Abstract
The biosynthesis of sterols is a major route for the development of antitrypanosomals. Squalene synthase (SQS) catalyzes the first step committed to the biosynthesis of sterols within the isoprenoid pathway, and several inhibitors of the enzyme have selective antitrypanosomal activity both in vivo and in vitro. The enzyme from Trypanosoma cruzi is a 404-amino-acid protein with a clearly identifiable membrane-spanning region. In an effort to generate soluble recombinant enzyme, we have expressed in Escherichia coli several truncated versions of T. cruzi SQS with a His tag attached to the amino terminus. Deletions of both the amino- and carboxyl-terminal regions generated active and soluble forms of the enzyme. The highest levels of soluble protein were achieved when 24 and 36 amino acids were eliminated from the amino and carboxyl regions, respectively, yielding a protein of 41.67 kDa. The Michaelis-Menten constants of the purified enzyme for farnesyl diphosphate and NAD (NADPH) were 5.25 and 23.34 μM, respectively, whereas the Vmax was 1,428.56 nmol min−1mg−1. Several quinuclidine derivatives with antiprotozoal activity in vitro were found to be selective inhibitors of recombinant T. cruzi SQS in comparative assays with the human enzyme, with 50% inhibitory concentration values in the nanomolar range. These data suggest that selective inhibition of T. cruzi SQS may be an efficient strategy for the development of new antitrypanosomal agents.
Trypanosoma (Schizotrypanum) cruzi, is an intracellular protozoan parasite that infects many wild mammals and humans, being the etiological agent of Chagas' disease, one of the major public health problems in many countries of Central and South America (20). Acute infections can be lethal, but the disease usually evolves into a chronic stage, accompanied in 25 to 30% of cases by severe debilitation and ultimately death due to irreversible lesions of the heart and gastrointestinal tract. It is estimated that 16 to 18 million people are infected with T. cruzi, primarily in Central and South America, with 21,000 deaths reported each year (27). Currently available chemotherapy has low efficacy (particularly in chronic infections), frequent toxic side effects, and drug resistance (3, 22). Studies have shown that protozoan parasites such as T. cruzi and different species of the Leishmania genus require the de novo synthesis of specific endogenous sterols (ergosterol and analogs), which act as essential growth factors for survival (5, 22, 23). These parasites are highly susceptible, in vivo and in vitro, to sterol biosynthesis inhibitors such as antifungal azoles, quinuclidine derivatives, allylamines, statins, and azasterols (5, 26). Indeed, sterol biosynthesis is a major route for intervention in the development of antitrypanosomals.
The enzyme squalene synthase (SQS; EC 2.5.1.21) catalyzes the condensation of two molecules of farnesyl diphosphate (FPP) to produce squalene, the first committed step of the sterol pathway (Fig. 1). FPP is a major branching point in isoprenoid biosynthesis: it can be converted by SQS to squalene and sterols, or it can be used for the production of other essential isoprenoids, such as dolichols, coenzyme Q, heme, and prenylated proteins. Hence, considerable effort has been devoted to the development of specific inhibitors of SQS, since this should prevent the biosynthesis of sterols while not affecting the production of other essential isoprenoids (14). The inhibition of SQS should also prevent the buildup of sterol intermediates that may occur should later steps of sterol biosynthesis be inhibited.
FIG. 1.
Chemical reaction catalyzed by SQS.
Published evidence has shown that several inhibitors of mammalian SQS have potent antitrypanosomal activity both in vitro and in animal models (16, 24). Inhibition of parasite growth was associated with a depletion of the parasite's endogenous sterols strongly, suggesting that the main mode of action of these compounds is through the inhibition of SQS. Rational drug design has been utilized in the development of mimetics of several substrates, intermediates, and transition states in the transformation of FPP to squalene (1). One class of compounds of particular interest is the arylquinuclidines, which are protonated at physiological pH and are thought to mimic a high-energy intermediate of the SQS reaction.
Several attempts have been made to express the soluble and active SQS from different organisms in Escherichia coli. Different truncated versions of the enzyme have been generated by molecular biology methods (12, 18, 21), based on the previous evidence that a soluble and active form of SQS purified from rat liver microsomes could be obtained after limited proteolysis with trypsin (11, 18) and genetic truncation to remove membrane binding regions (12, 28).
