (D. Zheng, pers. comm.). Binding of TFs to the 3′ untranslated region of genes has previously been observed. In a recent study of unbiasedly mapped TFBS regions of SP1, MYC, and TP53 along human chromosomes 21 and 22, Cawley et al. (2004) discovered that while only 22% of the identified TFBS regions are located at the 5′ termini of well-characterized protein-coding genes, 36% of them lie within or immediately 3′ to such genes. Based on the observation that the TFBS regions located at 3′ end of well-characterized genes are significantly correlated with noncoding RNAs, they argued such TFBS regions function as distal regulatory elements or promoters for noncoding transcripts.
Multivariate analysis of TREs in the ENCODE regions
Multivariate analysis of the coassociation of TFs enables many novel biological observations. Two distinct groups emerged from both biplot clustering and hierarchical clustering of the 18 TFs under consideration. They are clusters of sequence-specific and sequence-nonspecific TFs. In Figure 6B, however, group assignments of several TFs are particularly interesting and thus merit further consideration. MYC is commonly viewed as a sequence-specific TF since its function is mediated by binding to a particular DNA consensus sequence (the E-box) for transcriptional activation (Blackwell et al. 1990, 1993) and through certain distinct DNA elements for transcriptional inhibition (Facchini and Penn 1998; Claassen and Hann 1999). Early experimental results, however, suggest that MYC may modulate transcription via histone acetylation (Cole and McMahon 1999; Grandori et al. 2000; Amati et al. 2001; Frank et al. 2001), a discovery that is corroborated by its close association with H3ac and H4ac shown in Figure 6B. These findings together suggest that MYC may behave, at least under certain circumstances, more like a sequence-nonspecific TF.
Mediated by a Polycomb group (PcG) protein complex composed of EED, EZH2, and SUZ12, trimethylation of histone H3 on lysine 27 (H3K27me3) is integral in the process of differentiation, stem cell self-renewal, and tumorigenesis and has been implicated in transcriptional silencing (Cao et al. 2002; Czermin et al. 2002; Muller et al. 2002; Cao and Zhang 2004; Kuzmichev et al. 2004; Pasini et al. 2004). This close functional association between H3K27me3 and SUZ12 is reflected (and thus affirmed) by their high correlation shown in Figures 6B and 7A. Moreover, since H3K27me3 was clustered with sequence-specific TFs, it has small correlation with other types of histone methylation, such as H3K4me1, H3K4me2, and H3K4me3. H3K27me3 has been assumed to function like these sequence-nonspecific histone modifications. Our result, however, suggests otherwise: Instead of serving as a constitutive part of the basal transcriptional machinery, H3K27me3 functions in a transcriptional regulatory process in a rather sequence-specific manner.
The high correlations between POLR2A, TAF1, H3ac, H4ac, H3K4me1, H3K4me2, and H3K4me3 in the first cluster indicate that the regulation of these types of histone modification is tightly linked to POLR2A activity in human. A recent study showed that H3ac, H4ac, H3K4 methylation, and transcriptional activity across the majority of yeast genes are all correlated (Pokholok et al. 2005). A similar phenomena have been observed in fly, mouse, and human cells as well (Schubeler et al. 2004; Bernstein et al. 2005).
Systematic variation in the ENCODE ChIP–chip data sets
There is significant systematic variation in current ENCODE ChIP–chip data sets, as they were generated by different laboratories using different cell lines and array platforms. It is not immediately clear whether there are any discernible biological signals in such noisy data. However, with experimental variations carefully controlled, we find that ChIP–chip experiments are generally reproducible between different laboratories and microarray platforms, as there is significant (but not perfect) agreement between ChIP–chip experimental results produced by different laboratories or on different microarray platforms. The validity of the data is also demonstrated by the corroboration between some of our findings and observations made in other studies using different data. We also observe that the data sets containing more genomic locations tend to be more reproducible than data sets that contain fewer genomic locations.
