Abstract
The magnitude of the problems of drug abuse and Neuro-AIDS warrants the development of novel approaches for testing hypotheses in diagnosis and treatment ranging from cell culture models to developing databases. In this study, cultured neurons were treated with/without HIV-TAT, ENV, or cocaine in a 2x2x2 expression study design. RNA was purified, labeled, and expression data were produced and analyzed using ANOVA. Thus, we identified 35 genes that were significantly expressed across treatment conditions. A diagram is presented showing examples of molecular relationships involving a significantly expressed gene in the current study (SOX2). Also, we use this information to discuss examples of gene expression interactions as a means to portray significance and complexity of gene expression studies in Drug Abuse and Neuro-AIDS. Furthermore, we discuss here that critical interactions remain undetected, which may be unravelled by developing robust database systems containing large datasets and gleaned information from collaborating scientists . Hence, we are developing a public domain database we named The Agora database , that will served as a shared infrastructure to query, deposit, and review information related to drug abuse and dementias including Neuro-AIDS. A workflow of this database is also outlined in this paper.
Keywords: cocaine, HIV-1 gp120, HIV-1 tat, Neuro-AIDS, neuron, Gene Expression, public international database, The Agora
Background
HIV-1 is frequently detected in the brain of HIV-1 infected individuals. [1] The exposure of neural cells to some drugs, HIV-1 proteins or HIV-1 infection may cause brain damage and perturb gene expression . In neural cells, the toxicity of the HIV-1 proteins gp120 and TAT was shown to involve Ca+2 and glutamate channels, and it could be reversed by memantine. Furthermore, cocaine stimulates HIV-1 replication through the chemokine, cytokine, and signaling pathways. [2–3] Drug abuse and HIV-1 infection most likely enhance neuropsychiatric disease, HAD (HIV associated Dementia), and HIVE (HIV encephalitis) as shown elsewhere. [4–5] During the early course of HIV-1 infection, the virus penetrates the blood-brain barrier (BBB). [6 ] HIV-1 invasion of the brain most likely occurs via macrophage and microglial cells and this leads to HIVE. [2–7] Recent work describes the influx of CD16-positive macrophages from bone marrow into the brain. [8] Several inflammatory molecules including cytokines, chemokines (CCKs), growth factors, lipids, and excitatory compounds are associated with brain inflammation and damage. [9–10] Here, we provide a preliminary description of the molecular mechanisms underlying these processes using expression and gene annotation data. This information has been stored in a user friendly, online database which will be made available to the public domain.
Methodology
Cultured human fetal neurons were isolated, cultured, and treated with HIV-1 TAT, HIV-1 ENV and cocaine in a 2x2x2 study design. [5–11–12] The Affymetrix HGU95A oligonucleotide chip was used to measure the expression of 12,565 probes. The data were analyzed using a one-way ANOVA (analysis of variance) among the eight treatment groups across genes (p < 0.0005). The expression of 35 genes was statistically significant. [5] We then used Pathways Assist (Stratagene, Inc.) to portray pathways among genes through common and divergent relationships. We modeled the neurological processes involved in Drug Abuse and Neuro-AIDS by computational simulation of physiological processes using experimentally derived information.
Our model uses the representation of biochemical networks named Moirai that is described elsewhere. [13] The clinical and physiological features are represented as sub-networks that recursively expand to their known component reactions, recorded pertinent data based on functional assays, and information on cell/tissue types. These features are treated as containers for cellular compartments in anatomical structures. Thus, our model mixes information known at varying levels occurring at different spatial and logical scales. We then use a prototype version of The Agora to populate the computational model where the information entered in forms goes into the model after appropriate review. [14] The development of The Agora is in progress and the completed version when made available in the public domain will enable scientists to retrieve biochemical and physiological information on Neuro-AIDS. We use the language semantics in Glossa for gene to pathway mapping in this development. [15]
Results
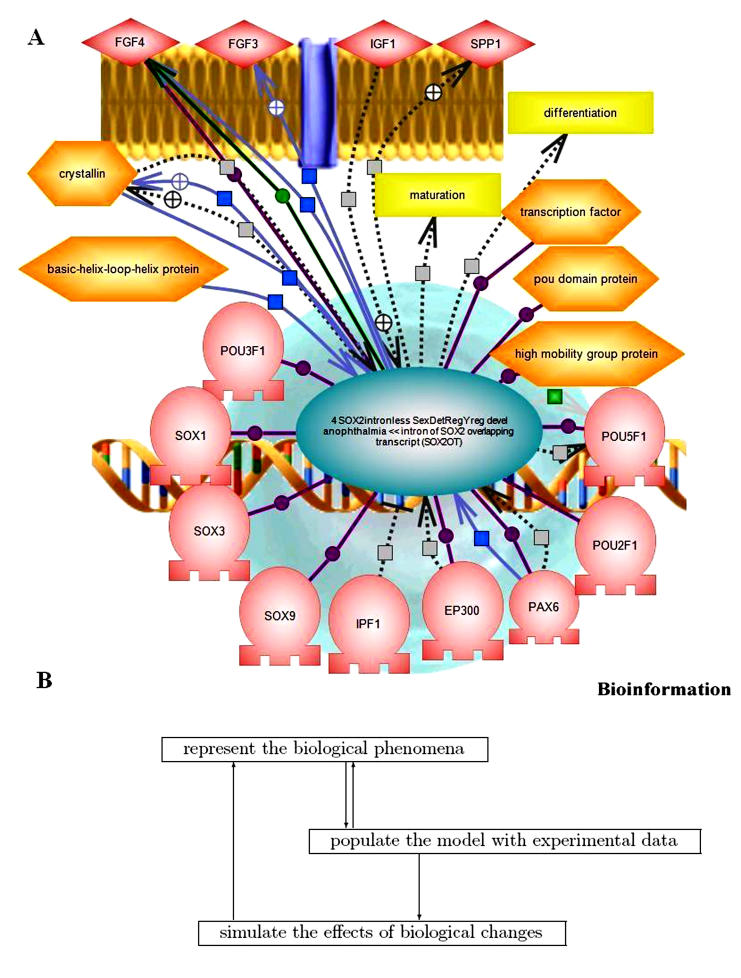