To avoid problems that arise when expressing and purifying a membrane-bound protein such as SQS, we generated here a recombinant T. cruzi enzyme that was truncated at both the amino- and the carboxyl-terminal regions to create a soluble, active protein amenable to kinetic characterization and inhibition studies. We also present a kinetic characterization of the purified soluble enzyme and show that several quinuclidine derivatives exhibit selective inhibition of T. cruzi SQS. This information could be exploited in the development of compounds with reduced toxicity for the etiological treatment of Chagas disease.
MATERIALS AND METHODS
Materials.
The triammonium salt of [3H]farnesyl diphosphate (15.0 Ci/mmol) was obtained from Amersham Biosciences. Restriction enzymes and protease inhibitors cocktail were from Roche. T4 DNA ligase and Taq polymerase from Invitrogen. The pET28(a) expression system and E. coli BL21(DE3)RP were purchased from Novagen (Madison, WI).
Cloning of the full-length T. cruzi SQS gene and generation of truncated versions.
The SQS gene was amplified by PCR using the oligonucleotide primers TcSQS N-term (CAT ATG GAG TCA ATG GAG GAG TTG) and TcSQS C-term (GAA TTA CTT CCC AAG ATA TCC AAC AAC), which were designed taking into account the T. cruzi sequence present in the GeneDB database, CDS: Tc00.1047053507897.20 (the restriction NdeI and EcoRI sites are underlined). The PCR was performed using genomic T. cruzi strain Y DNA as a template. A 1,215-bp fragment containing the entire T. cruzi SQS open reading frame gene (encoding 404 amino acids) was cloned into pGEM-T to generate the plasmid pSQS-ORF. Different constructs were made in order to obtain soluble and active protein. Additional primers were designed to generate truncated proteins by removing 13, 16, 17, and 24 amino acids from the N terminus and 36 and 46 amino acids from the C terminus. NdeI and EcoRI restriction sites were introduced for directional cloning in the pET28a(+) expression vector (Novagen) to yield pETTcSQS13/46, pETTcSQS16/46, pETTcSQS17/46, pETTcSQS13/36, pETTcSQS17/36, and pETTcSQS24/36. Double-stranded DNA sequencing was performed to confirm that the correct reading frame was used, with the polyhistidine tag placed in the N-terminal position.
Expression of truncated T. cruzi SQS.
For expression in E. coli, the expression plasmids pET28a T. cruzi double-truncated SQSs were transfected into the BL21(DE3)RP strain (Novagen). Bacteria cells were grown in Luria broth (LB) medium containing kanamycin (30 μg/ml) and chloramphenicol (34 μg/ml) and were incubated at 37°C overnight. When induction was performed, cells were incubated at 37°C and 120 rpm until reaching an optical density at 600 nm of 0.3, and then the flask was transferred to an incubator at 24°C to induce the expression of an optical density at 600 nm of 0.4 with 50 μM concentrations of IPTG (isopropyl-β-d-thiogalactopyranoside). Cells were induced for 4 h, pelleted by centrifugation at 4,000 rpm for 10 min at 4°C, and stored at −80°C until use.
SQS assay and product analysis.
The catalytic activity of SQS was assayed by measuring the conversion of [3H]FPP to [3H]squalene. Final assay concentrations were 50 mM morpholinepropanesulfonic acid-NaOH buffer (pH 7.4), 20 mM MgCl2, 5 mM CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}, 1% Tween 80, 10 mM dithiothreitol, 0.025 mg of bovine serum albumin ml−1, 0.25 mM NADPH, mg of purified recombinant protein ml−1, and different concentrations of FPP. For specific activity determinations, a saturating concentration of FPP (20 μM) was used. To determine the Km values for FPP, concentrations between 1 and 20 μM were used, while NADPH was maintained at saturation (2 mM). To determine the Km for NADPH, concentrations ranged from 5 to 400 μM, while the FPP concentration was 20 μM. 50% inhibitory concentration (IC50) determinations were determined at an FPP concentration of 0.5 μM (1 μCi per assay). The final volume of the reaction was 200 μl. After incubation at 37°C for 5 min, 40 μl of 10 M NaOH was added, followed by 10 μl of a mixture (50:1) of 70% ethanol and squalene. The resulting mixtures were mixed vigorously by vortexing, and then 10-μl aliquots were applied to 2.5-by-10-cm channels of a silica gel thin-layer chromatogram, and the newly formed squalene was separated from unreacted substrates by chromatography in toluene-ethyl acetate (9:1). The region of each chromatogram from 2 cm below the squalene band (Rf = 0.74) to the top of the chromatogram was removed and immersed in Hydrofluor liquid scintillation fluid and assessed for radioactivity, allowing the measure of the amount of conversion of FPP to squalene using a Pharmacia LKB liquid scintillation counter. The protein concentration was measured by the Bradford method with bovine serum as a standard.