The ChIP–chip data sets generated by the ENCODE consortium fall into two categories: histone modifications and DNA binding of TFs. Histone modifications tend to occur in many more genomic locations than the binding sites of most TFs. Some TFs also tend to have more binding sites than others. Specifically, data sets of the same histone modification assayed by different laboratories or on different platforms have higher global correlations than data sets for DNA binding of the same TF, reflecting higher signal-to-noise ratio of histone modification experiments. Similarly, the data sets from TFs with more binding sites such as E2F4 and MYC studied by different laboratories or on different platforms also have higher global correlations than those of TFs with fewer binding sites (e.g., STAT1), even after the global correlation is corrected for the number of genomic regions.
Currently, the ChIP–chip experiment is the most widely used high-throughput method for in vivo identification of TREs. It has been applied successfully in numerous studies of TFs. Since it is an in vivo technique, a biologically relevant cell line should be used. At present, an investigator can use either traditional PCR arrays or high-density tiling arrays. The current trend is to migrate from PCR arrays to tiling arrays for a much higher resolution and a comprehensive genomic coverage. Data reproducibility of the ChIP–chip experiment has been accessed (Euskirchen et al. 2007) and is currently under further investigation by several laboratories as part of the effort to standardize the experimental protocols for the next phase of the ENCODE Project. Our study shows that by using appropriate statistical methods it is possible to control data noise to a certain degree to aid biological discoveries. We also need to point out that many of the data sets on different platforms and/or by laboratories have been validated by quantitative PCRs, and we believe it is crucial to carry out multiple technical replications for each ChIP–chip experiment and result validation following ChIP–chip experiments as an internal data quality assessment.
Conclusion
The initial analysis of TREs identified in the ENCODE regions affords new insights into and raises new questions about the genomic distribution of these functional elements, the relationship among TFs assayed, and the nature of the underlying ChIP–chip experiments that generated the data sets analyzed here.
By forming locally enriched and depleted regions in the genome, TREs are distributed in the ENCODE regions in a highly nonrandom fashion. One striking example is the TRE island at the HOXA locus on chromosome 7. Moreover, TREs have a similar genomic distribution as known genes and are enriched in the vicinity of both transcription start and end sites. The nature of TREs at the 3′ end of genes and how they regulate gene transcription await further experimental investigation. Moreover, by using biplot, a multivariate analysis technique, we were able to clearly separate the TFs into sequence-specific and sequence-nonspecific clusters. This analysis reveals many unusual associations among TFs. For example, one striking observation is the close association of SUZ12 and H3K27me3 in the sequence-specific cluster, which also suggests an unusual histone modification role for H3K27me3.
Methods
Data sets used in this study
The data set analyzed in this study is 105 lists of TREs in the ENCODE regions. It was released on December 13, 2005, by the Transcriptional Regulation Group. TRE lists made available after this data freeze were not included in this study. A total of 29 TFs (SMARCC1, SMARCC2, SMARCA4, CEBPE, CTCF, E2F1, E2F4, H3ac, H4ac, H3K27me3, H3K27me3, H3K4me1, H3K4me2, H3K4me3, H3K9K14me2, HisH4, JUN, MYC, P300 [EP300], P63 [TP73L], POLR2A, PU.1 [SPI1], RARecA, SIRT1, SP1, SP3, STAT1, SUZ12, and TAF1) were assayed by seven laboratories (Affymetrix, Sanger, Stanford, UCD, UCSD, UT, Yale) using ChIP-chip experiments on three different microarray platforms (Affymetrix tiling array, NimbleGen tiling array, and traditional PCR array) in nine cell lines (HL-60, HeLa, GM06990, K562, IMR90, HCT116, THP1, Jurkat, and fibroblasts) or at two different experimental time points (P0, before the gamma-interferon was added to the cell culture; P30, 30 min after the gamma-interferon was added). The raw data from these 105 ChIP–chip experiments were uniformly processed using a method based on the false discovery rate (Efron 2004). Three sets of TRE lists were generated at 1%, 5%, and 10% false discovery rate, respectively, and the list generated at the lowest (1%) false discovery rate was used in this study.