Thirty-five (35) genes were previously identified by ANOVA analysis of gene expression results from neuron cultures treated with cocaine/TAT/ENV. [5] The Ariadne Pathways Assist™ program was then used to generate annotated pathway interactions among gene products. The categories of genes thus generated and with which they interact include inflammation, focal contact, proliferation and growth, motility, morphogenesis, shape, mitogenesis, synaptogenesis, differentiation, cell survival, and neuropsychiatric disease. In Figure 1A, we see that SOX2, a transcription factor, interacts with other transcription factors (e.g. SOX1, SOX3, etc.) in the nucleus and also promotes the expression of cell surface growth factor receptors (e.g. FGF4, FGF3, etc). This is interpreted as a series of reparative processes attempted by the neuron under stress due to exposure to TAT, ENV and cocaine.
Figure 1.
(A) Gene interactions using Ariadne Pathways AssistTM. This figure shows relationships among genes expressed in neurons in culture as an example of possible gene expression network connections. Connections from SOX-2, a central transcription factor/regulator are shown to nuclear (e.g. SOX1, SOX3, etc.) and membrane associated proteins (e.g. FGF4, FGF3, etc). (B) Modeling drug abuse and neurological phenomena. This figure illustrates the iterative process of adumbrating a representation (top box), populating it with experimental data (middle box), and using the populated representation to develop and test hypotheses (lowest box). Each of these steps interacts, but one cannot proceed to simulation until the first two have advanced sufficiently to provide an adequate initial model.
Interpreting results such as those reported here in a physiological and clinical context requires building and refining computational models for specific clinical conditions. There are three steps in the modeling process that continuously and cyclically interact (Figure 1B).
First, one must computationally represent the phenomena in a way that accurately captures the biology, biochemical and physiological data, including information on cells, compartments, and tissues, and supports simulations of biological phenomena. Our current model focuses on phenomena relevant to drug abuse and several dementias including NeuroAIDS. Second, one must populate that representation with experimental data to form the basic model. We are using a prototype version of The Agora, a planned public resource for the scientific community to curate and use their biochemical and physiological databases to populate our computational model. We enrich the model with new relationships and types of information as the data entry process proceeds. For example, we initially did not include information on molecular biology kits, but these turned out to be important ways of specifying certain experimental protocols. Third, one develops and tests hypotheses about the phenomena by modifying the model and simulating the results. Until the basic model satisfactorily captures both the biological ideas (the representation) and the data, one cannot embark on this stage. Once simulations commence, the representations are likely to change in response to the results.
Discussion
Drug abuse is a complicating factor in the diagnosis and treatment of HAD and related diseases. [4–16] The preliminary gene expression study in cultured neurons treated with cocaine and HIV-1 proteins is described as a useful controlled model. We previously analyzed gene expression related to AIDS directly in gross brain tissue (without cell fractionation or micro-dissection) and in cell culture. [ 5–17] In these brain gene expression studies, statistical hypothesis testing was used to identify genes that are differentially expressed between two groups, HIV+ cases and HIV- controls. [17] However, we did not use traditional hypothesis testing methods as these are insufficient given the extremely large number of tests involved. Since there are over 12,000 genes to be tested, the expected number of Type I errors (false positives) becomes unacceptable.
The preliminary analysis of the human neuron gene expression using the one-way ANOVA produced a list of 35 genes that were significantly expressed across treatments. Application of a GLM analysis for significant interactions indicated most of these genes were significant due to TAT, some due to TAT-ENV, TAT-cocaine, and ENV-cocaine interactions, and none due to TAT-ENV-cocaine interactions. [5] During the course of generating Figure 1A, 113,944 sequence objects were examined in the Ariadne database in order to identify the genes and their interactions and connections indicated in the figure. In this figure we see examples of extensive interactions that occur among transcription factors in the nucleus (SOX1, SOX2, SOX3, etc) as well as with cell surface growth factor receptors (e.g. FGF4, FGF3, etc). This is interpreted as a series of reparative processes attempted by the neuron under stress due to exposure to tat, env, and cocaine.