Enzyme kinetic parameters and IC50 values were estimated by utilizing the software SigmaPlot 2002 for Windows version 8.0. The Km/Vmax values were determined from nonlinear hyperbolic fits.
For the analysis of inhibitor interaction with human SQS, extracts from E. coli cells transformed with the expression plasmid pHSS16 (21) were used as the enzyme source. IC50 values were obtained by using the same procedures described above for T. cruzi SQS.
Purification of double-truncated T. cruzi SQS.
E. coli BL21(DE3)RP/pETTcSQS24/36 cells were resuspended in binding buffer (20 mM NaH2PO3 [pH 7.4], 10 mM CHAPS, 2 mM MgCl2, 10% glycerol, 10 mM β-mercaptoethanol, 500 mM NaCl, 10 mM imidazole, and protease inhibitor cocktail [Roche]), disrupted by sonication, and centrifuged at 14,000 rpm for 20 min. The soluble extract was applied to a HiTrap Nickel-Chelating HP column (Amersham Biosciences). The purification was performed according to the manufacturer's instructions in a Pharmacia FPLC system. The unbound protein was washed with 60 mM imidazole, and the His6-TcSQS24/36 was eluted with 500 mM imidazole. The enzyme purity was confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Coomassie brilliant blue R-250 (Sigma) was used for staining. Fractions containing the enzyme were pooled and dialyzed against buffer A (25 mM sodium phosphate [pH 7.4] 20 mM NaCl, 2 mM dithiothreitol, 1 mM EDTA, 10% glycerol, 10% methanol, 1 mM phenylmethylsulfonyl fluoride, 10 μM leupeptin, 1 μM pepstatin) prior to MonoQ ion-exchange chromatography. A 1-ml MonoQ HR5/5 anion-exchange column (Pharmacia) was equilibrated with 15 ml of buffer A. This chromatographic step was performed by using the Pharmacia HPLC system AKTA P900. The sample from immobilized metal affinity chromatography (IMAC) was applied to a MonoQ HR5/5 column at a flow rate of 0.5 ml/min. The sample was then eluted from the column at a flow rate of 0.5 ml/min by first washing with 15 ml of buffer A, followed by a linear 30-ml salt gradient up to 500 mM NaCl. Fractions containing SQS were identified by enzyme activity assays and SDS-PAGE, concentrated, and stored at −80°C.
Assay of quinuclidine derivatives against intracellular T. cruzi amastigotes cultured in vitro.
Rat skeletal myoblasts (L-6 cells) were seeded in 96-well microtiter plates at 2,000 cells/well/100 μl in RPMI 1640 medium with 10% fetal bovine serum and 2 mM l-glutamine. After 24 h, 5,000 T. cruzi trypomastigotes (Tulahuen strain C2C4 containing the β-galactosidase lacZ gene) (2) were added in aliquots of 100 μl per well with a twofold serial drug dilution. The plates were incubated at 37°C in 5% CO2 for 4 days. The substrate CPRG/Nonidet was then added to the wells. The color reaction, which developed during the following 2 to 4 h, was read spectrophotometrically at 540 nm. IC50 values were calculated from the sigmoidal inhibition curve by using Microsoft Excel.
Quinuclidine derivatives.
The quinuclidine derivatives used in the present study were synthesized and characterized by nuclear magnetic resonance and mass spectrometry. The full details of their chemical synthesis will be published elsewhere (S. B. Cammerer, C. Jimenez, S. Jones, L. Gros, S. Orenes-Lorente, C. Rodrigues, J. C. F. Rodrigues, A. Caldera, L. M. Ruiz-Perez, W. da Souza, M. Kaiser, R. Brun, J. A. Urbina, D. Gonzalez Pacanowska, and I. H. Gilbert, unpublished data), except for ER119884 and E5700, which were supplied by Tsukuba Research Laboratories, Eisai Co., Ltd., Ibaraki, Japan.