TRE distribution analysis
Several TFs were assayed at different time points or in different cell lines. To prevent inflated counting of TREs of each TF, a composite list of TREs was used. To assess the distribution of TREs in ENCODE regions, 44 ENCODE regions were partitioned into 227 150-kb subregions, and the number of TREs in each of them was counted. The null model of the distribution was generated by randomly dispersing TREs in the nonrepetitive sequences of the ENCODE regions. Similar partitioning and counting followed. The random TRE distribution was derived from 10 combined randomization procedures. To study how the number of subregions affects the difference between the actual TRE distribution and the null model, a series of different subregion sizes were examined.
The nonredundant factor-specific TRE lists were mapped onto the ENCODE regions. Uninterrupted genomic regions that are covered by one or more TREs were identified as TRE groups. Neighboring groups that are <1 kb apart are collected into TRE clusters. Unclustered groups that are covered by more than three TREs were promoted into clusters.
The list of composite genomic elements and a list of nonredundant known genes in the ENCODE regions were used to study the relationship between their genomic distributions. Forty-four ENCODE regions were again partitioned into 227 150-kb subregions, and the numbers of TREs and known genes in these subregions were counted and correlated. To generate the null distributions of the correlation coefficient, TREs were first randomly dispersed in the nonrepetitive sequences in the ENCODE regions while the locations of known genes were kept unchanged. Then the numbers of randomized TREs and known genes in these 150-kb subregions were correlated. This randomization-and-correlation procedure was repeated 1000 times.
For the study of the enrichment of TREs in the vicinities of the TSSs, the TSEs, and the TMSs, we used the integrated composite TRE list to minimize data redundancy as before. We first counted the total numbers of different TREs in 1-kb windows around TSSs, TSEs, TMSs of 701 genes from the UCSC genome browser known gene collection, and then generated corresponding (null) distributions of such counts with TREs randomly dispersed in the nonrepetitive ENCODE sequences. This procedure was performed on each ENCODE region separately and also on 44 regions combined.
Whole track correlation
To calculate the correlation between two lists (tracks) of TREs from two ChIP–chip experiments, a binary ∼30,000,000 × 105 data matrix A was first generated. Its rows correspond to genomic locations (observations) in the encode regions, and its columns correspond to ChIP–chip experiments (variables). Matrix element aij = 1 if genomic location i is in a TRE on track j, and aij = 0 otherwise. Pearson’s correlation coefficient r, calculated from two column binary vectors of A, is used to quantify the correlation between two corresponding TRE tracks. This method treats each genomic location as an independent entity and thus disregards the spatial distribution of TREs. To incorporate TRE information at neighboring genomic locations, a 3-kb sliding window with 1.5-kb overlap was used, and the number of nucleotides that are covered by TREs in each window is counted. The data matrix Ā generated by this sliding-window counting procedure is a 19,982 × 105 contingency table, whose element Ãij is the count of genomic locations covered by TREs in sliding window i on track j. The correlation between two TRE tracks can be calculated from Ā in a similar fashion as before. By drastically reducing the size of the data matrix, the sliding-window procedure also enables some of the downstream analyses.
Correspondence analysis
To study how TFs bind to different ENCODE subregions, a biplot was used to show the joint interrelationships between TF and ENCODE subregions by graphing the former as lines and the latter as points together within a common space (Gabriel 1971). First the 5996 × k count matrix à was prepared from the binary data matrix A (see above) using a 10-kb sliding window with 5-kb overlap. Here k = 105 if the original 105 ChIP-chip experimental results were considered, and k = 18 if the merged 64 non-Affy ChIP–chip experimental results were used. à was then column-centered and standardized. To obtain the coordinates for ChIP–chip experiments (or TFs) and genomic bins within the common space used for the biplot, the singular value decomposition was used to factorize à into three component matrices, à = USVT = (US1/2)(VS1/2)T, in which US1/2 and VS1/2 give the coordinates for genomic bins and ChIP–chip experiments (or TFs) in the same space respectively.
Hierarchical clustering of TRE lists was generated from the correlation distance matrix using the neighbor joining algorithm. The consensus dendrogram with bootstrap values at branching points was used to assess the robustness of the topology of the dendrogram. To do so, we first randomly sampled with replacement the count matrix à to produce 1000 new count matrices Ã(1), . . . , Ã(1000), and then generated one dendrogram from each Ã(i) using “neighbor” of the PHYLIP software package. These 1000 dendrograms were then combined to produce a consensus tree with bootstrap values using “consensus” of PHYLIP.