The preliminary analyses described here are part of a wider scope needed to defeat drug abuse, Neuro-AIDS, and other Dementias. [3] Computational modeling of biological phenomena serves three main purposes including to express what is known, to help formulate hypotheses, and to test those hypotheses. Large amounts of data are produced in diverse studies including molecular biology, drug abuse, virology, immunology, physiology, computational biology, and statistics. The vast majority of data needed to explore hypotheses and to interpret the results of these efforts for the most part remains locked in the print literature and posted web-sites. 18– 19–20– 21–22–23– 24–25– 26] We are currently using The AGORA to record this data related to Drug Abuse and Neuro-AIDS and other neurological diseases. [4] We are developing methods to structure anatomical information (location where specific biochemical reactions occur; e.g. basal ganglia of the brain) so that essential structural features are included along with the topological attributes.
Acknowledgments
We acknowledge NIH support (NIDA DA 14533, DA 12580, and NIGMS GM 056529)
Footnotes
Citation:Shapshak et al., Bioinformation 1(3): 86-88 (2006)
References
- 1.Speth C, et al. Mol Immunol. 2005;42:213. doi: 10.1016/j.molimm.2004.06.018. [DOI] [PubMed] [Google Scholar]
- 2.Bell JE, et al. Brain. 1998;121:2043. doi: 10.1093/brain/121.11.2043. [DOI] [PubMed] [Google Scholar]
- 3.Kuhar MJ, et al. Drug Alcohol Depend. 2001;62:157. doi: 10.1016/s0376-8716(00)00231-3. [DOI] [PubMed] [Google Scholar]
- 4.Nath A, et al. J Psychopharmacol. 2000;14:222. doi: 10.1177/026988110001400305. [DOI] [PubMed] [Google Scholar]
- 5.Shapshak P, et al. Front Biosci. 2006;11:1774. doi: 10.2741/1922. [DOI] [PubMed] [Google Scholar]
- 6.Resnick L, et al. Neurology. 1988;38:9. doi: 10.1212/wnl.38.1.9. [DOI] [PubMed] [Google Scholar]
- 7.Achim CL, et al. J Neuropathol Exp Neurol. 1994;53:284. doi: 10.1097/00005072-199405000-00010. [DOI] [PubMed] [Google Scholar]
- 8.Gartner S. Science. 2000;287:602. doi: 10.1126/science.287.5453.602. [DOI] [PubMed] [Google Scholar]
- 9.Heyes MP, et al. J Ann Neurol. 1991;29:202. doi: 10.1002/ana.410290215. [DOI] [PubMed] [Google Scholar]
- 10.Tyor WR, et al. Ann Neurol. 1992;31:349. doi: 10.1002/ana.410310402. [DOI] [PubMed] [Google Scholar]
- 11.Nath A, et al. Ann Neurol. 2000;47:186. [PubMed] [Google Scholar]
- 12.Turchan J, et al. BMC Neurosci. 2001;2:3. doi: 10.1186/1471-2202-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. http://www.biocheminfo.org/repository/sci2002.ps.
- 14. https://www.the-AGORA.org.
- 15.Kazic T. Bioinformatics. 2000;16:1129. doi: 10.1093/bioinformatics/16.12.1129. [DOI] [PubMed] [Google Scholar]
- 16.Goodkin K, et al. J Neuroimmunol. 1998;83:88. doi: 10.1016/s0165-5728(97)00225-7. [DOI] [PubMed] [Google Scholar]
- 17.Shapshak P, et al. Front Biosci. 2004;9:2935. doi: 10.2741/1449. [DOI] [PubMed] [Google Scholar]
- 18.Galey D, et al. J Neurovirol. 2003;9:358. doi: 10.1080/13550280390201119. [DOI] [PubMed] [Google Scholar]
- 19.Geiss GK, et al. Virology. 2000;266:8. doi: 10.1006/viro.1999.0044. [DOI] [PubMed] [Google Scholar]
- 20.Gekker G, et al. J Pharm & Exp Therap. 2004;309:600. doi: 10.1124/jpet.103.060160. [DOI] [PubMed] [Google Scholar]
- 21.Gendelman HE, et al. J Leuk Biol. 1994;56:389. doi: 10.1002/jlb.56.3.389. [DOI] [PubMed] [Google Scholar]
- 22.Masliah E, et al. Ann Neurol. 1992;32:321. doi: 10.1002/ana.410320304. [DOI] [PubMed] [Google Scholar]
- 23.Minagar A, et al. J Neurosci Res. 2005;224:3. [Google Scholar]
- 24.Nath A, et al. Ann Neurol. 2000;47:186. [PubMed] [Google Scholar]
- 25.Roberts ES, et al. Am J Pathol. 2003;162:2041. doi: 10.1016/S0002-9440(10)64336-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vahey MT, et al. AIDS Res & Hum Retrovirol. 2003;19:369. doi: 10.1089/088922203765551728. [DOI] [PubMed] [Google Scholar]