RESULTS
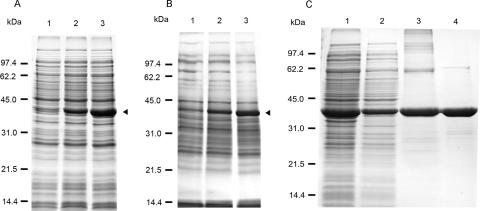
The amino acid sequence of T. cruzi SQS is conserved relative to other eukaryotic SQSs and has 55 to 58% identity and 65 to 73% similarity with other representatives of the Trypanosomatidae family (T. brucei and Leishmania major). As shown in Fig. 2, all of the conserved residues described to be involved in catalysis (17) are present in the T. cruzi enzyme such as the aspartate rich motifs involved in substrate binding (82DTVED and 229CFYED). Previous reports have shown the development of SQS expression systems for yeast, rat, and human enzyme (12, 18, 21). For the production of soluble protein, partial deletion of the N-terminal, C-terminal, or both regions was required. Likewise, initial attempts to produce a full-length soluble T. cruzi enzyme were unsuccessful. Based on a comparative analysis of other recombinant soluble SQS enzymes several constructs were designed in order to eliminate 13, 16, 17, or 24 amino acids from the amino terminus combined with the elimination of 36 or 46 amino acids from the carboxyl-terminal regions. All of the constructs were cloned into the pET28a(+) expression vector and transformed in E. coli BL21(DE3)RP for analysis of the level of expression and solubility on SDS-polyacrylamide gels (data not shown). After analysis for soluble protein and activity measurements, we found that a soluble active double truncated form was only obtained after the elimination of 17 or 24 amino acids from the amino terminus and the elimination of 36 residues from the carboxyl terminus. Panels A and B of Fig. 3 represent the SDS-PAGE analysis after the induction of expression with IPTG for the constructs pETTcSQS24/36 and pETTcSQS17/36, respectively, showing high levels of soluble and active recombinant SQS.
FIG. 2.
Alignment of the amino acid sequences of SQS from different organisms: T. cruzi (XP806809), L. major (CAJ08546), human (I52090), and yeast (CAA42583). Sequences were aligned by using the AlignX module of Vector NTI (Invitrogen). Conserved regions are highlighted. The arrows indicate the site of the different truncations.
FIG. 3.
SDS-PAGE analysis of expression and purification of double-truncated SQS. (A) Soluble fractions of E. coli BL21(DE3)RP/pETTcSQS24/36 cells. Lanes 1 to 3 contain the soluble fraction of at 0, 1, and 4 h after induction. (B) Soluble fractions of E. coli BL21(DE3)RP/pETTcSQS17/36 cells. Lanes 1 to 3 contain the soluble fraction at 0, 1, and 4 h after induction. The arrows show the position of the T. cruzi SQS recombinant protein. (C) Purification of truncated SQS (TcSQS24/36). Lane 1, cell extract; lane 2, soluble fraction; lane 3, IMAC; lane 4, after MonoQ HR5/5 chromatography.
Purification of truncated soluble T. cruzi SQS (TcSQS24/36).
The TcSQS24/36 truncated protein (lacking 24 and 36 amino acids from the amino- and carboxyl-terminal regions, respectively) was the construct that showed the strongest band at approximately 41 kDa and exhibited high SQS activity. The isolation and purification of truncated T. cruzi SQS from E. coli cells was accomplished with two purification steps as described in Materials and Methods: IMAC and MonoQ ion-exchange chromatography. The purification data are summarized in Table 1. IMAC analysis yielded a 1.7-fold purification. This material was loaded directly into a MonoQ ion-exchange chromatography column and eluted with a salt gradient. In this step, SQS eluted with a NaCl gradient ranging from 250 to 350 mM NaCl. The MonoQ ion-exchange chromatography gave a further 4.5-fold purification. Pure truncated SQS was stable in buffer A with 40% of glycerol and could be stored at −80°C for extended periods of time. The purity of the SQS was determined by SDS-PAGE analysis (Fig. 3C). The enzyme migrates as a single band with an estimated molecular mass for the truncated enzyme of 41.67 kDa. All kinetic experiments were performed with purified recombinant enzyme.
TABLE 1.
Purification of TcSQS24/36a
| Step | Vol (ml) | Protein concn (mg ml−1) | Total protein (mg) | Sp act (nmol min−1mg−1) | Total U (nmol min−1) | % Yield | Fold purification |
|---|---|---|---|---|---|---|---|
| Soluble extract | 8.50 | 5.70 | 49.08 | 100.29 | 4,922.6 | 100 | 1 |
| IMAC-His | 11 | 0.67 | 7.20 | 178.26 | 1,283.5 | 26 | 1.7 |
| MonoQ HR5/5 | 1.50 | 1.60 | 2.40 | 459.97 | 1,103.9 | 22 | 4.5 |
The truncated protein was purified using a combination of IMAC and MonoQ ion-exchange chromatography.