To study how TFs relate to each other by their TREs including or excluding TSSs, each TRE track was first filtered for regions within 2 kb of or at least 2 kb away from TSSs.
The TRE islands and deserts identified in this study are available in the Database for Active Regions with Tools (DART) at http://dart.gersteinlab.org/ENCODE/TR/.
Acknowledgments
Z.D.Z. thanks Prof. John A. Hartigan for his generous help and many lively discussions. Z.D.Z. was funded by an NIH grant (T15 LM07056) from the National Library of Medicine. This work was also supported by an ENCODE grant (1U01HG003156-01) from NHGRI/NIH to M.S. and M.G. Y.F. and Z.W. were funded by another ENCODE grant (R01HG03110) from NHGRI/NIH.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article is online at http://www.genome.org/cgi/doi/10.1101/gr.5573107
References
- Amati B., Frank S.R., Donjerkovic D., Taubert S., Frank S.R., Donjerkovic D., Taubert S., Donjerkovic D., Taubert S., Taubert S. Function of the c-Myc oncoprotein in chromatin remodeling and transcription. Biochim. Biophys. Acta. 2001;1471:M135–M145. doi: 10.1016/s0304-419x(01)00020-8. [DOI] [PubMed] [Google Scholar]
- Bell A.C., West A.G., Felsenfeld G., West A.G., Felsenfeld G., Felsenfeld G. Insulators and boundaries: Versatile regulatory elements in the eukaryotic. Science. 2001;291:447–450. doi: 10.1126/science.291.5503.447. [DOI] [PubMed] [Google Scholar]
- Berger S.L. Histone modifications in transcriptional regulation. Curr. Opin. Genet. Dev. 2002;12:142–148. doi: 10.1016/s0959-437x(02)00279-4. [DOI] [PubMed] [Google Scholar]
- Bernstein B.E., Kamal M., Lindblad-Toh K., Bekiranov S., Bailey D.K., Huebert D.J., McMahon S., Karlsson E.K., Kulbokas E.J., 3rd, Gingeras T.R., Kamal M., Lindblad-Toh K., Bekiranov S., Bailey D.K., Huebert D.J., McMahon S., Karlsson E.K., Kulbokas E.J., 3rd, Gingeras T.R., Lindblad-Toh K., Bekiranov S., Bailey D.K., Huebert D.J., McMahon S., Karlsson E.K., Kulbokas E.J., 3rd, Gingeras T.R., Bekiranov S., Bailey D.K., Huebert D.J., McMahon S., Karlsson E.K., Kulbokas E.J., 3rd, Gingeras T.R., Bailey D.K., Huebert D.J., McMahon S., Karlsson E.K., Kulbokas E.J., 3rd, Gingeras T.R., Huebert D.J., McMahon S., Karlsson E.K., Kulbokas E.J., 3rd, Gingeras T.R., McMahon S., Karlsson E.K., Kulbokas E.J., 3rd, Gingeras T.R., Karlsson E.K., Kulbokas E.J., 3rd, Gingeras T.R., Kulbokas E.J., 3rd, Gingeras T.R., Gingeras T.R., et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Blackwell T.K., Kretzner L., Blackwood E.M., Eisenman R.N., Weintraub H., Kretzner L., Blackwood E.M., Eisenman R.N., Weintraub H., Blackwood E.M., Eisenman R.N., Weintraub H., Eisenman R.N., Weintraub H., Weintraub H. Sequence-specific DNA binding by the c-Myc protein. Science. 1990;250:1149–1151. doi: 10.1126/science.2251503. [DOI] [PubMed] [Google Scholar]
- Blackwell T.K., Huang J., Ma A., Kretzner L., Alt F.W., Eisenman R.N., Weintraub H., Huang J., Ma A., Kretzner L., Alt F.W., Eisenman R.N., Weintraub H., Ma A., Kretzner L., Alt F.W., Eisenman R.N., Weintraub H., Kretzner L., Alt F.W., Eisenman R.N., Weintraub H., Alt F.W., Eisenman R.N., Weintraub H., Eisenman R.N., Weintraub H., Weintraub H. Binding of myc proteins to canonical and noncanonical DNA sequences. Mol. Cell. Biol. 1993;13:5216–5224. doi: 10.1128/mcb.13.9.5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer L.A., Plath K., Zeitlinger J., Brambrink T., Medeiros L.A., Lee T.I., Levine S.S., Wernig M., Tajonar A., Ray M.K., Plath K., Zeitlinger J., Brambrink T., Medeiros L.A., Lee T.I., Levine S.S., Wernig M., Tajonar A., Ray M.K., Zeitlinger J., Brambrink T., Medeiros L.A., Lee T.I., Levine S.S., Wernig M., Tajonar A., Ray M.K., Brambrink T., Medeiros L.A., Lee T.I., Levine S.S., Wernig M., Tajonar A., Ray M.K., Medeiros L.A., Lee T.I., Levine S.S., Wernig M., Tajonar A., Ray M.K., Lee T.I., Levine S.S., Wernig M., Tajonar A., Ray M.K., Levine S.S., Wernig M., Tajonar A., Ray M.K., Wernig M., Tajonar A., Ray M.K., Tajonar A., Ray M.K., Ray M.K., et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Butler J.E., Kadonaga J.T., Kadonaga J.T. The RNA polymerase II core promoter: A key component in the regulation of gene expression. Genes & Dev. 2002;16:2583–2592. doi: 10.1101/gad.1026202. [DOI] [PubMed] [Google Scholar]
- Cao R., Zhang Y., Zhang Y. SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED–EZH2 complex. Mol. Cell. 2004;15:57–67. doi: 10.1016/j.molcel.2004.06.020. [DOI] [PubMed] [Google Scholar]
- Cao R., Wang L., Wang H., Xia L., Erdjument-Bromage H., Tempst P., Jones R.S., Zhang Y., Wang L., Wang H., Xia L., Erdjument-Bromage H., Tempst P., Jones R.S., Zhang Y., Wang H., Xia L., Erdjument-Bromage H., Tempst P., Jones R.S., Zhang Y., Xia L., Erdjument-Bromage H., Tempst P., Jones R.S., Zhang Y., Erdjument-Bromage H., Tempst P., Jones R.S., Zhang Y., Tempst P., Jones R.S., Zhang Y., Jones R.S., Zhang Y., Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- Cawley S., Bekiranov S., Ng H.H., Kapranov P., Sekinger E.A., Kampa D., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., Bekiranov S., Ng H.H., Kapranov P., Sekinger E.A., Kampa D., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., Ng H.H., Kapranov P., Sekinger E.A., Kampa D., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., Kapranov P., Sekinger E.A., Kampa D., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., Sekinger E.A., Kampa D., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., Kampa D., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., Sementchenko V., Cheng J., Williams A.J., Cheng J., Williams A.J., Williams A.J., et al. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell. 2004;116:499–509. doi: 10.1016/s0092-8674(04)00127-8. [DOI] [PubMed] [Google Scholar]
- Claassen G.F., Hann S.R., Hann S.R. Myc-mediated transformation: The repression connection. Oncogene. 1999;18:2925–2933. doi: 10.1038/sj.onc.1202747. [DOI] [PubMed] [Google Scholar]
- Cole M.D., McMahon S.B., McMahon S.B. The Myc oncoprotein: A critical evaluation of transactivation and target gene regulation. Oncogene. 1999;18:2916–2924. doi: 10.1038/sj.onc.1202748. [DOI] [PubMed] [Google Scholar]
- Czermin B., Melfi R., McCabe D., Seitz V., Imhof A., Pirrotta V., Melfi R., McCabe D., Seitz V., Imhof A., Pirrotta V., McCabe D., Seitz V., Imhof A., Pirrotta V., Seitz V., Imhof A., Pirrotta V., Imhof A., Pirrotta V., Pirrotta V. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell. 2002;111:185–196. doi: 10.1016/s0092-8674(02)00975-3. [DOI] [PubMed] [Google Scholar]
- Efron B. Large-scale simultaneous hypothesis testing: The choice of a null hypothesis. J. Am. Stat. Assoc. 2004;99:96–104. [Google Scholar]