Kinetic properties of soluble truncated SQS: determination of Km and kcat values.
Standard procedures were used to determine kinetic parameters. Km and Vmax values were obtained by nonlinear regression fit of the data to the Michaelis-Menten equation (Sigmaplot 2002 for Windows, version 8.0). The Km and the Vmax for FPP were 5.25 μM and 1,428.56 nmol min−1 mg−1 respectively, and for NADPH these values were 23.34 μM and 1,853.24 nmol min−1 mg−1 (Fig. 4). The calculated kcat values were 1.05 s−1 and 1.29 s−1 for FPP and NADPH, respectively. These values are of the same order of magnitude as those described for other truncated recombinant enzymes, as shown in Table 2.
FIG. 4.
Michaelis-Menten and Lineweaver-Burk (inset) plots for the calculation of the Km and Vmax values of T. cruzi SQS. (A) Plots for calculation of the Km for FPP. The concentration range of FPP was 0.5 to 20 μM. (B) Plots for calculation of the Km for NADPH. The concentration range of NADPH was 5 to 400 μM.
TABLE 2.
Kinetic constants of squalene synthase from different species and sources
| Species and/or sourcea | SQS type | FPP
|
NADPH
|
kcat (s−1) | Reference | ||
|---|---|---|---|---|---|---|---|
| Km (μM) | Vmax (nmol min−1 mg−1) | Km (μM) | Vmax (nmol min−1 mg−1) | ||||
| Human pHSS12/pHSS16 | Double-truncated SQS | 2.8 | 1.44 | 21 | |||
| T. cruzi | Glycosomal SQS | 2.8 | 0.97 | 40 | 0.43 | 25 | |
| T. cruzi | Microsomal/mitochondrial SQS | 2.3 | 1.18 | 33 | 0.76 | 25 | |
| L. mexicana | Glycosomal SQS | 3.2 | 0.75 | 62 | 0.58 | 25 | |
| L. mexicana | Microsomal/mitochondrial SQS | 2.8 | 0.98 | 57 | 0.84 | 25 | |
| Human | Full-length SQS expressed in a baculoviral system | 2.3 ± 0.5 | 4,800 ± 460 | 430 ± 60 | 3,800 ± 150 | 19 | |
| Yeast | C-terminal truncated SQS | 2.5 ± 0.46 | 530 ± 77 | 0.53 ± 0.03 | 12 | ||
| T. cruzi | |||||||
| TcSQS24/36 | 5.25 ± 1.2 | 1,428.56 ± 317 | 23.34 ± 4.5 | 1,853.24 ± 435 | 1.05 ± 0.16 | ||
Inhibition by quinuclidine derivatives.
A series of quinuclidine derivatives were tested against the purified T. cruzi recombinant enzyme, the human recombinant SQS, and against the intracellular form of the parasite cultured in vitro. When tested against purified recombinant T. cruzi enzyme, the analogs gave IC50 values in the low micromolar or nanomolar range (Table 3). Compound 1 exhibited an IC50 value of 50 nM, whereas the Eisai compounds E5700 and ER119884 gave values of 0.84 and 3.52 nM, respectively. We sought to analyze the interaction of some of these compounds with a soluble truncated form of recombinant human SQS. As shown in Table 3, some of the compounds appeared to be less active against the human enzyme and, when IC50 values were compared, a selectivity index of 140 was obtained in the case of compound 1. Selectivity indexes higher than 27 and 13 were obtained for compounds 2 and 4, respectively. In the case of E5700 and ER-119884, both compounds appear to be equally active on the human and T. cruzi enzymes.
TABLE 3.
Interaction of SQS from different sources with quinuclidine analogsa
SIenz is the ratio of IC50 values for human SQS to those for T. cruzi SQS, and SIgrowth is the ratio of IC50 values for growth inhibition against L6 cells to those against intracellular T. cruzi amastigotes.
b Values to be reported elsewhere (Cammerer et al., unpublished).
c Values for inhibition of amastigote growth as reported by Urbina et al. (24).
d ND, not determined.
When these compounds were assayed against intracellular T. cruzi amastigotes in vitro, they exhibited potent and selective antitrypanosomal activity, with IC50 values in the low micromolar range (Table 3). There was not a direct correlation between inhibition of the enzyme and inhibition of the growth of the parasite, but this may not be totally unexpected, since T. cruzi is an intracellular parasite, and hence the activity of the compounds against the parasite will depend not only on the inhibition of the parasite's SQS but also on other factors such as drug penetration through the several permeability barriers involved and other properties of the molecules. However, although there is not a direct correlation between enzyme inhibition and growth inhibition of the intracellular parasites, these studies are proof of principle that inhibitors of T. cruzi SQSs also show activity against the clinically relevant form of the parasite. Study of a larger range of analogues will provide more information.