- The ENCODE Project Consortium, The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium, Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007 doi: 10.1038/nature05874. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euskirchen G.M., Rozowsky J., Wei C.-L., Lee W.H., Zhang Z.D., Hartman S., Emanuelsson O., Stolc V., Weissman S., Gerstein M., Rozowsky J., Wei C.-L., Lee W.H., Zhang Z.D., Hartman S., Emanuelsson O., Stolc V., Weissman S., Gerstein M., Wei C.-L., Lee W.H., Zhang Z.D., Hartman S., Emanuelsson O., Stolc V., Weissman S., Gerstein M., Lee W.H., Zhang Z.D., Hartman S., Emanuelsson O., Stolc V., Weissman S., Gerstein M., Zhang Z.D., Hartman S., Emanuelsson O., Stolc V., Weissman S., Gerstein M., Hartman S., Emanuelsson O., Stolc V., Weissman S., Gerstein M., Emanuelsson O., Stolc V., Weissman S., Gerstein M., Stolc V., Weissman S., Gerstein M., Weissman S., Gerstein M., Gerstein M., et al. Mapping of transcription factor binding regions in mammalian cells by ChIP: Comparison of array- and sequencing-based technologies. Genome Res. 2007 doi: 10.1101/gr.5583007. (this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Facchini L.M., Penn L.Z., Penn L.Z. The molecular role of Myc in growth and transformation: Recent discoveries lead to new insights. FASEB J. 1998;12:633–651. [PubMed] [Google Scholar]
- Frank S.R., Schroeder M., Fernandez P., Taubert S., Amati B., Schroeder M., Fernandez P., Taubert S., Amati B., Fernandez P., Taubert S., Amati B., Taubert S., Amati B., Amati B. Binding of c-Myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation. Genes & Dev. 2001;15:2069–2082. doi: 10.1101/gad.906601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel K.R. The biplot graphical display of matrices with application to principal component analysis. Biometrika. 1971;58:453–467. [Google Scholar]
- Geserick C., Meyer H.A., Haendler B., Meyer H.A., Haendler B., Haendler B. The role of DNA response elements as allosteric modulators of steroid receptor function. Mol. Cell. Endocrinol. 2005;236:1–7. doi: 10.1016/j.mce.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Grandori C., Cowley S.M., James L.P., Eisenman R.N., Cowley S.M., James L.P., Eisenman R.N., James L.P., Eisenman R.N., Eisenman R.N. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu. Rev. Cell Dev. Biol. 2000;16:653–699. doi: 10.1146/annurev.cellbio.16.1.653. [DOI] [PubMed] [Google Scholar]
- Horak C.E., Snyder M., Snyder M. ChIP–chip: A genomic approach for identifying transcription factor binding sites. Methods Enzymol. 2002;350:469–483. doi: 10.1016/s0076-6879(02)50979-4. [DOI] [PubMed] [Google Scholar]
- Kadonaga J.T. Regulation of RNA polymerase II transcription by sequence-specific DNA binding factors. Cell. 2004;116:247–257. doi: 10.1016/s0092-8674(03)01078-x. [DOI] [PubMed] [Google Scholar]
- Kuhn E.J., Geyer P.K., Geyer P.K. Genomic insulators: Connecting properties to mechanism. Curr. Opin. Cell Biol. 2003;15:259–265. doi: 10.1016/s0955-0674(03)00039-5. [DOI] [PubMed] [Google Scholar]
- Kuzmichev A., Jenuwein T., Tempst P., Reinberg D., Jenuwein T., Tempst P., Reinberg D., Tempst P., Reinberg D., Reinberg D. Different EZH2-containing complexes target methylation of histone H1 or nucleosomal histone H3. Mol. Cell. 2004;14:183–193. doi: 10.1016/s1097-2765(04)00185-6. [DOI] [PubMed] [Google Scholar]