DISCUSSION
Genes encoding SQS have been isolated from many sources, such as fungi, plants and animals (4, 6-10, 12, 13, 15, 28). The enzyme is monomeric and has been reported to be associated with the endoplasmic reticulum at least in most eukaryotes. The T. cruzi enzyme is considerably conserved and a comparative analysis of the amino acid sequences reveals an overall high degree of similarity. The generation of high quantities of soluble enzyme for inhibitor screening was attempted using a strategy that proved to be successful with other eukaryotic SQSs. Thus far, truncated soluble and active recombinant enzymes have been generated for yeast, rat, and human SQSs. In the yeast protein, the 24 carboxyl-terminal residues were removed via genetic manipulation, and a soluble, active enzyme was produced that could account for up to 20% of the total soluble protein when expressed in E. coli (12). The production of an active human enzyme was attained after truncation of both the carboxy (47 amino acids) and the amino terminus (30 amino acids) (31 to 371) that yielded a soluble protein with catalytic properties similar to the native enzyme (21). In the present study we generated a soluble enzyme by elimination of 24 amino acids of the amino terminus and 36 amino acids of the carboxyl terminus. The kinetic parameters were compared to those previously reported for preparations of T. cruzi glycosomal and microsomal SQS (25) and other recombinant enzymes. The resulting enzyme proved to be catalytically active and exhibited kinetic parameters highly similar to those obtained with the native enzyme in purified glycosomes and mitochondria from T. cruzi epimastigotes (25), albeit the Km for FPP was slightly higher. Likewise, the Km and kcat values were highly similar to those obtained for the truncated recombinant enzyme from yeast (12).
The first indication of the antitrypanosomal activity of SQS inhibitors came from studies by Urbina et al. with 3-(biphenyl-4-yl)-3-hydroxyquinuclidine (BPQ-OH), a potent and specific inhibitor of mammalian SQS (25). It was found that BPQ-OH induced a dose-dependent reduction of the proliferation of extracellular stages (epimastigotes) of these parasites with MICs of 30 μM. Growth inhibition and cell lysis induced by BPQ-OH in both parasites was associated with complete depletion of endogenous squalene and sterols. BPQ-OH was able to eradicate intracellular T. cruzi amastigotes from Vero cells with an MIC of 30 μM, with no deleterious effects on host cells at up to 100 μM.
Several other analogues have been tested for activity against the L. major enzyme and the parasite in vitro (16). An analog of BPQ-OH, where the 3-OH group has been removed by dehydration, leaving a Δ2(3) bond, was clearly more potent than BPQ-OH, since cell lysis was observed in the presence of 1 μM in L. mexicana promastigotes and exhibited an IC50 for recombinant SQS from L. major of 0.24 μM (16).
Recent studies (24) with the compounds E5700 and ER-119884 (Eisai Chemical Company, Tokyo, Japan) showed that they were very potent noncompetitive or mixed-type inhibitors of native T. cruzi SQS with Ki values in the low nanomolar or subnanomolar range in the absence or presence of 20 μM inorganic pyrophosphate. Their antiproliferative IC50s against extracellular epimastigotes and intracellular amastigotes were ca. 10 nM and 0.4 to 1.6 nM, respectively, with no effects on host cells. These compounds are among the most potent antitrypanosomals ever tested in vitro.
We now show that E5700 and ER-119884 are also highly potent inhibitors of purified recombinant T. cruzi SQS, but they have no selectivity toward the parasite's enzyme in comparative assays with the recombinant human enzyme. On the other hand, we have identified analogs such as compound 2, which is a good inhibitor of intracellular amastigote growth in vitro but also a selective inhibitor of T. cruzi SQS. Intracellular amastigotes are the clinically relevant form of the parasite. Compound 1 is also selective for the trypanosomal enzyme, although it was was less active against the parasite in vitro. These observations offer a new approach for the design of SQS inhibitors with a potential application as antitrypanosomal compounds. Specific inhibitors of the T. cruzi enzyme would allow for increased efficacy and the minimization of possible adverse effects due to inhibition of human sterol biosynthesis. Studies are currently under way to determine the structural requirements for specific inhibition.