- Martin D.I. Transcriptional enhancers—on/off gene regulation as an adaptation to silencing in higher eukaryotic nuclei. Trends Genet. 2001;17:444–448. doi: 10.1016/s0168-9525(01)02368-x. [DOI] [PubMed] [Google Scholar]
- Muller J., Hart C.M., Francis N.J., Vargas M.L., Sengupta A., Wild B., Miller E.L., O’Connor M.B., Kingston R.E., Simon J.A., Hart C.M., Francis N.J., Vargas M.L., Sengupta A., Wild B., Miller E.L., O’Connor M.B., Kingston R.E., Simon J.A., Francis N.J., Vargas M.L., Sengupta A., Wild B., Miller E.L., O’Connor M.B., Kingston R.E., Simon J.A., Vargas M.L., Sengupta A., Wild B., Miller E.L., O’Connor M.B., Kingston R.E., Simon J.A., Sengupta A., Wild B., Miller E.L., O’Connor M.B., Kingston R.E., Simon J.A., Wild B., Miller E.L., O’Connor M.B., Kingston R.E., Simon J.A., Miller E.L., O’Connor M.B., Kingston R.E., Simon J.A., O’Connor M.B., Kingston R.E., Simon J.A., Kingston R.E., Simon J.A., Simon J.A. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell. 2002;111:197–208. doi: 10.1016/s0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- Ng P., Wei C.L., Sung W.K., Chiu K.P., Lipovich L., Ang C.C., Gupta S., Shahab A., Ridwan A., Wong C.H., Wei C.L., Sung W.K., Chiu K.P., Lipovich L., Ang C.C., Gupta S., Shahab A., Ridwan A., Wong C.H., Sung W.K., Chiu K.P., Lipovich L., Ang C.C., Gupta S., Shahab A., Ridwan A., Wong C.H., Chiu K.P., Lipovich L., Ang C.C., Gupta S., Shahab A., Ridwan A., Wong C.H., Lipovich L., Ang C.C., Gupta S., Shahab A., Ridwan A., Wong C.H., Ang C.C., Gupta S., Shahab A., Ridwan A., Wong C.H., Gupta S., Shahab A., Ridwan A., Wong C.H., Shahab A., Ridwan A., Wong C.H., Ridwan A., Wong C.H., Wong C.H., et al. Gene identification signature (GIS) analysis for transcriptome characterization and genome annotation. Nat. Methods. 2005;2:105–111. doi: 10.1038/nmeth733. [DOI] [PubMed] [Google Scholar]
- Nikolov D.B., Burley S.K., Burley S.K. RNA polymerase II transcription initiation: A structural view. Proc. Natl. Acad. Sci. 1997;94:15–22. doi: 10.1073/pnas.94.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D., Bracken A.P., Jensen M.R., Lazzerini Denchi E., Helin K., Bracken A.P., Jensen M.R., Lazzerini Denchi E., Helin K., Jensen M.R., Lazzerini Denchi E., Helin K., Lazzerini Denchi E., Helin K., Helin K. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J. 2004;23:4061–4071. doi: 10.1038/sj.emboj.7600402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokholok D.K., Harbison C.T., Levine S., Cole M., Hannett N.M., Lee T.I., Bell G.W., Walker K., Rolfe P.A., Herbolsheimer E., Harbison C.T., Levine S., Cole M., Hannett N.M., Lee T.I., Bell G.W., Walker K., Rolfe P.A., Herbolsheimer E., Levine S., Cole M., Hannett N.M., Lee T.I., Bell G.W., Walker K., Rolfe P.A., Herbolsheimer E., Cole M., Hannett N.M., Lee T.I., Bell G.W., Walker K., Rolfe P.A., Herbolsheimer E., Hannett N.M., Lee T.I., Bell G.W., Walker K., Rolfe P.A., Herbolsheimer E., Lee T.I., Bell G.W., Walker K., Rolfe P.A., Herbolsheimer E., Bell G.W., Walker K., Rolfe P.A., Herbolsheimer E., Walker K., Rolfe P.A., Herbolsheimer E., Rolfe P.A., Herbolsheimer E., Herbolsheimer E., et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- Pozzoli U., Sironi M., Sironi M. Silencers regulate