Acknowledgments
We acknowledge the EU INCO-DEV program (ICA4-2000-10028), the Spanish Plan Nacional (SAF2004-03828), the Junta de Andalucía CVI-199, and the Howard Hughes Medical Institute (grant 55000620 to M.S.-C. and J.A.U.) for financial support.
We thank Sofia Vargas for technical assistance and John F. Thompson (Pfizer) for kindly providing the expression system for human SQS.
Footnotes
Published ahead of print on 19 March 2007.
REFERENCES
- 1.Biller, S. A., K. Neuenschwander, M. M. Ponpipom, and C. D. Poulter. 1996. Squalene synthase inhibitors. Curr. Pharm. Design 2:1-40. [Google Scholar]
- 2.Buckner, F. S., A. J. Wilson, and W. C. Van Voorhis. 1999. Detection of live Trypanosoma cruzi in tissues of infected mice by using histochemical stain for beta-galactosidase. Infect. Immun. 67:403-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Andrade, A. L., F. Zicker, R. M. de Oliveira, S. Almeida Silva, A. Luquetti, L. R. Travassos, I. C. Almeida, S. S. de Andrade, J. G. de Andrade, and C. M. Martelli. 1996. Randomised trial of efficacy of benznidazole in treatment of early Trypanosoma cruzi infection. Lancet 348:1407-1413. [DOI] [PubMed] [Google Scholar]
- 4.Devarenne, T. P., D. H. Shin, K. Back, S. Yin, and J. Chappell. 1998. Molecular characterization of tobacco squalene synthase and regulation in response to fungal elicitor. Arch. Biochem. Biophys. 349:205-215. [DOI] [PubMed] [Google Scholar]
- 5.Docampo, R., and G. A. Schmunis. 1997. Sterol biosynthesis inhibitors: potential chemotherapeutics against Chagas disease. Parasitol. Today 13:129-130. [DOI] [PubMed] [Google Scholar]
- 6.Fegueur, M., L. Richard, A. D. Charles, and F. Karst. 1991. Isolation and primary structure of the ERG9 gene of Saccharomyces cerevisiae encoding squalene synthetase. Curr. Genet. 20:365-372. [DOI] [PubMed] [Google Scholar]
- 7.Hanley, K. M., O. Nicolas, T. B. Donaldson, C. Smith-Monroy, G. W. Robinson, and G. M. Hellmann. 1996. Molecular cloning, in vitro expression and characterization of a plant squalene synthetase cDNA. Plant Mol. Biol. 30:1139-1151. [DOI] [PubMed] [Google Scholar]
- 8.Hata, S., K. Sanmiya, H. Kouchi, M. Matsuoka, N. Yamamoto, and K. Izui. 1997. cDNA cloning of squalene synthase genes from mono- and dicotyledonous plants, and expression of the gene in rice. Plant Cell Physiol. 38:1409-1413. [DOI] [PubMed] [Google Scholar]
- 9.Jennings, S. M., Y. H. Tsay, T. M. Fisch, and G. W. Robinson. 1991. Molecular cloning and characterization of the yeast gene for squalene synthetase. Proc. Natl. Acad. Sci. USA 88:6038-6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee, J. H., Y. H. Yoon, H. Y. Kim, D. H. Shin, D. U. Kim, I. J. Lee, and K. U. Kim. 2002. Cloning and expression of squalene synthase cDNA from hot pepper (Capsicum annuum L.). Mol. Cells 13:436-443. [PubMed] [Google Scholar]
- 11.Lindsey, S., and H. J. Harwood, Jr. 1995. Inhibition of mammalian squalene synthetase activity by zaragozic acid A is a result of competitive inhibition followed by mechanism-based irreversible inactivation. J. Biol. Chem. 270:9083-9096. [DOI] [PubMed] [Google Scholar]
- 12.LoGrasso, P. V., D. A. Soltis, and B. R. Boettcher. 1993. Overexpression, purification, and kinetic characterization of a carboxyl-terminal-truncated yeast squalene synthetase. Arch. Biochem. Biophys. 307:193-199. [DOI] [PubMed] [Google Scholar]
- 13.McKenzie, T. L., G. Jiang, J. R. Straubhaar, D. G. Conrad, and I. Shechter. 1992. Molecular cloning, expression, and characterization of the cDNA for the rat hepatic squalene synthase. J. Biol. Chem. 267:21368-21374. [PubMed] [Google Scholar]