both constitutive and alternative splicing events in mammals. Cell. Mol. Life Sci. 2005;62:1579–1604. doi: 10.1007/s00018-005-5030-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubeler D., MacAlpine D.M., Scalzo D., Wirbelauer C., Kooperberg C., van Leeuwen F., Gottschling D.E., O’Neill L.P., Turner B.M., Delrow J., MacAlpine D.M., Scalzo D., Wirbelauer C., Kooperberg C., van Leeuwen F., Gottschling D.E., O’Neill L.P., Turner B.M., Delrow J., Scalzo D., Wirbelauer C., Kooperberg C., van Leeuwen F., Gottschling D.E., O’Neill L.P., Turner B.M., Delrow J., Wirbelauer C., Kooperberg C., van Leeuwen F., Gottschling D.E., O’Neill L.P., Turner B.M., Delrow J., Kooperberg C., van Leeuwen F., Gottschling D.E., O’Neill L.P., Turner B.M., Delrow J., van Leeuwen F., Gottschling D.E., O’Neill L.P., Turner B.M., Delrow J., Gottschling D.E., O’Neill L.P., Turner B.M., Delrow J., O’Neill L.P., Turner B.M., Delrow J., Turner B.M., Delrow J., Delrow J., et al. The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes & Dev. 2004;18:1263–1271. doi: 10.1101/gad.1198204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinklein N.D., Karaöz U., Wu J., Halees A., Force Aldred S., Collins P.J., Zheng D., Zhang Z.D., Gerstein M., Snyder M., Karaöz U., Wu J., Halees A., Force Aldred S., Collins P.J., Zheng D., Zhang Z.D., Gerstein M., Snyder M., Wu J., Halees A., Force Aldred S., Collins P.J., Zheng D., Zhang Z.D., Gerstein M., Snyder M., Halees A., Force Aldred S., Collins P.J., Zheng D., Zhang Z.D., Gerstein M., Snyder M., Force Aldred S., Collins P.J., Zheng D., Zhang Z.D., Gerstein M., Snyder M., Collins P.J., Zheng D., Zhang Z.D., Gerstein M., Snyder M., Zheng D., Zhang Z.D., Gerstein M., Snyder M., Zhang Z.D., Gerstein M., Snyder M., Gerstein M., Snyder M., Snyder M., et al. Integrated analysis of experimental data sets reveals many novel promoters in 1% of the human genome. Genome Res. 2007 doi: 10.1101/gr.5716607. (this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner B.M. Cellular memory and the histone code. Cell. 2002;111:285–291. doi: 10.1016/s0092-8674(02)01080-2. [DOI] [PubMed] [Google Scholar]
- Wang W., Cote J., Xue Y., Zhou S., Khavari P.A., Biggar S.R., Muchardt C., Kalpana G.V., Goff S.P., Yaniv M., Cote J., Xue Y., Zhou S., Khavari P.A., Biggar S.R., Muchardt C., Kalpana G.V., Goff S.P., Yaniv M., Xue Y., Zhou S., Khavari P.A., Biggar S.R., Muchardt C., Kalpana G.V., Goff S.P., Yaniv M., Zhou S., Khavari P.A., Biggar S.R., Muchardt C., Kalpana G.V., Goff S.P., Yaniv M., Khavari P.A., Biggar S.R., Muchardt C., Kalpana G.V., Goff S.P., Yaniv M., Biggar S.R., Muchardt C., Kalpana G.V., Goff S.P., Yaniv M., Muchardt C., Kalpana G.V., Goff S.P., Yaniv M., Kalpana G.V., Goff S.P., Yaniv M., Goff S.P., Yaniv M., Yaniv M., et al. Purification and biochemical heterogeneity of the mammalian SWI–SNF complex. EMBO J. 1996a;15:5370–5382. [PMC free article] [PubMed] [Google Scholar]
- Wang W., Xue Y., Zhou S., Kuo A., Cairns B.R., Crabtree G.R., Xue Y., Zhou S., Kuo A., Cairns B.R., Crabtree G.R., Zhou S., Kuo A., Cairns B.R., Crabtree G.R., Kuo A., Cairns B.R., Crabtree G.R., Cairns B.R., Crabtree G.R., Crabtree G.R. Diversity and specialization of mammalian SWI/SNF complexes. Genes & Dev. 1996b;10:2117–2130. doi: 10.1101/gad.10.17.2117. [DOI] [PubMed] [Google Scholar]