- 14.McTaggart, F., G. R. Brown, R. G. Davidson, S. Freeman, G. A. Holdgate, K. B. Mallion, D. J. Mirrlees, G. J. Smith, and W. H. Ward. 1996. Inhibition of squalene synthase of rat liver by novel 3′ substituted quinuclidines. Biochem. Pharmacol. 51:1477-1487. [DOI] [PubMed] [Google Scholar]
- 15.Nakashima, T., T. Inoue, A. Oka, T. Nishino, T. Osumi, and S. Hata. 1995. Cloning, expression, and characterization of cDNAs encoding Arabidopsis thaliana squalene synthase. Proc. Natl. Acad. Sci. USA 92:2328-2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Orenes Lorente, S., R. Gomez, C. Jimenez, S. Cammerer, V. Yardley, K. de Luca-Fradley, S. L. Croft, L. M. Ruiz Perez, J. Urbina, D. Gonzalez Pacanowska, and I. H. Gilbert. 2005. Biphenylquinuclidines as inhibitors of squalene synthase and growth of parasitic protozoa. Bioorg. Med. Chem. 13:3519-3529. [DOI] [PubMed] [Google Scholar]
- 17.Pandit, J., D. E. Danley, G. K. Schulte, S. Mazzalupo, T. A. Pauly, C. M. Hayward, E. S. Hamanaka, J. F. Thompson, and H. J. Harwood, Jr. 2000. Crystal structure of human squalene synthase. A key enzyme in cholesterol biosynthesis. J. Biol. Chem. 275:30610-30617. [DOI] [PubMed] [Google Scholar]
- 18.Shechter, I., E. Klinger, M. L. Rucker, R. G. Engstrom, J. A. Spirito, M. A. Islam, B. R. Boettcher, and D. B. Weinstein. 1992. Solubilization, purification, and characterization of a truncated form of rat hepatic squalene synthetase. J. Biol. Chem. 267:8628-8635. [PubMed] [Google Scholar]
- 19.Soltis, D. A., G. McMahon, S. L. Caplan, D. A. Dudas, H. A. Chamberlin, A. Vattay, D. Dottavio, M. L. Rucker, R. G. Engstrom, S. A. Cornell-Kennon, et al. 1995. Expression, purification, and characterization of the human squalene synthase: use of yeast and baculoviral systems. Arch. Biochem. Biophys. 316:713-723. [DOI] [PubMed] [Google Scholar]
- 20.Tanowitz, H. B., L. V. Kirchhoff, D. Simon, S. A. Morris, L. M. Weiss, and M. Wittner. 1992. Chagas' disease. Clin. Microbiol. Rev. 5:400-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thompson, J. F., D. E. Danley, S. Mazzalupo, P. M. Milos, M. E. Lira, and H. J. Harwood, Jr. 1998. Truncation of human squalene synthase yields active, crystallizable protein. Arch. Biochem. Biophys. 350:283-290. [DOI] [PubMed] [Google Scholar]
- 22.Urbina, J. A. 2002. Chemotherapy of Chagas disease. Curr. Pharm. Des. 8:287-295. [DOI] [PubMed] [Google Scholar]
- 23.Urbina, J. A. 2003. World class parasites, vol. American trypanosomiasis. Kluwer Academic Publishers, Boston, MA.
- 24.Urbina, J. A., J. L. Concepcion, A. Caldera, G. Payares, C. Sanoja, T. Otomo, and H. Hiyoshi. 2004. In vitro and in vivo activities of E5700 and ER-119884, two novel orally active squalene synthase inhibitors, against Trypanosoma cruzi. Antimicrob. Agents Chemother. 48:2379-2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Urbina, J. A., J. L. Concepcion, S. Rangel, G. Visbal, and R. Lira. 2002. Squalene synthase as a chemotherapeutic target in Trypanosoma cruzi and Leishmania mexicana. Mol. Biochem. Parasitol. 125:35-45. [DOI] [PubMed] [Google Scholar]
- 26.Urbina, J. A., and R. Docampo. 2003. Specific chemotherapy of Chagas disease: controversies and advances. Trends Parasitol. 19:495-501. [DOI] [PubMed] [Google Scholar]
- 27.WHO. 2002. The world health report. World Health Organization, Geneva, Switzerland.
- 28.Zhang, D., S. M. Jennings, G. W. Robinson, and C. D. Poulter. 1993. Yeast squalene synthase: expression, purification, and characterization of soluble recombinant enzyme. Arch. Biochem. Biophys. 304:133-143. [DOI] [PubMed] [Google Scholar]